Abstract
In the thymus, several steps of proliferative expansion and selection coordinate the maturation of precursors into antigen-specific T cells. Here we identify the transcriptional repressor Gfi1 as an important regulator of this maturation process. Mice lacking Gfi1 show reduced thymic cellularity due to an increased cell death rate, lack of proliferation, and a differentiation block in the very early uncommitted CD4−/CD8−/c-Kit+ cytokine-dependent T cell progenitors that have not yet initiated VDJ recombination. In addition, Gfi1-deficient mice show increased major histocompatibility complex class I–restricted positive selection and develop significantly more CD8+ cells suggesting a requirement of Gfi1 for a correct CD4/CD8 lineage decision. Absence of Gfi1 correlates with high level expression of the genes for lung Krüppel-like factor (LKLF), inhibitor of DNA binding (Id)1 and Id2, suggesting the existence of new regulatory pathways in pre-T cell development and thymic selection in which Gfi1 acts upstream of LKLF as well as the E-proteins, which are negatively regulated by Id1 and Id2.
Keywords: zinc finger protein, T cell differentiation, apoptosis, lineage skewing, helix-loop-helix proteins
Introduction
Gfi1 is a 55-kD nuclear protein and a member of a protein family that includes Gfi1b (1, 2) as well as the murine proteins Snail and Slug (3, 4). All proteins of the Gfi1 family share six carboxyterminal C2-H2 zinc finger domains and the characteristic “SNAG” domain that comprises the first amino terminal 20 amino acid residues (4). It has been demonstrated that Gfi1 is able to bind to specific DNA target sequences in vitro and that DNA binding depends on the presence of the zinc finger domains. Experiments with reporter genes driven by synthetic promoters containing Gfi1 binding sites suggested a transcriptional repressor activity of Gfi1 (3, 4). An alternative activity of Gfi1 has been discovered through its interaction with the protein inhibitor of activated signal transducer and activator of transcription (STAT) (PIAS)*3 (5). PIAS3 can bind to the activated form of the latent STAT3 transcription factor and is able to down-regulate the activity of STAT3 as a transcriptional transactivator (6). By virtue of its interaction with PIAS3, Gfi1 is able to relieve STAT3 from PIAS3-mediated inhibition with the consequence of an enhanced STAT3 response (5). This suggested a role of Gfi1 in a set of specific cytokine-signaling pathways because STAT3 is activated in response to a number of cytokines, among them IL-2, IL-6, IL-10, or G-CSF.
A role of Gfi1 in cytokine or growth factor signaling had already been suggested earlier because the Gfi1 gene was found to be activated by proviral insertion in Nb2 rat lymphoma cell clones selected for IL-2–independent growth after infection with Moloney murine leukemia virus (7). Indeed, several studies with cultured cells indicated that a constitutive Gfi1 expression can relieve peripheral mature T cells from a requirement of IL-2 to overcome a G1 arrest (3) or could help sustain cell proliferation of IL-2–dependent cells in the absence of the cytokine (8), indicating a role of Gfi1 in IL-2–dependent cell cycle progression of T cells. Similarly, the Gfi1 gene is transcriptionally activated by retroviral insertion in T lymphoid tumors that arise in mice after infection with Moloney murine leukemia virus (8–10). In addition, we have previously shown that Gfi1 acts as a dominant oncogene when overexpressed and cooperates strongly with pim-1 and myc genes in T cell lymphomagenesis (11).
Gfi1 is expressed in T cell precursors from the early stages of pre-T cell development up to the point when cells express both CD4 and CD8 surface markers (12). In mature CD4 or CD8 single positive (SP) T cells, Gfi1 is absent but rapidly up-regulated after TCR stimulation (5). The T cell progenitors that lack CD4 and CD8 surface marker (double negative [DN] cells) can be subdivided according to the surface markers CD44 and CD25 (13). The earliest precursor cells that enter the thymus from the bone marrow, which express CD44 (DN1) or CD44 and CD25 (DN2), are still uncommitted and depend on the presence of cytokines such as stem cell factor (SCF; c-Kit ligand) and IL-7 and the functionality of their respective receptors (14–18). These cells can still develop into other lineages (Lins) such as thymic dendritic cells, NK cells, and B cells, and maintain an unrearranged germline configuration of all TCR loci (16, 19). Therefore, deletion of both c-Kit and IL-7R entirely block pre-T cell development beyond this very early stage as well as the formation of a T cell repertoire. In contrast, deletion of only c-Kit or SCF or of the IL-7R still allows the development of all T cell Lins albeit at reduced rates (17, 20, 21).
