Abstract
Immunization with vaccinia virus resulted in long-lasting protection against smallpox and was the approach used to eliminate natural smallpox infections worldwide. Due to the concern about the potential use of smallpox virus as a bioweapon, smallpox vaccination is currently being reintroduced. Severe complications from vaccination were associated with congenital or acquired T cell deficiencies, but not with congenital agammaglobulinemia, suggesting the importance of T cell immunity in recovery from infection. In this report, we identified two CD8+ T cell epitopes restricted by the most common human major histocompatibility complex (MHC) class I allele, HLA-A*0201. Both epitopes are highly conserved in vaccinia and variola viruses. The frequency of vaccinia-specific CD8+ T cell responses to these epitopes measured by interferon (IFN)-γ enzyme-linked immunospot (ELISPOT) assay and HLA/peptide tetramer staining peaked 2 wk after primary immunization and then declined, but were still detectable 1 to 3 yr after primary immunization. 2 wk after immunization, IFN-γ–producing cells specific to these two epitopes were 14% of total vaccinia virus-specific IFN-γ–producing cells in one donor, 35% in the second donor, and 6% in the third donor. This information will be useful for studies of human T cell memory and for the design and analyses of the immunogenicity of experimental vaccinia vaccines.
Keywords: immunologic memory, T lymphocyte epitopes, cytotoxic T lymphocytes, smallpox vaccine, major histocompatibility complex
Introduction
Immunization with vaccinia virus resulted in long-lasting protection against smallpox and was the successful approach used to eliminate natural smallpox infections worldwide. This accomplishment was achieved without a detailed understanding of human T cell responses to poxviruses. Severe and life threatening complications from vaccination were associated with congenital or acquired T cell deficiencies, but not with congenital agammaglobulinemia (1). In addition to emphasizing the importance of T cells in the immunity to smallpox, this finding suggests a critical need to develop new vaccines safe for use in T cell–deficient populations.
Due to the concern about the potential use of smallpox virus as a bioweapon, smallpox vaccination is currently being reintroduced. Considering the high incidence of side effects from current smallpox vaccine, the development of a safer, but equally effective vaccine is very important (2). To analyze T cell responses to licensed and experimental smallpox vaccines at the single cell level, it is essential to identify CD8+ T cell epitopes. We detected vaccinia-specific CD4+ and CD8+ T cells in humans and have measured the number of vaccinia virus-specific T cell responses to smallpox vaccine (3–7). An intracellular cytokine staining assay was applied to quantitate and characterize vaccinia-specific T cells in mice (8). However, no T cell epitopes have been identified in humans or mice systems. One major obstacle is the size of the virus. Vaccinia is a large virus with an ∼200-kbp DNA genome that has ∼200 open reading frames (9).
Among the highly polymorphic human MHC class I genes we chose HLA-A*0201 to identify CD8+ T cell epitopes because of the commonality of this allele among most ethnic groups (10). We first established several vaccinia-specific HLA-A*0201–restricted CTL lines, and then identified CD8+ T cell epitopes recognized by these CTL lines in cytotoxicity assays using synthetic vaccinia peptides designed based on the HLA-A*0201 binding motif (11).
In this report we identified two CD8+ T cell epitopes that are highly-conserved between vaccinia and variola (smallpox) viruses. The induction of the T cell responses after primary smallpox vaccination is demonstrated by the kinetics of epitope specific CD8+ T cells in three HLA-A*0201–positive individuals. This information will be useful for the design and analyses of the immunogenicity of experimental vaccinia vaccines, and for basic studies of human T cell memory.
Materials and Methods
Donors.
Donors in this study were three HLA-A*0201–positive laboratory workers received primary immunization by scarification with the licensed smallpox vaccine, Dryvax®, as recommended by the Centers for Disease Control and Prevention for laboratory personnel working with vaccinia viruses. The HLA-A and B alleles of donor 1 were A2 (A*0201), B15, B18; those of donor 2 were A2 (A*0201), B15, B44; and those of donor 3 were A2 (A*0201), A31, B40, B51.
Viruses.
Vaccinia virus New York City Board of Health (NYCBH), the same strain used to produce Dryvax®, was provided by Gail Mazzara and Dennis Panicali of Applied Biotechnology, Inc., and propagated and titrated in CV-1 cells (American Type Culture Collection no. CCL-70) as previously described (3, 12). Modified vaccinia virus Ankara strain (MVA) was supplied by Bernard Moss of National Institute of Allergy and Infectious Diseases/National Institutes of Health (Bethesda, MD), and was propagated and titrated in BHK-21 cells (American Type Culture Collection no. CCL-10) following published methods (13).
