Abstract
In addition to their CD1d-restricted T cell receptor (TCR), natural killer T (NKT) cells express various receptors normally associated with NK cells thought to act, in part, as modulators of TCR signaling. Immunoreceptor-tyrosine activation (ITAM) and inhibition (ITIM) motifs associated with NK receptors may augment or attenuate perceived TCR signals respectively, potentially influencing NKT cell development and function. ITIM-containing Ly49 family receptors expressed by NKT cells are proposed to play a role in their development and function. We have produced mice transgenic for the ITAM-associated Ly49D and ITIM-containing Ly49A receptors and their common ligand H2-Dd to determine the importance of these signaling interplays in NKT cell development. Ly49D/H2-Dd transgenic mice had selectively and severely reduced numbers of thymic and peripheral NKT cells, whereas both ligand and Ly49D transgenics had normal numbers of NKT cells. CD1d tetramer staining revealed a blockade of NKT cell development at an early precursor stage. Coexpression of a Ly49A transgene partially rescued NKT cell development in Ly49D/H2-Dd transgenics, presumably due to attenuation of ITAM signaling. Thus, Ly49D-induced ITAM signaling is incompatible with the early development of cells expressing semi-invariant CD1d-restricted TCRs and appropriately harmonized ITIM–ITAM signaling is likely to play an important role in the developmental program of NKT cells.
Keywords: NKT cell development, ITAM-containing receptors, ITIM-containing receptors, TCR repertoire, NK receptor repertoire
Introduction
Murine natural killer T (NKT) cells may be defined as T cells expressing diverse inhibitory and activating receptors normally found on NK cells as well as an αβTCR selected for reactivity to CD1d, a nonpolymorphic MHC Ib molecule (1, 2). Activated NKT cells rapidly produce cytokines capable of influencing a wide variety of innate and adaptive immune responses. CD1d-restricted NKT cells express a very limited, “semi-invariant,” TCR repertoire. In the mouse, this consists of a germline-encoded Vα14–Jα18 chain paired with β chains containing a small range of Vβ segments, the most commonly used being Vβ8.2 (3). One widely held hypothesis is that this TCR repertoire specializes in the recognition of self-lipid antigens in contexts where their presentation is aberrant. The development of NKT cells would thus require the selection of a potentially autoreactive TCR repertoire, accompanied by the establishment of tight controls upon this autoreactivity in the mature population.
Expression of MHC class I-specific inhibitory receptors by NKT cells and subsets of T cells provides a possible mechanism for regulating perceived TCR signals (4). Murine NKT cells express many members of the Ly49 family of lectin-like receptors which recognize H2-encoded ligands (5) and Ly49 ligation influences NKT cell responsiveness to antigen (6, 7). Ly49 family receptors exist in inhibitory and activating forms that signal through immunoreceptor-tyrosine inhibition motifs (ITIMs) and immunoreceptor-tyrosine activation motifs (ITAMs) respectively. Ligated inhibitory Ly49 receptors, such as Ly49A, recruit and activate SH2-domain containing phosphatases, whereas Ly49D and other activating Ly49 receptors form complexes with an ITAM-containing signaling subunit, DAP12 (8). Upon ligation of its associated receptor, DAP12 mediates recruitment and phosphorylation of the syk and ZAP 70 kinases. As phosphorylated ZAP 70 is an important participant in the TCR/CD3 signaling cascade, ITAM-mediated signaling by activating Ly49 receptors could have additive effects on TCR signaling. Curiously however, expression of Ly49D and Ly49H, the only activating Ly49 receptors for which specific antibodies are available, is not detected on CD3+ cells (9–11). This might indicate a general lack of activating Ly49 receptor expression on T and NKT cells, a situation which would raise the possibility that activating (but not inhibitory) Ly49 signaling is in some way specifically incompatible with T/NKT cell development and/or function.
Transgenic expression of inhibitory Ly49 receptors on T and NKT cells has provided much insight into possible roles for Ly49 signaling in T and NKT cell selection and development. Studies of mice expressing transgenic Ly49A on thymocytes have demonstrated the ability of inhibitory Ly49 signaling to affect the outcome of positive and negative selection of T cells (12, 13). Other Ly49A transgenic mice exhibit a defect late in the development of NKT cells (14). While these mice have near normal numbers of thymic Vβ8.2+ (i.e., CD1d-restricted) NKT cells, numbers of hepatic Vβ8.2+ NKT cells are significantly reduced in a ligand-dependent fashion.
These studies of Ly49A transgenic mice as well as in vitro studies indicating a role for Ly49 signaling in regulating NKT cell responses (6, 7), indicate that Ly49 expression may influence NKT cell development. To investigate the effects of activating Ly49 signaling on the selection and development of CD1d-restricted NKT cells, we created transgenic mice coexpressing Ly49D and its associated DAP12 signaling subunit. Ligation of this transgenic receptor complex is predicted to induce ITAM signaling, thereby adding to the TCR signals perceived by developing NKT cells. We report here that mice expressing a transgenic Ly49D receptor complex and H2-Dd, one of its ligands, have a selective defect in the generation of CD1d-restricted NKT cells that can be partially reversed by concomitant inhibitory Ly49A signaling.
Materials and Methods
Preparation of Transgenic Mice.