The early CD44+ CD25− (DN1) precursor cells up-regulate CD25 expression and start to proliferate in a first wave of cytokine-dependent pre-T cell expansion and give rise to the DN2 subset. When these cells enter G1/G0 and start to rearrange their TCRβ, TCRγ, and TCRδ chain genes (22, 23), CD44 expression decreases to form the CD25+/CD44−lo DN3 subset. This DN3 population is proliferating as well as the main source of precursor cells that populate the thymus. DN3 cells are subject to a process called pre-TCR selection or “β selection” (13, 16, 24–26). Only those DN3 cells that have productively rearranged their TCRβ chain gene segments survive this selection step (“L cells”). CD25−/CD44−lo (DN4) cells, which directly emerge from DN3, express functional TCRβ chains and are termed “β selected” (26, 27). They quickly up-regulate the surface markers CD4 and CD8 and give rise to the major double positive (DP) T cell population of the thymus (28, 29). High levels of constitutively expressed Gfi1 can inhibit the passage of DN3 cells that have not yet completed VDJ recombination (“E cells”) to L cells during β selection, suggesting a role of Gfi1 in this process (12). Once DN cells up-regulate the CD4 and CD8 surface markers and express functional α/β TCR molecules, a process called positive/negative selection ensures the elimination of self-reactive T cells and the propagation of self-tolerant T cells (30–32). Positive/negative selection overlaps with CD4/CD8 Lin decision leading to the exclusive expression of either CD4 on MHC class II– or CD8 on MHC class I–restricted T cells, respectively.
Here we show that Gfi1−/− mice have a severely decreased thymic cellularity due to defects in cell survival and proliferation of the very early DN1 and DN2 c-Kit+ pre-T cells associated with a block of differentiation from DN1 cells into the DN2 stage and a reduced DN3 population. We also show accelerated passage from E to L cells during pre-TCR selection. In addition, Gfi1 deficiency is associated with accelerated positive selection of MHC I–restricted DP thymocytes and seems to affect the CD4/CD8 Lin decision. Finally, the disruption of thymic T cell maturation in mice lacking Gfi1 is associated with the deregulated expression of a restricted set of effector genes, among them the helix-loop-helix transcription factors inhibitor of DNA binding (Id)1 and Id2 (33).
Materials and Methods
Generation of Gfi1-deficient Mice.
Gfi1-deficient mice were generated by homologous recombination in R1 embryonic stem cells. Transfection of embryonic stem cells and selection of clones was performed essentially as previously described (34). Mice were housed at the animal facility of the University of Essen Medical School in single ventilated cages under specific pathogen-free conditions. Mice that were used for analyses were healthy 4–6-wk-old animals from an 8–10 generation backcross with C57BL/6 animals.
Antibody Staining Procedures and Cell Cycle Analysis.