CTL Lines.
Vaccinia virus-specific CTL lines were isolated from PBMCs of immunized donors by limiting dilution cloning (4). Vaccinia virus NYCBH strain was used to stimulate PBMCs for cloning and to infect target cells for cytotoxicity assays. Cytotoxicity assays were performed as previously described (7). Hmy C1R A2.1 cells (gift from William E. Biddison of NIH/NINDS), which express only HLA-A*0201 at normal levels, were used as targets in cytotoxicity assay to confirm the HLA-A*0201 restriction. Surface expression of CD4 and CD8 was determined by flow cytometry using FITC-conjugated antibodies (Becton Dickinson). Cross-reactivity of CTL lines was determined using autologous B-LCLs (B-lymphoblastoid cell lines), that were infected with MVA as target cells in cytotoxicity assays.
Screening Peptides in Cytotoxicity Assay.
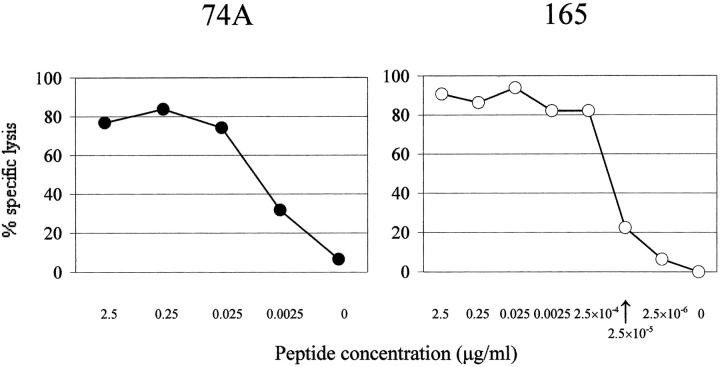
Peptides were synthesized with a Symphony automated peptide synthesizer at the Protein Core Facility in the University of Massachusetts Medical School or purchased from Mimotopes Pty. Ltd. When predicted epitopes overlapped, we made them as a longer peptide fragment. For technical reasons some of these screening peptides were made as a 13 mer instead of a 9 mer. In screening cytotoxicity assays, mixtures of five peptides were used and the concentration of each peptide was 5 μg/ml. When the peptide recognized was longer than 9 amino acids, truncated 9 mer peptide epitopes were constructed and analyzed in cytotoxicity assays. All peptides recognized were tested in dose–response experiments (Fig. 1) .
Figure 1.
Recognition of peptides 74A and 165 by cytotoxic T cell lines. CTL lines, VA55 3.13 (for 74A peptide) and VA49 3.12 (for 165 peptide), were tested against Hmy C1R-A2.1 cells pulsed with peptides at the indicated concentrations. Effector/target ratio was 5.
Tetramer Staining.
Peptide/HLA-A*0201 tetramers were made in the Tetramer Core Facility in the University of Massachusetts Medical School following the protocol published previously (14). Each lot of tetramer was titrated using CTL lines specific to the peptide mixed with autologous PBMCs at a 1 to 10 (or 20) ratio to determine the optimal concentration for staining.
IFN-γ ELISPOT Assay.
IFN-γ ELISPOT assays were performed as described previously (6). For stimulation, PBMCs were incubated with vaccinia virus NYCBH strain at an MOI of 1 or with peptide at a final concentration of 10 μg/ml for 16 h.
Results
Vaccinia virus-specific CD8+ CTL lines were established by limiting dilution cloning from the PBMCs of HLA-A*0201–positive donors who received primary immunization with the licensed smallpox vaccine, Dryvax®. We decided to identify T cell epitopes restricted by HLA-A*0201 and cross-reactive to MVA, which is being proposed for use as an attenuated smallpox vaccine and as a vector for vaccination against other infectious agents (15). We performed HLA-A*0201 peptide binding motif searches on all of the protein sequences of the MVA strain (16). We used the computer algorithm “HLA Peptide Binding Predictions” (http://bimas.dcrt.nih.gov/molbio/hla_bind/) to calculate the binding affinity (score) of 9 mer peptides to the HLA-A*0201 molecule (11). We hypothesized that early gene products may be more likely to have CD8+ T cell epitopes. In both humans and mice all of the known CD8+ T cell epitopes to cytomegalovirus are encoded by immediate-early phase proteins (17, 18). The early, early and late, and late genes in vaccinia were categorized by nucleotide sequence motifs, such as a late promoter or an early termination motif (19). For initial screening we synthesized all peptides with; (a) a binding score of more than a 1,000 (70 peptides); or (b) a binding score of 100 to 999 and encoded by a gene expressed early or both early and late (125 peptides). A total of 195 peptides were screened using the fifteen CTL lines.