A Ly49D cDNA identical to that previously described (GenBank/EMBL/DDBJ accession no. U10090) was isolated by RT-PCR using the primers AATAGTCGA 50C TCTACATACTCCCGAGATG and AATAGATCT A863CTCACTGGAGAGTCAATG (italics indicate restriction sites added to facilitate cloning, numbers indicate nucleotide positions in the Ly49D sequence). SalI, BglII-cut PCR product was ligated into SalI, BamHI-cut H2-Kb promoter/Ig enhancer expression cassette (15). A murine DAP12 cDNA identical to that reported previously (GenBank/EMBL/DDBJ accession no. AF024637) was obtained by RT-PCR using the primers CGGAATTCCACCA1TGGGGCTCTGGAGCCCTCC and CGGAATTCT345CATCTGTAATATTGCCTCTG. A DAP12 cDNA was isolated from the plasmid containing the cloned PCR product as an XhoI, BamHI fragment and ligated into SalI, BamHI-cut expression cassette. Double transgenic mice were generated by coinjection of Ly49D and DAP12 constructs into (C57BL/6 × DBA/2)F2 eggs. Founders were identified by Southern blotting and expression of transgenic Ly49D was confirmed by FACS® analysis of PBL. Expression of the DAP12 transgene in PBL was verified by RT-PCR using the 5′ primer shown above and a 3′ transgene-specific primer CTGGTGGGGTGAATTCTTTGCC. Founders were backcrossed to C57BL/6 at least six times.
C57BL/6 and DBA/2 mice were obtained from Harlan. C57BL/6 mice transgenic for Ly49A (line #2; reference 16) and for a large genomic fragment of H2-Dd (17) have been described previously.
DAP12 Expression Analysis.
cDNA was prepared from 105 sorted thymocytes or liver mononuclear cells by standard methods. DAP12 transcripts were amplified using the exon-spanning primers 70CCGTACAGGCCCAGAGTG and 224CTTGACCTCGGGAGACCAG. β2-microglubulin transcripts were amplified using the primers: GTGTATGCTATCCAGAAAACCC and TCACATGTCTCGATCCCAGTAG. Cycling conditions for both primer pairs were: 95°C for 3 min, then 40 cycles of 94°C for 30 s, 60°C for 30 s, 72°C 40 s, then 72°C for 10 min.
Cell Preparation.
Nylon wool nonadherent spleen and bone marrow cells were isolated as described previously (18). Liver mononuclear cells were prepared on 40/70% percoll gradients by standard methods (18). Erythrocytes in spleen, bone marrow, and liver cell preparations were lysed using 0.83% ammonium chloride solution before FACS® analysis. HSA-/CD8-depleted thymocytes were prepared using hybridoma supernatants 3.168.8.1 (anti-CD8α) and B2A2 (anti-HSA) followed by treatment with rabbit complement and purification on a Lympholyte M (Cedarlane) gradient as described previously (19).
Antibodies and Flow Cytometry.
mAb conjugates purchased from BD Biosciences were: Ly49D-FITC (4E5), γδTCR-FITC (GL3), NK1.1-PE and -biotin (PK136), TCRβ-Cychrome (H57–597). CD4-APC (RM4–5) was purchased from eBioscience. mAb conjugates produced at our institute were: CD44-FITC (1M.781), Vβ8.2-biotin (F23.2). Streptavidin–APC was purchased from Molecular Probes. Murine CD1d tetramers complexed using PE-streptavidin and loaded (or not) with α-galactosyl–ceramide were a kind gift of Dr. Mitchell Kronenberg (La Jolla Institute for Allergy and Immunology, La Jolla, CA). Tetramer stainings were performed before other staining steps as recommended (20). Blockage of Fc receptors with the hybridoma supernatant 2.4G2, antibody staining of cells and FACS® analysis were performed as described previously (18).
Results and Discussion
Although Ly49D may be capable of associating with and signaling via CD3ζ (21), the DAP12 signaling subunit is essential for the function and efficient surface expression of Ly49D on NK cells (22). As DAP12 does not appear to be expressed to any significant extent by CD4+ CD8+ (DP) thymocytes, NKT, or T cells from normal C57BL/6 mice (Fig. 1 C), we created mice doubly transgenic for Ly49D and DAP12 to ensure optimal Ly49D signaling activity. Coinjection of constructs using the same H2-Kb promoter/Ig enhancer expression cassette (Fig. 1 A; reference 15) produced three founders that had cointegrated both constructs and expressed transgenic Ly49D. These were backcrossed six or more times to C57BL/6 mice. Each line was then crossed with C57BL/6 mice transgenic for a large genomic fragment of the H2-Dd gene (17). Regulation of H2-Dd expression in these transgenic mice is comparable in all respects examined to that of the endogenous H2-Dd gene in H2d mice (17, and unpublished data). Use of these H2-Dd transgenic mice permitted the physiological expression of a single, defined Ly49D ligand on an otherwise genetically identical C57BL/6 background. For simplicity, Ly49D/DAP12 double transgenic mice are hereafter referred to as being “Ly49D transgenic.”
Figure 1.