Single cell suspensions were prepared as previously described (35) at the time of autopsy from the thymus or spleen in PBS supplemented with 0.5% FCS (staining solution). Cell numbers were calculated using a CASY-1 cell counter (Schärfe System). Cells were washed in this solution and incubated at 4°C for 30 min with antibodies directly conjugated with fluorochromes. Cells were washed twice in staining solution after the incubation and examined with a FACSCalibur® (Becton Dickinson). To analyze the DN population, Lin marker negative cells (Lin−) were selected by staining thymocytes with the biotinylated antibodies against CD3, CD4, CD8, B220, Gr-1, Mac-1, Ter-119, and DX5 followed by streptavidin-PerCP or –tri-color and APC- or PE-labeled anti-CD25 and FITC- or PE-labeled anti-CD44. PerCP or tri-color–negative cells (Lin−) were analyzed for expression of CD44 versus CD25. Staining for intracellular TCRβ chains was performed as previously described (36). Antibodies against TCRα and TCRβ chain subtypes and all tri-color–conjugated antibodies were purchased from Medac, antibodies against γ/δ TCR and CD62L were obtained from Immunotech, and all other antibodies used in FACS® analysis were from BD Biosciences. For the cell cycle analysis, thymocytes were stained as described above, washed, and then incubated in 2 μg/ml Hoechst 33382 (Sigma-Aldrich) in PBS for 30 min at 37°C. FACS® analysis on the gated DN subsets was performed by flow cytometry with an ultra violet laser at 424 nm. For the detection of apoptotic cells, an annexin V staining kit from BD Biosciences was used according to the manufacturer's instructions.
Bromo-deoxy-uridine (BrdU) Incorporation.
Three groups of six mice (three WT and three Gfi1−/− for each group) were initially injected intraperitoneally with 1.8 mg/200 μl BrdU (Roth) in saline and then continuously given BrdU at 1 mg/ml in the drinking water that was changed daily. At different time points (24, 48, and 72 h), three WT and three Gfi1−/− animals were killed and their thymi were removed. Thymocytes were first stained for extracellular markers and then washed, fixed, and stained again with FITC-conjugated anti-BrdU antibodies (Caltag) using the Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer's instructions.
Fetal Thymic Organ Culture (FTOC).
Pregnant mice were killed at day 13 after coitum and fetal thymi were prepared and incubated on isopore membranes (0.8 μm ATTP; Millipore) that had been equilibrated on FTOC medium (95% medium-119, 5% FBS, 1,000 U penicillin/streptomycin) for 1 h. Incubation of FTOC was performed at 5% CO2 for 5 d. The medium was changed after a period of 2 d. Typical cell recovery was between 2.5 and 3 × 104 cells per lobe.
DNA Array Analysis.
Total RNA from thymocytes was used without enrichment of the polyA+ fraction to prepare a labeled RNA target as previously described (37). In brief, the RNA was used to synthesize a single stranded cDNA via oligo dT priming and RT. The cDNA was made double stranded and then transcribed into a cRNA in an in vitro transcription reaction. The in vitro transcription reaction results in a linear amplification of the mRNA molecules originally isolated. This target RNA was hybridized to the Murine 11K Affymetrix high density DNA probe array. Signal intensities for the known and defined genes in this array omitting the EST sequences were recorded with the Microarray Suite 4.0 Software.
RT-PCR.
RT-PCR was performed as previously described (38). In brief, DP thymocytes were sorted by FACSVantage™ and DN and CD8+ thymocytes were isolated by using magnetic beads on autoMACS (Miltenyi Biotec). RNA was prepared with the use of RNeasy Kit (QIAGEN) according to the manufacturer's instructions. The synthesis of cDNA was performed as described above. The sequences of primers used are as follows: Id1 sense, 5′-TCAGGATCATGAAGGTCGCCAGTG-3′; Id1 antisense, 5′-TGAAGGGCTGGAGTCCATCTGGT-3′; Id2 sense, 5′-TCTGAGCTTATGTCGAATGATAGC-3′; Id2 antisense, 5′-CACAGCATTCAGTAGGCTCGTGTC-3′; β actin sense, 5′-TCCTGTGGCATCCATGAAACT-3′; and β actin antisense, 5′-GAAGCACTTGCGGTGCACGAT-3′.
Results
Gfi1-deficient Mice Have Low Thymic Cellularity and a Bias toward the Development of CD8 SP Cells.