One CTL line, VA55 3.13, established from donor 2, recognized a peptide 74A, CLTEYILWV, in a 21.7K protein encoded by a putative early and late gene, “189R,” of the MVA strain with a calculated binding score of 3607. Another CTL line, VA49 3.12, established from donor 1, recognized a peptide 165, KVDDTFYYV, which is in a host range protein encoded by a putative early and late gene, “018L,” with a calculated binding score of 365. Fig. 1 demonstrates the high level of specific recognition by these CTL lines of their respective epitope peptides in a dose response CTL experiment. These epitope sequences are highly-conserved in vaccinia and variola viruses (Table I).
Table I.
Conservation of Epitopes Among Poxviruses Causing Infection in Human
| GenBank/EMBL/DDBJ accession no. |
Gene name | 74A peptide | Gene name | 165 peptide | |
|---|---|---|---|---|---|
| Vaccinia | |||||
| MVA | U94848 | 189R | CLTEYILW V | 018L | KVDDTFYYV |
| Copenhagen | M35027 | B22R & C16Lb | * * * * * * * * * | C7L | * * * * * * * * * |
| Tian Tana | AF095689 | TC7L | * * * * * * * * * | ||
| Variola major | |||||
| Bangladesh-1975 | L22579 | D2L | * * * * * * * * * | D11L | * * * * * * * * * |
| India-1967 | X69198 | D1L | * * * * * * * * * | D8L | * * * * * * * * * |
| Variola minor | |||||
| Garcia-1966 | Y16780 | B1L | * * * * * * * * * | B14L | * * * * * * * * * |
| Cowpox | |||||
| Brighton Red | AF482758 | V212 | * * * * * * * * * | V028 | * * * * * * * * * |
| Monkeypox | |||||
| Zaire-96-I-16 | AF380138 | N1R | * * * * * * * * * | D10L | * * * Y * L * * * |
Only strains of which complete genome has been sequenced are listed.
indicate identical amino acid.
Tian Tan strain does not have 189R ortholog according to the nucleotide sequence.
Both genes are located within the inverted terminal repeats and the gene sequences are identical.
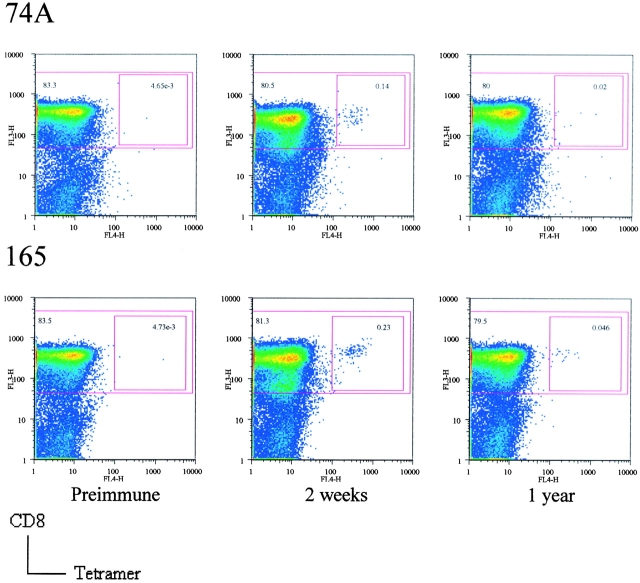
CD8+ T cells specific to these epitopes were measured at several time points after primary immunization by peptide/HLA-A*0201 tetramer staining using the PBMCs of three HLA-A*0201–positive donors. Fig. 2 shows representative FACS® plots of donor 1 PBMC. In Figs. 3 and 4 , “preimmune” means before primary first immunization, and “two weeks” means two weeks after the second immunization for donor 3 who failed to develop a “take” after primary immunization, and twenty days after primary immunization was immunized for the second time. In all three donors the frequency of vaccinia-specific CD8+ T cells peaked 2 wk after primary immunization and then declined, but were still detectable 1 to 3 yr after primary immunization (Fig. 3). 2 wk after vaccination the IFN-γ–producing cells specific to these two epitopes were 14% of total vaccinia virus-specific IFN-γ–producing cells in donor 1, 35% in donor 2, and 6% in donor 3 (Fig. 4).