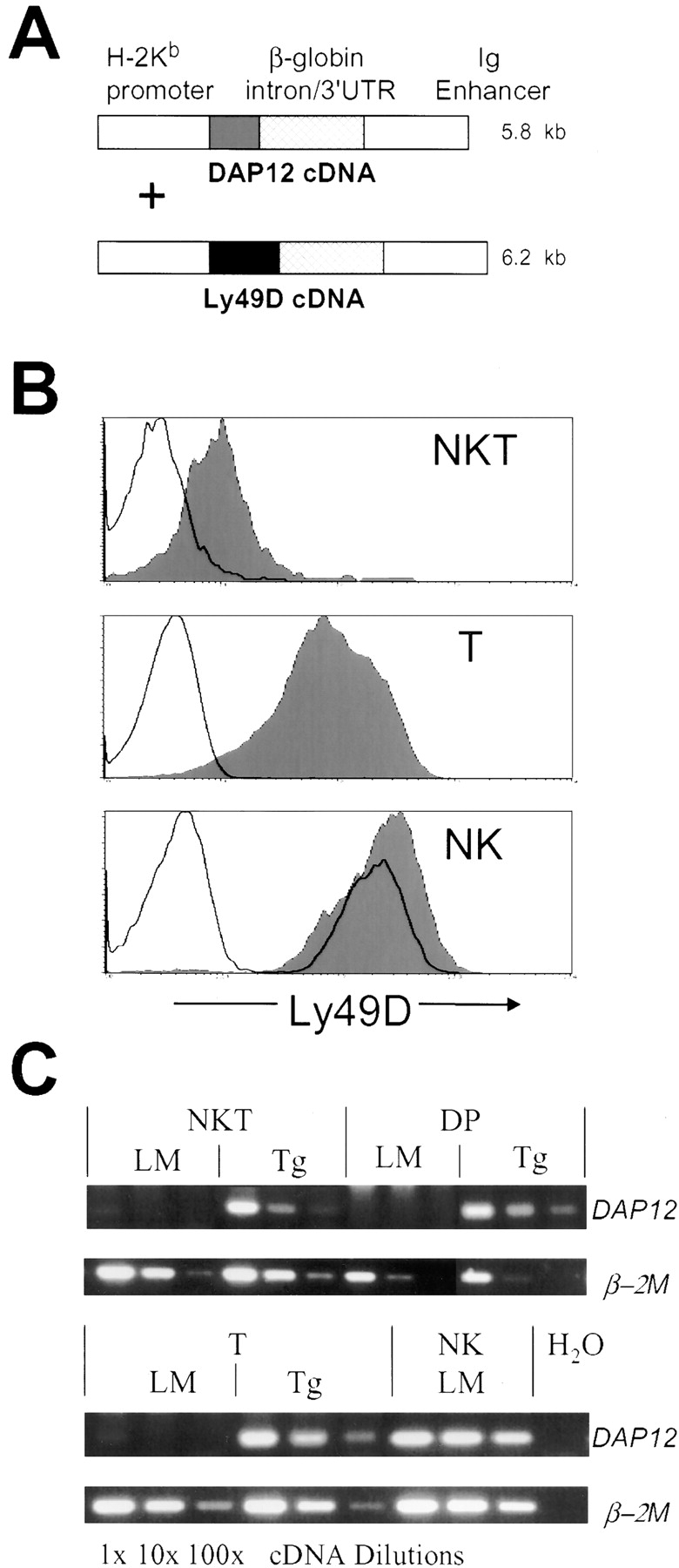
Transgene expression in Ly49D/DAP12 transgenic (“Ly49D transgenic”) mice. (A) Schematic of coinjected transgene expression constructs. (B) Ly49D expression on thymic NKT, T, and splenic NK cells. Expression in nontransgenic littermate (unfilled histogram) and Ly49D transgenic (filled histogram) mice is shown. NKT cells are gated as NK1.1+, TCRβInt, T cells are gated as NK1.1−, TCRβHi, and NK cells as NK1.1+, TCRβ−, γδTCR−. (C) RT-PCR analysis of DAP12 and control β-2 microglobulin (β-2M) transcript expression by sorted cell populations (DP thymocytes were gated as CD4+, CD8+, TCRβLo, other populations were gated as above) from nontransgenic littermate (LM) and Ly49D transgenic (Tg) mice. The cDNA was used neat, as well as diluted 10-fold and 100-fold to titrate expression levels. The undiluted cDNA reactions contained ∼5 × 103 cell equivalents from a cDNA pool synthesized from 105 cells.
All three transgenic lines expressed Ly49D on all thymocytes from the CD4−, CD8−, CD25+ (DN3) stage onward (unpublished data). The levels of Ly49D expressed by mature NKT, T and NK cells were examined in all three transgenic lines (Fig. 1 B, Table I, and unpublished data). Levels of transgenic Ly49D expression were highest on NK cells but were still comparable to endogenous Ly49D levels on NK cells in littermate mice. T cells and NKT cells both showed lower levels of transgene expression, with NKT cells expressing less Ly49D than T cells in all tissues examined. Analysis of DAP12 transgene expression in DP thymocytes, NKT and T cells by RT-PCR determined that levels of transgenic DAP12 transcripts expressed by these populations were similar to, although somewhat lower than, the levels of endogenous DAP12 transcripts expressed by NK cells (Fig. 1 C).
Table I.
Transgenic and Endogenous Ly49D Expression on NKT, T, and NK Cells
| NKT cellsa | T cellsb | NK cells | |
|---|---|---|---|
| Ly49Dtg | 18 ± 5 | 78 ± 10 | 126 ± 23c |
| Ly49D/H2-Ddtg | 14 ± 2 | 25 ± 8 | 108 ± 28c |
| H2-Ddtg | 6 ± 1 | 4 ± 1 | 134 ± 4d |
| C57BL/6 | 6 ± 1 | 4 ± 1 | 142 ± 3d |
Transgenic and endogenous Ly49D expression is expressed as mean fluorescence intensity ± SD.
Gated as NK1.1+, TCRβInt thymocytes.