The generation of Gfi1-deficient mice has been described elsewhere (34). Animals were generated in a mixed F1 (129×C57BL/6) background and then backcrossed with C57BL/6 mice for six to eight generations. All mice used in this study were healthy animals from this intercross held under specific pathogen-free conditions at 4–6 wk of age except when otherwise indicated. The thymi of Gfi1-deficient (Gfi1−/−) mice were very small and total cell numbers were reduced on average to only 10% of levels found in WT or Gfi1 heterozygous animals (Gfi1+/−; Fig. 1 A) whereas histological sections revealed a largely normal thymic architecture (unpublished data). Accordingly, the absolute numbers of the DN, DP, and SP populations and of α/β TCR– and γ/δ TCR–bearing T cells were significantly reduced in Gfi1−/− mice compared with WT controls (Fig. 1, B and C). The relative numbers of DP cells were decreased whereas DN and CD4+ SP cell frequencies remained unchanged in Gfi1−/− mice (Fig. 1 D). However, the ratio of CD4 to CD8 percentages were altered in favor of the CD8 SP population from 4:1 in WT to equal levels (1:1) in Gfi1-deficient mice (Fig. 1 D). An additional CD4+/CD8lo population was detected in Gfi1-deficient animals that is absent from the corresponding WT control (Fig. 1 D, small round gate).
Figure 1.
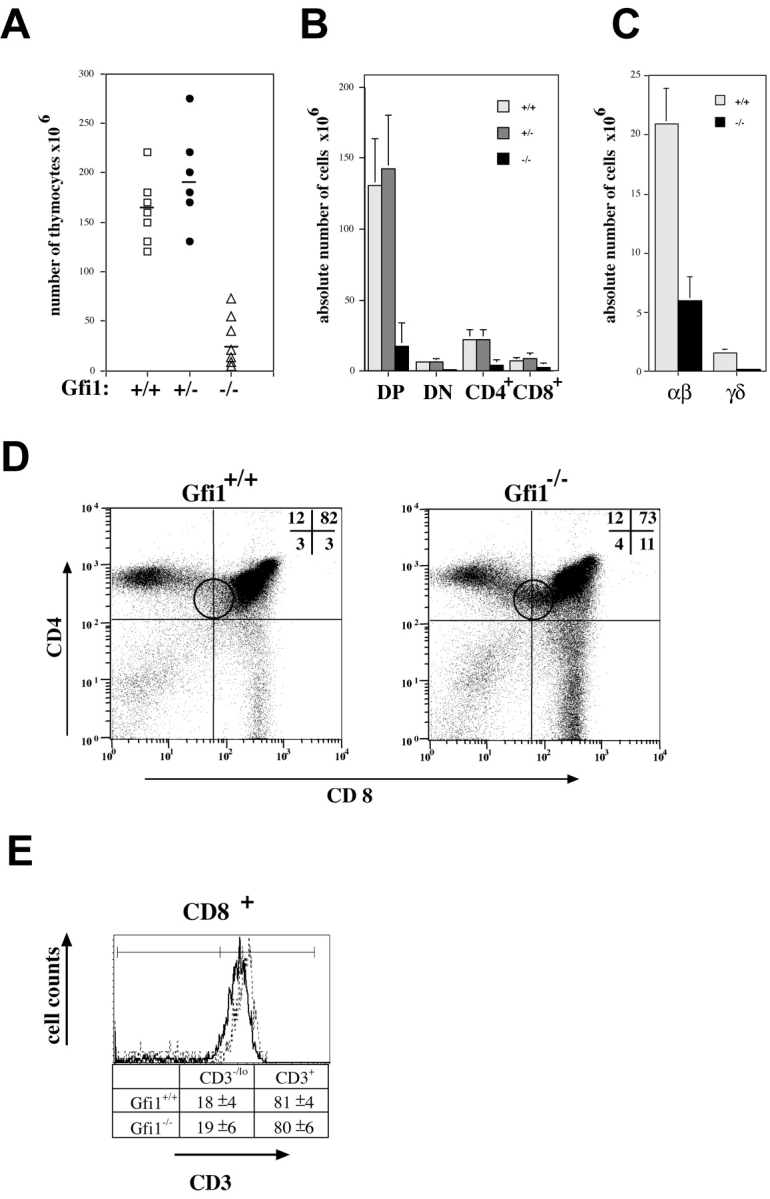
Gfi1−/− mice show impaired thymocyte development and reduced thymic cellularity. (A) Absolute thymocyte numbers from Gfi1-deficient mice (▵) are reduced compared with those from their WT (□) or heterozygous littermates (•). The average cell number is indicated by a solid line. (B and C) Thymocytes from 4–6-wk-old WT, Gfi1+/−, and Gfi1−/− mice were isolated, stained with anti-CD4, anti-CD8 antibodies, or with antibodies recognizing TCRβ chain or γ/δ chains and were analyzed by flow cytometry. Absolute numbers of all four CD4/CD8 subsets and of TCR-bearing cells representative of several animals are documented. (D) Cytofluorimetric analysis showing the relative percentages of cells bearing CD4 or CD8 surface markers of normal control littermates (left) and Gfi1-deficient mice (right). The percentage of CD8 SP cells is increased from 3 to 11% whereas the percentage of CD4 SP cells