Figure 2.
Quantitation of vaccinia virus epitope-specific CD8+ T cells by HLA-A*0201/peptide 74A tetramer staining (top) and HLA-A*0201/peptide 165 tetramer staining (bottom) of PBMCs of donor 1. Cells were gated on CD3+ and CD4− cells with tetramer staining on the x-axis and CD8 on the y-axis. The larger squares show CD8+ cells and the smaller squares show CD8+ and tetramer+ cells.
Figure 3.
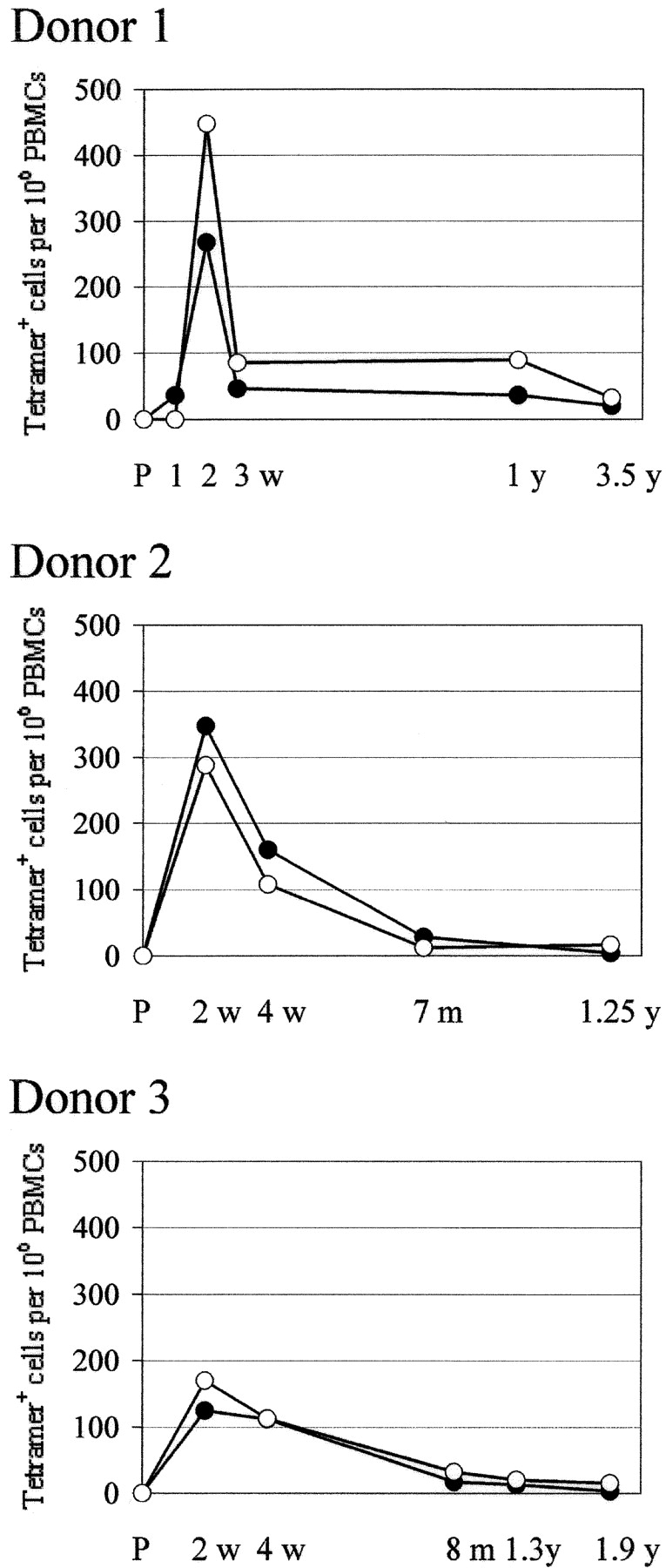
Kinetics of the frequency of CD8+ T cells specific for each epitope quantitated by tetramer staining in PBMCs of three donors after primary immunization. Closed circle, 74A-specific; open circle, 165-specific; P, preimmunization. Frequency is calculated per million PBMCs for comparison with the data from epitope-specific IFN-γ ELISPOT assays.