Gated as NK1.1−, TCRβHi thymocytes.
Gated as NK1.1+, TCRβ−, γδTCR− splenocytes.
Gated as Ly49D+ (endogenously expressed), NK1.1+, TCRβ−, γδTCR− splenocytes.
Examination of Ly49D × H2-Dd transgenic intercross progeny revealed that Ly49D/H2-Dd transgenic mice had smaller thymi and fewer peripheral T cells than littermate mice of other genotypes. The reduction in thymic cellularity was approximately fivefold (Fig. 2 B) and numbers of peripheral αβT cells were reduced about threefold (Fig. 2, A and B). Strongly reduced thymocyte numbers were first seen at the DP stage (unpublished data). The nature of this general T cell developmental phenotype is being examined and will be reported elsewhere.
Figure 2.
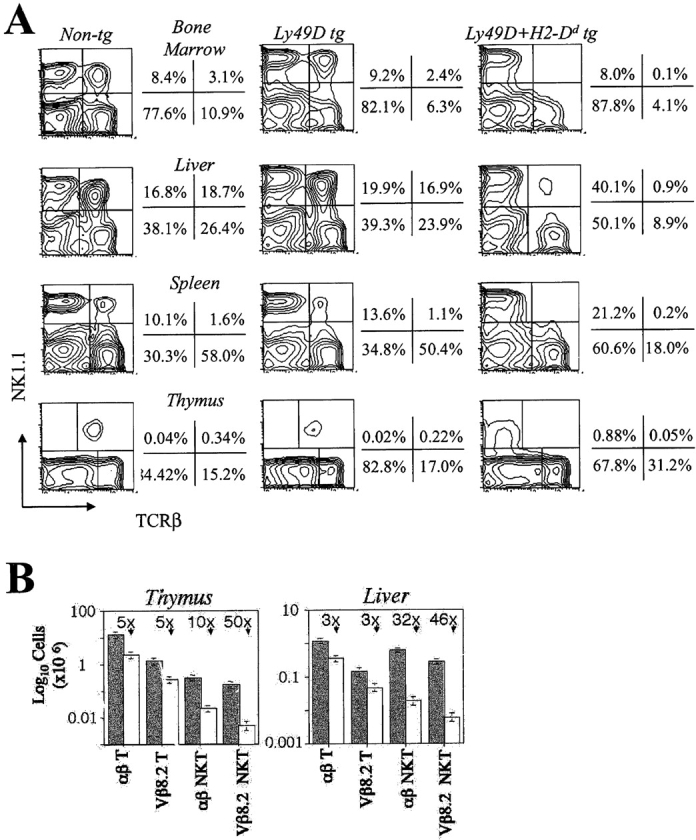
Selective impairment of NKT cell development in Ly49D transgenic mice is ligand-dependent. (A) FACS® analysis of αβNKT cells in various tissues of nontransgenic littermate, Ly49D transgenic and Ly49D/H2-Dd transgenic mice. Contour plots are gated on lymphoid cells among nylon wool nonadherent cells from bone marrow and spleen, among lymphoid-gated liver mononuclear cells, and among viable untreated thymocytes. (B) Histograms showing absolute numbers of thymic and hepatic T and NKT cells in Ly49D transgenic (shaded bars) and Ly49D/H2-Dd transgenic (unfilled bars) mice with standard deviations calculated from three independent experiments. Approximate fold reductions of cell numbers in Ly49D/H2-Dd transgenic mice are also indicated. All mice used were 5 wk old.
When NKT cell numbers in the thymi and peripheral tissues of these transgenic intercross progeny were assessed by NK1.1, TCRβ double staining, a striking reduction in the proportion of αβNKT cells present in Ly49D/H2-Dd transgenic mice was noted in all organs examined (Fig. 2 A). Thus, absolute numbers of NK1.1+, TCRβ+ cells were reduced by 10-fold in the thymus and 32-fold in the liver when compared with Ly49D or H2-Dd transgenic mice (Fig. 2 B). The presence of normal numbers of NK1.1+, TCRβ+ cells in both Ly49D and H2-Dd transgenic mice suggested that this reduction depended upon ligation of the Ly49D/DAP12 complex by H2-Dd. In the thymi of Ly49D/H2-Dd transgenic mice, a large population of NK1.1+, TCRβ– cells was evident (Fig. 2 A). These were found to be γδTCR+ NKT cells (unpublished data), a population of cells which also expands in CD1d-deficient mice (unpublished data) and other mice with defects in CD1d-restricted NKT cell development (23).
Mice derived from all three Ly49D transgenic lines displayed similar phenotypes with respect to NKT and T cell development when crossed to H2-Dd transgenic mice (unpublished data). H2-Dd molecules expressed from the endogenous gene also selectively impaired NKT cell development in Ly49D transgenic mice since αβNKT cell numbers in Ly49D transgenic B6:DBA/2 F1 offspring were reduced to levels similar to those found in Ly49D/H2-Dd transgenic mice (unpublished data).