remained unchanged in Gfi1−/− mice. In addition, the percentage of DP cells is decreased to 73% in the absence of Gfi1 and a new CD4+ CD8lo population appears in Gfi1−/− mice (round gate). These alterations are consistently found in all analyses. Representative examples of ∼10 6-wk-old animals for each genotype are shown. (E) Flow cytometric analysis of CD3 expression of electronically gated CD8+ cells of both WT and Gfi1null mice. A representative histogram for each subset and compiled percentages of CD3+- and CD3−/lo-expressing cells is shown. The number represents means with standard deviations from six individual WT and six individual Gfi1-deficient mice. The dotted line represents WT animals and the dark line represents the Gfi1-deficient cells.
To test whether the higher percentage of CD8+ cells was due to an overrepresentation or a delayed development of immature CD8 SP cells, we analyzed electronically gated CD8+ cells from six individual WT and six individual Gfi1-deficient mice for the expression level of CD3. Immature SP (ISP) cells bear low levels of CD3 whereas mature selected CD8+ cells express high levels. We find the same percentages of CD8+ CD3hi and CD8+ CD3lo populations in WT as in Gfi1-deficient mice (Fig. 1 E) excluding overrepresentation of ISP cells in Gfi1 KO mice.
Expression of different variable TCR chains (Vα8, Vβ4, Vβ7, Vβ8.2, Vβ10, and Vβ12b) was generally detected on thymocytes of Gfi1−/− mice (unpublished data) suggesting that the lack of Gfi1 does not affect VDJ recombination during or after pre-TCR selection nor grossly alters the TCR repertoire. Moreover, spleen and lymph nodes of Gfi1−/− animals contained mature T cells indicating that the loss of Gfi1 does not generally affect T lymphocyte homing nor exit from the thymus (unpublished data). The low thymic cellularity, the bias toward CD8+ cells, and the emergence of a new CD4+/CD8lo population could be reproduced in mixed bone marrow chimeras by transplantation of Gfi1−/− cells (unpublished data). This indicated that the phenotype of Gfi1-deficient thymocytes is intrinsic and cell autonomous.
Gfi1 Deficiency Negatively Affects the Generation of CD4 SP Cells and Accelerates MHC I–restricted Positive Selection of CD8 SP Cells.