Figure 4.
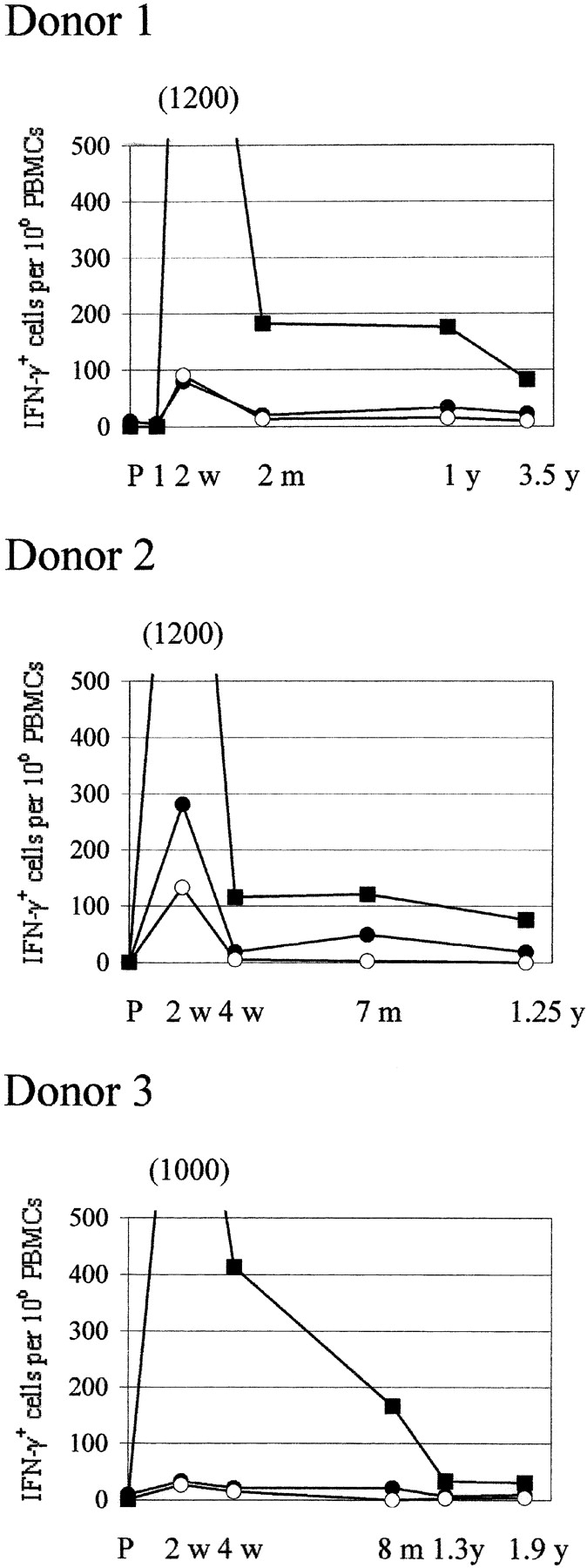
Kinetics of the frequency of IFN-γ–producing cells specific to these peptides and to whole vaccinia virus quantitated by IFN-γ ELISPOT assay using the PBMCs of the same donors shown in Fig. 3. Closed circle, peptide 74A–specific cells; open circle, peptide 165–specific cells; closed square, vaccinia virus–specific cells; P, preimmunization. Frequency is calculated per million PBMC.
Discussion
In this report we identified two CD8+ T cell epitopes restricted by HLA-A*0201, the most common MHC class I allele in humans. These are the first T cell epitopes that have been reported for vaccinia virus. IFN-γ–producing cells specific to these two epitopes represented 6 to 35% of total number of IFN-γ–producing cells specific to vaccinia virus. The frequency of epitope-specific T cells was always higher by peptide/HLA tetramer staining than by IFN-γ–ELISPOT assay, although post-vaccination kinetics for each epitope-specific T cell were similar using both methods.