Murine CD1d-restricted NKT cells most commonly utilize Vβ8.2 in the TCRβ chain that pairs with Vα14-Jα18 (3). Vβ8.2 expression in an NK1.1+, TCRβ+ population is thus a useful indicator of CD1d specificity. Proportions of NKT cells expressing Vβ8.2 were also reduced approximately twofold, from 48 ± 5% to 21 ± 6% (see also Fig. 4 B), in the thymi of Ly49D/H2-Dd transgenic mice and absolute numbers were reduced ∼50-fold in thymus and liver (Fig. 2 B). This reduction in numbers of Vβ8.2+ cells was selective for NKT cells since the proportion of Vβ8.2+ cells among NK1.1− conventional T cells in Ly49D/H2-Dd transgenic mice was unaffected (unpublished data and see Fig. 4 B) and their numbers were not selectively reduced (Fig. 2 B). The few NK1.1+, TCRβ+ cells found in Ly49D/H2-Dd transgenic mice thus had a significantly different TCR repertoire to NK1.1+, TCRβ+ cells found in Ly49D or H2-Dd transgenic mice, suggesting that development of NKT cells expressing semiinvariant CD1d-restricted TCRs was selectively impaired.
Figure 4.

Partial rescue of NKT cell development in Ly49D/H2-Dd transgenic mice by a Ly49A transgene. (A) Thymocytes stained for NK1.1 and TCRβ from 9-wk-old mice expressing the indicated transgenes (top panels, mice on C57BL/6 background; bottom panels, mice on H2-Dd transgenic C57BL/6 background). The percentages of cells in the upper two quadrants are indicated with corresponding absolute numbers (in thousands) of cells per thymus in brackets. In Ly49D/H2-Dd and Ly49A/Ly49D/H2-Dd transgenic thymi, cells in the top left quadrants are predominantly γδTCR+ NKT cells (unpublished data). (B) Percentages of Vβ8.2+ NKT and T cells in the thymi of mice in panel A. Gating is as for Fig. 1 B. Representative data from one of three similar experiments are shown.
Little is known of the origin and functional significance of the CD1d-independent NKT cell population which expresses a diverse TCR repertoire and is particularly abundant in the bone marrow and spleen (18). It is noteworthy, however, that this population was also less abundant in Ly49D/H2-Dd transgenic mice since absolute numbers of NK1.1+, TCRβ+ cells were reduced in bone marrow and spleen by significantly more than the 30–40% that would be expected (20, 24) if only the CD1d-dependent NKT cells in these tissues were affected (Fig. 2 A). Proportions of Vβ8.2+ cells among residual NK1.1+ T cells in Ly49D/H2-Dd transgenic mice were, nonetheless, severely reduced (12 ± 2% in bone marrow and 10 ± 3% in spleen, compared with 22 ± 3% and 33 ± 2% in the same tissues of Ly49D transgenic littermates). This suggested that the development of the CD1d-dependent NKT cell population was most severely affected in these tissues.
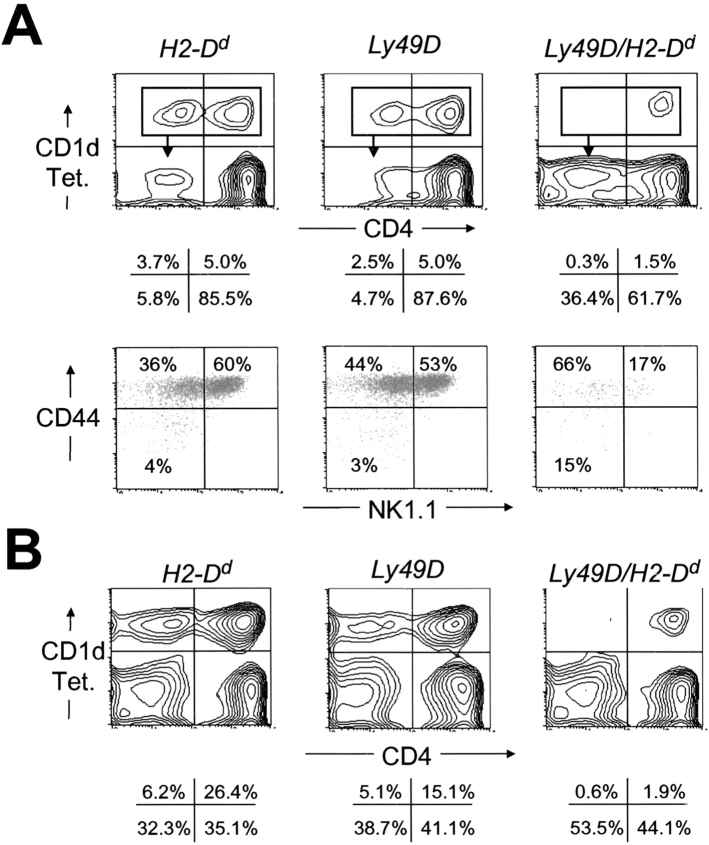
To directly confirm that CD1d-restricted NKT cells were selectively reduced in number in Ly49D/H2-Dd transgenic mice, we used multimeric CD1d complexes loaded with the artificial NKT cell ligand α-galactosyl-ceramide (reference 20; hereafter referred to as “tetramers”) to detect cells expressing CD1d-restricted TCRs. Whereas normal numbers of tetramer positive thymocytes were found in Ly49D transgenic and H2-Dd transgenic mice, very few tetramer positive cells were found in Ly49D/H2-Dd transgenic thymi (numerically ∼30-fold fewer; Fig. 3 A). Greatly reduced numbers of tetramer positive cells were also noted in the livers of Ly49D/H2-Dd transgenic mice (Fig. 3 B). Thus, there was a dramatic reduction in the numbers of CD1d-restricted NKT cells in Ly49D/H2-Dd transgenic mice that was of a similar order to that seen for NK1.1+, TCRβ+, Vβ8.2+ cells.
Figure 3.