To analyze in more detail the effect of Gfi1 deficiency on the ratio of CD4/CD8 cells, we have used the activation marker CD69, which is transiently expressed during the selection process and discriminates between cells before and after positive/negative selection (39). To distinguish between different maturation stages of DP and SP cells, we have stained thymocytes from both WT and Gfi1−/− mice with CD4, CD8, CD69, and TCRβ expression and analyzed the cells with four-color flow cytometry (40). This defined five sequential maturation stages: R2: TCRβ− CD69−; R3: TCRβint CD69−; R4: TCRβint CD69+; R5: TCRβhi CD69+; and R6: TCRβhi CD69− (Fig. 2 A). This analysis showed that the percentages of TCRβhi CD69+ cells are reduced from 13% in WT mice to 5% in Gfi1null mice (Fig. 2 A, R5) whereas the relative percentages of TCRβint CD69+ cells remained unchanged (Fig. 2 A, R4). In accordance with previously published experiments, CD4+ cells appear before CD8+ cells (Fig. 2 A, R4 and R5). In Gfi1-deficient mice, this sequential appearance seems to be unaffected but the relative proportion of CD4+ SP cells is decreased from 17 to 8% in R4 and from 69 to 40% in R5 compared with WT mice (Fig. 2 A). This suggests that during and after selection, the ratio of CD4/CD8 percentages is continuously altered in Gfi1−/− mice in favor to the CD8+ compartment (Fig. 2 A, R4, R5, and R6). An even more pronounced situation was found in FTOC experiments of d13 fetal thymocytes where CD4 SP cells were almost completely absent in thymi from Gfi1-deficient mice after 5 d of culture (Fig. 2 B) and in the IL-7Rα+ subpopulation of freshly isolated thymocytes where CD8 SP cells are also overrepresented. (Fig. 2 C). In addition, percentages of cells positive for the IL-7Rα chain were found to be significantly higher in DP and in CD8 SP subsets of Gfi1−/− mice than in the same subsets of WT mice (Fig. 2 D). The expression level of IL-7Rα chain itself was similar in WT and Gfi1−/− DP and SP subsets (Fig. 2 D).
Figure 2.
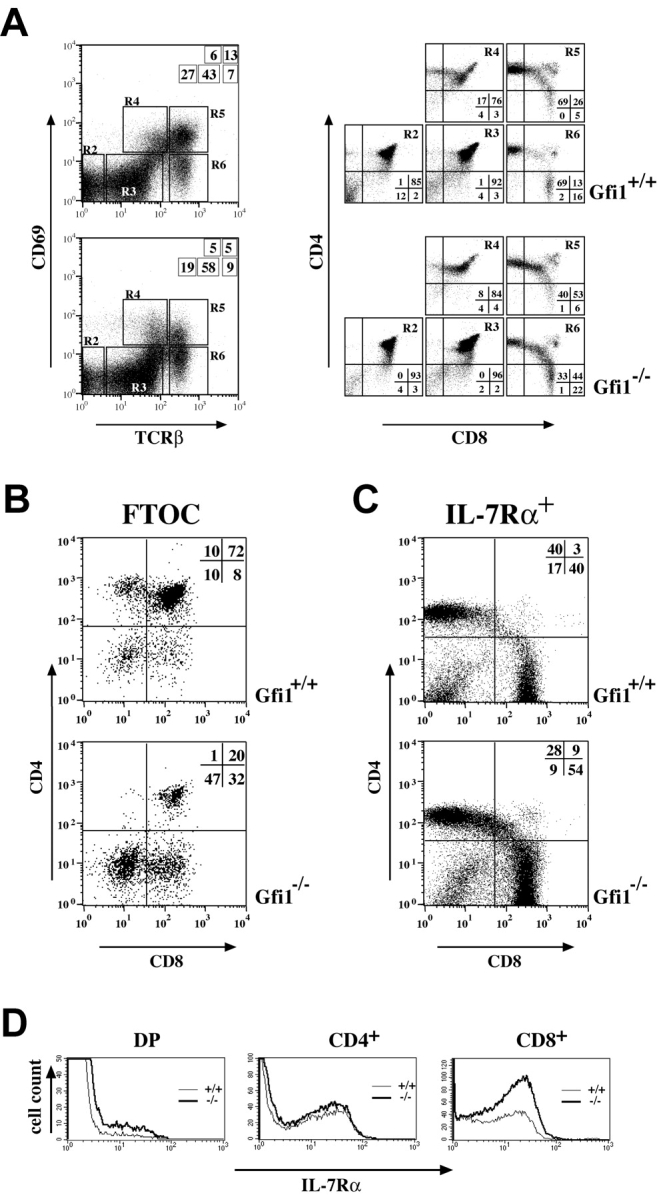
Gfi1−/− mice show an imbalance of CD4 and CD8 SP cells in positively selected subsets and show altered IL-7Rα expression in DN subsets. (A) Thymic subpopulation defined by the expression of CD69 and the TCRβ chain. Total thymocytes were stained for the presence of CD69, TCRβ, CD4, and CD8 and were analyzed by four-color flow cytometry. Gates in the CD69/TCRβ analysis are indicated (left, R2–R6). The subsets R2–R6 were reanalyzed for CD4 and CD8 expression (right). Relative percentages of cells are depicted as insets. The analysis was performed on four