As for epitope selection, peptide 74A was the 15th highest binding peptide to HLA-A*0201 of the 195 peptides we selected for screening and peptide 165 was the 95th highest binder. We did not experimentally measure binding affinity of these peptides, but they are not epitopes based solely on their HLA-A*0201 binding affinity. One common characteristic of these two peptides is that they are both encoded by genes with a late promoter and an early termination motif (19), which means they may be expressed at both early and late phases of infection. The 189R gene of MVA strain encoding peptide 74A is a nonessential gene (20) with unknown function (21). The 018L gene of MVA encoding peptide 165 is an ortholog of the host range protein, C7L, of the Copenhagen strain (22). Although our selection of peptides was biased toward genes expressed in the early phase of infection, viral proteins produced in the early phase of infection may be processed and presented more efficiently by infected cells than those produced only in late phase, as a result of vaccinia virus down-regulating host protein synthesis. These two epitopes are highly conserved among variola viruses, suggesting the CTLs recognizing these epitopes will recognize variola virus-infected cells.
The frequency of epitope-specific HLA-A*0201–restricted T cells varied among the three donors. Boon et al. reported that the HLA class I background of individuals has a major influence on the magnitude and specificity of the CTL response against influenza A virus. Even in individuals who share a certain HLA allele, the CTL response restricted by that HLA allele differs and may be affected by other nonmatching HLA molecules resulting in different CTL responses (23). To determine immunodominance we need to identify other CD8+ T cell epitopes, and analyze the PBMCs of more donors with different combinations of MHC class I molecules (24).
After submission of this paper for publication, Drexler et al. published a report of a vaccinia virus epitope that they found in HLA-A*0201-transgenic mice (25). Their epitope is different from the two we report in this publication. They did not report any experiments with quantitation of human T cells specific to their epitope. We failed to detect responses to that epitope by IFN-γ–ELISPOT assay in the three vaccinated individuals who developed responses to the two epitopes we now report. We conclude that the epitopes reported here are dominant to the one reported recently by Drexler et al.
Identification of these epitopes will enable the analysis and quantitation of vaccinia virus-specific CD8+ T cells in the acute and memory phases and to compare CD8+ T cell responses specific to different epitopes. We should also be able to monitor expansion and subsequent shrinkage of epitope-specific CD8+ T cells at the T cell receptor level. Definition of T cell epitopes will help us to better understand human T cell responses to vaccinia virus as a model of human infection. In addition, it will provide a quantitative measure of poxvirus T cell immunity when these viruses are used as viral vectors.
Acknowledgments
We thank Laura Orphins and Vera Hauptfeld for the assistance with the IFN-γ ELISPOT and HLA typing, respectively.
This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases grant PO1 AI-49320 and a subcontract, AI-46725.
References
- 1.Esposito, J.J., and F. Fenner. 2001. Poxviruses. Fields Virology. D.M. Knipe and P.M. Howley, editors. Lippincott Williams & Wilkins, Philadelphia, PA. 2885–2921.
- 2.Enserink, M. 2002. Bioterrorism. In search of a kinder, gentler vaccine. Science. 296:1594. [DOI] [PubMed] [Google Scholar]
- 3.Littaua, R.A., A. Takeda, J. Cruz, and F.A. Ennis. 1992. Vaccinia virus-specific human CD4+ cytotoxic T-lymphocyte clones. J. Virol. 66:2274–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demkowicz, W.E., Jr., and F.A. Ennis. 1993. Vaccinia virus-specific CD8+ cytotoxic T lymphocytes in humans. J. Virol. 67:1538–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demkowicz, W.E., Jr., R.A. Littaua, J. Wang, and F.A. Ennis. 1996. Human cytotoxic T-cell memory: long-lived responses to vaccinia virus. J. Virol. 70:2627–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ennis, F.A., J. Cruz, W.E. Demkowicz, Jr., A.L. Rothman, and D.J. McClain. 2002. Primary induction of human CD8+ cytotoxic T lymphocytes and interferon-gamma-producing T cells after smallpox vaccination. J. Infect. Dis. 185:1657–1659. [DOI] [PubMed] [Google Scholar]
- 7.Frey, S.E., F.K. Newman, J. Cruz, W.B. Shelton, J.M. Tennant, T. Polach, A.L. Rothman, J.S. Kennedy, M. Wolff, R.B. Belshe, and F.A. Ennis. 2002. Dose-related effects of smallpox vaccine. N. Engl. J. Med. 346:1275–1280. [DOI] [PubMed] [Google Scholar]
- 8.Harrington, L.E., R. van der Most, J.L. Whitton, and R. Ahmed. 2002. Recombinant vaccinia virus-induced T-cell immunity: quantitation of the response to the virus vector and the foreign epitope. J. Virol. 76:3329–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moss, B. 2001. Poxviridae: The viruses and their replication. Fields Virology. D.M. Knipe and P.M. Howley, editors. Lippincott Wiliams & Wilkins, Philadelphia, PA. 2849–2883.