Early impairment of CD1d-restricted NKT precursor development in Ly49D/H2-Dd transgenic mice. (A) Thymocytes from 5-wk-old H2-Dd transgenic, Ly49D transgenic, and Ly49D/H2-Dd transgenic mice were enriched for NKT cells and precursors by treatment with anti-CD8α, anti-HSA, and complement. Top panels show staining of viable cells with α-galactosyl-ceramide loaded mouse CD1d tetramers (CD1d Tet.) and CD4. Bottom panels show staining of tetramer positive cells (gated as indicated) for CD44 and NK1.1. Background staining of CD8-/HSA-depleted thymocytes with tetramers that had not been loaded with α-galactosyl-ceramide was negligible (unpublished data). (B) Staining of gated TCRβ+ liver mononuclear cells from the mice in panel A for tetramers and CD4. Data from one of two similar experiments are shown.
Recent studies have identified proliferating immediate precursors of NKT cells as tetramer positive cells lacking surface expression of NK1.1 that are most evident in the first few weeks after birth (25–27). The first tetramer positive cells to appear in young mice have been reported to be predominantly CD4+ and to express low or intermediate levels of CD44. Residual tetramer positive thymocytes in young Ly49D/H2-Dd transgenic mice were mostly CD4+ NK1.1− and a larger proportion of them were CD44 low or intermediate (Fig. 3 A), whereas tetramer positive thymocytes in Ly49D and H2-Dd transgenic mice were a mixture of CD4−, CD8– and CD4+ cells that were proportionately more NK1.1+ and CD44Hi (Fig. 3 A). This suggests that the few tetramer positive cells present in young Ly49D/H2-Dd transgenic thymi are more immature than the tetramer positive cells found in littermate thymi. The strong overall reduction in numbers of NKT cell precursors indicates that the blockade of NKT cell development in Ly49D/H2-Dd transgenic mice is first manifest at a very early stage, during establishment of the precursor pool. This blockade, while severe, is not absolute and development of small numbers of NKT precursors still occurs. However, these immature precursors would seem to progress and/or expand less efficiently during their subsequent development since proportions of mature phenotype NKT cells are selectively reduced (Fig. 3 A).
To study the signaling interactions between activating and inhibitory Ly49 receptors expressed on NKT and T cells, we crossed Ly49D transgenic mice with Ly49A transgenic mice. Ly49A expression in these transgenic mice is directed by the same H2-Kb/Ig enhancer expression cassette used to generate Ly49D transgenic mice and all T and NKT cells express Ly49A (14, 16). As H2-Dd is a ligand of both Ly49A and Ly49D, intercrossing of the Ly49A, Ly49D, and H2-Dd transgenic lines provides a genetically well defined in vivo model to study this signaling interplay. Expression levels of the Ly49D and H2-Dd transgenes were found to be equivalent on NKT and other hematopoietic cell populations of these intercross mice (unpublished data). Interestingly, mice transgenic for Ly49A/Ly49D/H2-Dd had a population of NK1.1+, TCRβ+ thymocytes, six times larger than that of Ly49D/H2-Dd transgenic littermates (Fig. 4 A). Mice of all other genotypes had normal numbers of NK1.1+, TCRβ+ thymocytes. Ly49A/Ly49D/H2-Dd transgenic mice also had an increased proportion of Vβ8.2+, NK1.1+, TCRβ+ thymocytes: (34%), compared with Ly49D/H2-Dd transgenic mice (22%; Fig. 4 B). Proportions of Vβ8.2+, NK1.1–, TCRβ+ mature thymic T cells, however, remained constant in mice of all genotypes (Fig. 4 B). This suggests that Vβ8.2+, CD1d-restricted NKT cells are being rescued by Ly49A-mediated inhibition of Ly49D and/or TCR signaling. Despite the apparent dominance of ITIM signaling (21), numbers of NKT cells found in Ly49A/Ly49D/H2-Dd transgenic mice were not increased to levels comparable to those found in Ly49A/H2-Dd transgenic mice, perhaps in part because the restoration of thymic cellularity was only partial (unpublished data).
Collectively, our data suggest that the development of CD1d-restricted NKT cells depends critically on the level of ITAM (i.e., TCR) signaling perceived by very early NKT precursors, as additional ITAM signaling by activating Ly49 receptors appears to interfere with establishment of the early precursor pool and impede their subsequent development. These findings are consistent with instructive models of NKT cell development where a common T/NKT cell precursor is recruited to the NKT lineage by expression of a TCR with a specific affinity range for CD1d (perceived primarily as a specific level of ITAM signaling). Other recent support for an instructive model of NKT cell development comes from the observation that intrathymic injection of DP thymocytes generates NKT cells in a CD1d-dependent fashion (28). It is thus possible that the primary blockade of NKT cell development in Ly49D/H2-Dd transgenic mice occurs at a DP precursor stage. It has been proposed that the semi-invariant TCRs expressed by NKT cells have intrinsically high affinities for self-ligands presented by CD1d that may approach or even exceed the affinity thresholds prompting negative selection of mainstream αβT cells (1). In this scenario, DP cells expressing a limited range of CD1d-restricted TCRs (3) would be exquisitely sensitive to negative selection due to additional ITAM-signaling provoked by Ly49D ligation. The selective loss of Vβ8.2+ NKT cells seen in our system might therefore indicate that the Vα14/Vβ8.2 TCRs have a relatively higher affinity for CD1d complexed with self-ligand when compared with other CD1d-restricted TCRs. Alternatively or additionally, excessive ITAM signaling could interfere with differentiation and expansion of early NKT cell precursors, as would be suggested by the higher proportions of immature NKT precursors seen in Ly49D/H2-Dd transgenic thymi. In any case, the selective loss of CD1d-restricted NKT cells seen in our study would seem to result from a failure to satisfy specific signaling criteria dictated by their unique developmental program.