independent sets of Gfi1-deficient (Gfi1−/−) and WT (Gfi1+/+) animals. (B) Two-color analysis of CD4- and CD8-stained cells from d13 fetal thymi after 5 d in culture showing a dramatically reduced number of CD4 SP thymocytes in cultures from Gfi1-deficient mice (+/+, n = 4 and −/−, n = 5). (C) Three-color flow cytometric analysis of CD4 and CD8 expression gated on IL-7Rα+ thymocytes. A significant decrease of relative percentages of CD4 SP cells and a relative increase of the CD8 SP population can be observed in Gfi1−/− mice compared with the WT control in all cases. All analyses are representative for at least six individual sets of animals. (D) Three-color analysis of IL-7Rα chain expression gated on DP and CD4 or CD8 SP cells. Thin lines indicate the WT (+/+) control animal and the thick line represents the Gfi1-deficient mouse (−/−).
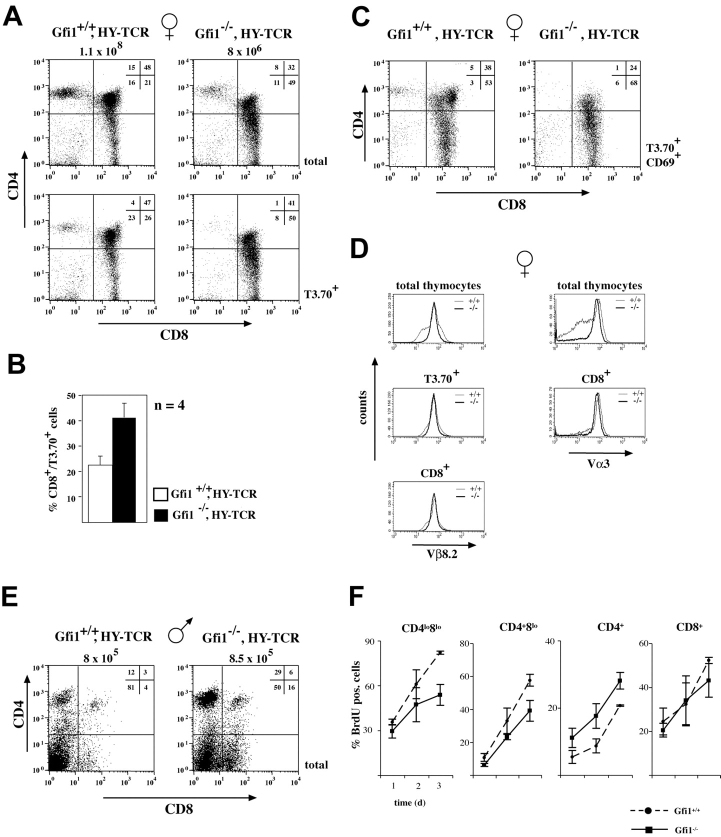
In the next step, we examined whether the lack of Gfi1 would directly affect positive selection of DP thymocytes. We crossed Gfi1−/− mice with animals expressing a transgenic T cell receptor recognizing the male specific HY antigen, which is presented by MHC I (41). Due to allelic exclusion, nearly all T cells of HY-TCR transgenic mice carry the T cell receptor directed against the HY antigen. Therefore, male mice that express this antigen delete DP cells because the HY-TCR is self-reactive. In female WT mice that lack the HY-antigen, the positive selection of DP cells to CD8+ SP cells is enhanced resulting in an increased relative proportion of CD8 SP cells of ∼21% (Fig. 3 A) compared to WT mice that display around 3% CD8+ cells (Fig. 1 D). In female Gfi1−/−/HY-TCR mice, the percentage of CD8 SP cells strongly increased from 21 to 49% of all thymocytes and from 26 to 50% on thymocytes selected for the expression of the HY-TCR Vα3 chain (detected with the clonotypic monoclonal T3.70 antibody; Fig. 3 A). The compiled analysis of four individual WT and four individual Gfi1-deficient mice showed 22.7% (±3.5) CD8+ T3.70+ cells versus 41.3% (±5.6), respectively (Fig. 3 B). This overrepresentation of CD8+ SP cells is also evident when only the T3.70+ CD69+ subset is analyzed (Fig. 3 C). Both CD8+ cells and total thymocytes from Gfi1-deficient HY-TCR or WT HY-TCR transgenic mice expressed identical peak levels of the HY-TCR Vβ8 and Vα3 chains (Fig. 3 D) excluding a perturbation of the HY-TCR expression in Gfi1−/− mice. Strikingly, the percentage of CD8+ or total thymocytes expressing high levels of HY-TCR α and β chains was almost 100% in HY-TCR females negative for Gfi1, which represents a significant increase