- 10.Imanishi, T., T. Akaza, A. Kimura, K. Tokunaga, and T. Gojobori. 1997. Allele and haplotype frequencies for HLA and complement loci in various ethnic groups. Proceedings of the Twelfth International Histocompatibility Workshop and Conference. D. Charron, editor. Medical and Scientific Publisher, Paris, France. 1065–1074.
- 11.Parker, K.C., M.A. Bednarek, and J.E. Coligan. 1994. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 152:163–175. [PubMed] [Google Scholar]
- 12.Terajima, M., H.L. Van Epps, D. Li, A.M. Leporati, S.E. Juhlin, J. Mustonen, A. Vaheri, and F.A. Ennis. 2002. Generation of recombinant vaccinia viruses expressing Puumala virus proteins and use in isolating cytotoxic T cells specific for Puumala virus. Virus Res. 84:67–77. [DOI] [PubMed] [Google Scholar]
- 13.Carroll, M.W., and B. Moss. 1997. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a nonhuman mammalian cell line. Virology. 238:198–211. [DOI] [PubMed] [Google Scholar]
- 14.Catalina, M.D., J.L. Sullivan, K.R. Bak, and K. Luzuriaga. 2001. Differential evolution and stability of epitope-specific CD8(+) T cell responses in EBV infection. J. Immunol. 167:4450–4457. [DOI] [PubMed] [Google Scholar]
- 15.Carroll, M.W., and B. Moss. 1997. Poxviruses as expression vectors. Curr. Opin. Biotechnol. 8:573–577. [DOI] [PubMed] [Google Scholar]
- 16.Antoine, G., F. Scheiflinger, F. Dorner, and F.G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology. 244:365–396. [DOI] [PubMed] [Google Scholar]
- 17.Reddehase, M.J., and U.H. Koszinowski. 1984. Significance of herpesvirus immediate early gene expression in cellular immunity to cytomegalovirus infection. Nature. 312:369–371. [DOI] [PubMed] [Google Scholar]
- 18.Reddehase, M.J. 2000. The immunogenicity of human and murine cytomegaloviruses. Curr. Opin. Immunol. 12:390–396. [DOI] [PubMed] [Google Scholar]
- 19.Goebel, S.J., G.P. Johnson, M.E. Perkus, S.W. Davis, J.P. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virology. 179:247–266, 517–263. [DOI] [PubMed]
- 20.Perkus, M.E., S.J. Goebel, S.W. Davis, G.P. Johnson, E.K. Norton, and E. Paoletti. 1991. Deletion of 55 open reading frames from the termini of vaccinia virus. Virology. 180:406–410. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, G.P., S.J. Goebel, and E. Paoletti. 1993. An update on the vaccinia virus genome. Virology. 196:381–401. [DOI] [PubMed] [Google Scholar]
- 22.Perkus, M.E., S.J. Goebel, S.W. Davis, G.P. Johnson, K. Limbach, E.K. Norton, and E. Paoletti. 1990. Vaccinia virus host range genes. Virology. 179:276–286. [DOI] [PubMed] [Google Scholar]
- 23.Boon, A.C., G. de Mutsert, Y.M. Graus, R.A. Fouchier, K. Sintnicolaas, A.D. Osterhaus, and G.F. Rimmelzwaan. 2002. The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype. J. Virol. 76:582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Betts, M.R., J.P. Casazza, B.A. Patterson, S. Waldrop, W. Trigona, T.M. Fu, F. Kern, L.J. Picker, and R.A. Koup. 2000. Putative immunodominant human immunodeficiency virus-specific CD8(+) T-cell responses cannot be predicted by major histocompatibility complex class I haplotype. J. Virol. 74:9144–9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drexler, I., C. Staib, W. Kastenmuller, S. Stevanovic, B. Schmidt, F.A. Lemonnier, H.G. Rammensee, D.H. Busch, H. Bernhard, V. Erfle, and G. Sutter. 2003. Identification of vaccinia virus epitope-specific HLA-A*0201-restricted T cells and comparative analysis of smallpox vaccines. Proc. Natl. Acad. Sci. USA. 100:217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]