Based on our data and those obtained by others with Ly49A transgenic mice (12, 13, 29), ITAM- and ITIM-mediated signaling by Ly49 family molecules can have competing positive and negative effects on TCR signaling during thymocyte development. The detrimental effects of Ly49D signaling on NKT cell development are counteracted by coexpression of Ly49A, probably as a consequence of diminution of the perceived ITAM (Ly49D/TCR) signal by coactivation of the Ly49A ITIM. This might free cells expressing CD1d-restricted TCRs from the constraints upon their early development imposed by excessive ITAM signaling. If this is the case, it might have been expected that substantial numbers of Ly49D/H2-Dd transgenic NKT cells would be rescued in this way by endogenous expression of Ly49A and other inhibitory receptors specific for H2-Dd. However, there may have been no opportunity for this to occur since endogenous inhibitory Ly49 receptors are not expressed until the later stages of NKT cell development (25, 28).
Regulated expression of inhibitory NK cell-associated receptors, including Ly49 family members, plays a crucial role in controlling the development, maturation, and responsiveness of CD1d restricted NKT cells (6, 7, 14, 25–27). It is becoming apparent that NKT cells are subject to a particular and elaborate developmental program depending upon specific, balanced signaling outcomes. This program is likely to involve positive selection of rare DP precursors expressing semi-invariant, CD1d-restricted TCRs followed by their expansion and maturation into cells with potential self-reactivity, which may be partly counteracted by expression of inhibitory Ly49 receptors (6, 7). The deleterious effects of Ly49D signaling on NKT cell development reported here underline the importance of timely and appropriate NK receptor acquisition in this program. NK cells are known to undergo a step-wise program of receptor expression that plays an important role in their development, tolerance, and functional specification (5, 30). It now seems increasingly likely that NKT cells undergo a similar finely-orchestrated program of NK receptor acquisition which might also be critical for their selection, expansion, tolerance, and function.
Acknowledgments
We are grateful to Dr. Mitchell Kronenberg for his kind gift of murine CD1d tetramers. We thank Drs. Anne Wilson and Isabel Ferrero for many helpful discussions. Dr. Bente Lowin-Kropf is gratefully acknowledged for her gift of a murine DAP12 cDNA.
This work was supported in part by a grant (RG-00168/2000) to H.R. MacDonald from the Human Frontier Science Program. W. Held received grant support from the Swiss National Science Foundation.
References
- 1.Bendelac, A., M. Bonneville, and J.F. Kearney. 2001. Autoreactivity by design: innate B and T lymphocytes. Nat. Rev. Immunol. 1:177–186. [DOI] [PubMed] [Google Scholar]
- 2.Kronenberg, M., and L. Gapin. 2002. The unconventional lifestyle of NKT Cells. Nat. Rev. Immunol. 2:557–568. [DOI] [PubMed] [Google Scholar]
- 3.Park, S.H., A. Weiss, K. Benlagha, T. Kyin, L. Teyton, and A. Bendelac. 2001. The mouse CD1d-restricted repertoire is dominated by a few autoreactive T cell receptor families. J. Exp. Med. 193:893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ugolini, S., and E. Vivier. 2000. Regulation of T cell function by NK cell receptors for classical MHC class I molecules. Curr. Opin. Immunol. 12:295–300. [DOI] [PubMed] [Google Scholar]
- 5.Raulet, D.H., R.E. Vance, and C.W. McMahon. 2001. Regulation of the natural killer cell receptor repertoire. Annu. Rev. Immunol. 19:291–330. [DOI] [PubMed] [Google Scholar]
- 6.Ikarashi, Y., R. Mikami, A. Bendelac, M. Terme, N. Chaput, M. Terada, T. Tursz, E. Angevin, F.A. Lemonnier, H. Wakasugi, and L. Zitvogel. 2001. Dendritic cell maturation overrules H-2D-mediated natural killer T (NKT) cell inhibition: critical role for B7 in CD1d-dependent NKT cell interferon gamma production. J. Exp. Med. 194:1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maeda, M., S. Lohwasser, T. Yamamura, and F. Takei. 2001. Regulation of NKT cells by Ly49: analysis of primary NKT cells and generation of NKT cell line. J. Immunol. 167:4180–4186. [DOI] [PubMed] [Google Scholar]
- 8.Lanier, L.L., and A.B. Bakker. 2000. The ITAM-bearing transmembrane adaptor DAP12 in lymphoid and myeloid cell function. Immunol. Today. 21:611–614. [DOI] [PubMed] [Google Scholar]
- 9.Ortaldo, J.R., R. Winkler-Pickett, A.T. Mason, and L.H. Mason. 1998. The Ly-49 family: regulation of cytotoxicity and cytokine production in murine CD3+ cells. J. Immunol. 160:1158–1165. [PubMed] [Google Scholar]
- 10.Smith, H.R., H.H. Chuang, L.L. Wang, M. Salcedo, J.W. Heusel, and W.M. Yokoyama. 2000. Nonstochastic coexpression of activation receptors on murine natural killer cells. J. Exp. Med. 191:1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lees, R.K., I. Ferrero, and H.R. MacDonald. 2001. Tissue-specific segregation of TCRgamma delta+ NKT cells according to phenotype TCR repertoire and activation status: parallels with TCR alphabeta+NKT cells. Eur. J. Immunol. 31:2901–2909. [DOI] [PubMed] [Google Scholar]
- 12.Pauza, M., K.M. Smith, H. Neal, C. Reilly, L.L. Lanier, and D. Lo. 2000. Transgenic expression of Ly-49A in thymocytes alters repertoire selection. J. Immunol. 164:884–892. [DOI] [PubMed] [Google Scholar]
- 13.Fahlen, L., L. Oberg, T. Brannstrom, N.K. Khoo, U. Lendahl, and C.L. Sentman. 2000. Ly49A expression on T cells alters T cell selection. Int. Immunol. 12:215–222. [DOI] [PubMed] [Google Scholar]
- 14.MacDonald, H.R., R.K. Lees, and W. Held. 1998. Developmentally regulated extinction of Ly-49 receptor expression permits maturation and selection of NK1.1+ T cells. J. Exp. Med. 187:2109–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pircher, H., T.W. Mak, R. Lang, W. Ballhausen, E. Ruedi, H. Hengartner, R.M. Zinkernagel, and K. Burki. 1989. T cell tolerance to Mlsa encoded antigens in T cell receptor V beta 8.1 chain transgenic mice. EMBO J. 8:719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Held, W., D. Cado, and D.H. Raulet. 1996. Transgenic expression of the Ly49A natural killer cell receptor confers class I major histocompatibility complex (MHC)-specific inhibition and prevents bone marrow allograft rejection. J. Exp. Med. 184:2037–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ioannidis, V., J. Zimmer, F. Beermann, and W. Held. 2001. Cre recombinase-mediated inactivation of H-2Dd transgene expression: evidence for partial missing self-recognition by Ly49A NK cells. J. Immunol. 167:6256–6262. [DOI] [PubMed] [Google Scholar]
- 18.Eberl, G., R. Lees, S.T. Smiley, M. Taniguchi, M.J. Grusby, and H.R. MacDonald. 1999. Tissue-specific segregation of CD1d-dependent and CD1d-independent NKT cells. J. Immunol. 162:6410–6419. [PubMed] [Google Scholar]
- 19.Wolfer, A., T. Bakker, A. Wilson, M. Nicolas, V. Ioannidis, D.R. Littman, P.P. Lee, C.B. Wilson, W. Held, H.R. MacDonald, and F. Radtke. 2001. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat. Immunol. 2:235–241. [DOI] [PubMed] [Google Scholar]
- 20.Matsuda, J.L., O.V. Naidenko, L. Gapin, T. Nakayama, M. Taniguchi, C.R. Wang, Y. Koezuka, and M. Kronenberg. 2000. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J. Exp. Med. 192:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortaldo, J.R., R. Winkler-Pickett, J. Willette-Brown, R.L. Wange, S.K. Anderson, G.J. Palumbo, L.H. Mason, and D.W. McVicar. 1999. Structure/function relationship of activating Ly-49D and inhibitory Ly-49G2 NK receptors. J. Immunol. 163:5269–5277. [PubMed] [Google Scholar]
- 22.Bakker, A.B., R.M. Hoek, A. Cerwenka, B. Blom, L. Lucian, T. McNeil, R. Murray, L.H. Phillips, J.D. Sedgwick, and L.L. Lanier. 2000. DAP12-deficient mice fail to develop autoimmunity due to impaired antigen priming. Immunity. 13:345–353. [DOI] [PubMed] [Google Scholar]
- 23.Eberl, G., H.J. Fehling, H. von Boehmer, and H.R. MacDonald. 1999. Absolute requirement for the pre-T cell receptor alpha chain during NK1.1+ TCRalphabeta cell development. Eur. J. Immunol. 29:1966–1971. [DOI] [PubMed] [Google Scholar]
- 24.Benlagha, K., A. Weiss, A. Beavis, L. Teyton, and A. Bendelac. 2000. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J. Exp. Med. 191:1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benlagha, K., T. Kyin, A. Beavis, L. Teyton, and A. Bendelac. 2002. A thymic precursor to the NK T cell lineage. Science. 296:553–555. [DOI] [PubMed] [Google Scholar]
- 26.Pellicci, D.G., K.J. Hammond, A.P. Uldrich, A.G. Baxter, M.J. Smyth, and D.I. Godfrey. 2002. A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1.1(-)CD4(+) CD1d-dependent precursor stage. J. Exp. Med. 195:835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gadue, P., and P.L. Stein. 2002. NK T cell precursors exhibit differential cytokine regulation and require itk for efficient maturation. J. Immunol. 169:2397–2406. [DOI] [PubMed] [Google Scholar]
- 28.Gapin, L., J.L. Matsuda, C.D. Surh, and M. Kronenberg. 2001. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat. Immunol. 2:971–978. [DOI] [PubMed] [Google Scholar]
- 29.Oberg, L., M. Eriksson, L. Fahlen, and C.L. Sentman. 2000. Expression of Ly49A on T cells alters the threshold for T cell responses. Eur. J. Immunol. 30:2849–2856. [DOI] [PubMed] [Google Scholar]
- 30.Kim, S., K. Iizuka, H.S. Kang, A. Dokun, A.R. French, S. Greco, and W.M. Yokoyama. 2002. In vivo developmental stages in murine natural killer cell maturation. Nat. Immunol. 3:523–528. [DOI] [PubMed] [Google Scholar]