over HY-TCR animals with intact Gfi1. In contrast, the deletion of DP cells through negative selection was effective in male HY-TCR, leading to a reduction of total thymocyte numbers to 8 × 105 cells of which the majority (81%) lack CD4 or CD8 surface markers (Fig. 3 E). In HY-TCR/Gfi1−/− mice the total number of thymocytes was reduced to about the same number (8.5 × 105) but elevated percentages of DP (6%), CD4 SP (29%), or CD8 SP (16%) cells were observed with regards to HY-TCR animals with intact Gfi1 expression (Fig. 3 E).
Figure 3.
Lack of Gfi1 alters MHC class I–restricted positive selection. (A) Flow cytometric analysis of CD4 and CD8 expression of total or T3.70+ thymocytes from female WT (Gfi1+/+) or Gfi1−/− mice expressing the HY-TCR transgene. Total thymocyte numbers and the percentages of the respective DN, DP, and SP cells are indicated. Experiments are representative for five different sets of mice. (B) Compilation of percentages of CD8+/T3.70+ cells from seven female Gfi1+/+ HY-TCR animals (open bar) and from seven female Gfi1−/− HY-TCR animals (solid bar). (C) CD69+/T3.70+ thymocytes from female WT (Gfi1+/+) or Gfi1−/− mice expressing the HY-TCR transgene. Total thymocyte numbers and the percentages of the respective DN, DP, and SP cells are indicated. Experiments are representative for five different sets of mice. (D) CD8+ or total thymocytes of both HY-TCR transgenic mice and Gfi1−/− carrying the HY-TCR transgene were analyzed with an antibody against the Vβ8.2 or the Vα3 variable chain of the HY-TCR. The Vα3 chain was detected with the T3.70 clonotypic antibody. (E) Flow cytometric analysis of CD4 and CD8 expression on thymocytes from 8-wk-old male WT (Gfi1+/+) or Gfi1−/− mice expressing the HY-TCR transgene. Total thymocyte numbers and the percentages of the respective DN, DP, and SP cells are indicated. (F) Three WT and three Gfi1null mice were analyzed for BrdU incorporation for each time point over a period of 3 d. The indicated cell subsets were analyzed by flow cytometric measurements and the percentages of BrdU+ cells were determined. Values are means with standard deviations and are plotted against the time in days (d).
To gain more insight into the mechanism of how the loss of Gfi1 affects positive selection we have measured BrdU incorporation over a period of 3 d into different thymocytes subsets. For each time point, we have analyzed three individual control mice and three individual Gfi1 KO mice. Cell populations bearing the CD4 or CD8 coreceptors were gated according to Germain (reference 42; Fig. 4 B) and analyzed for BrdU incorporation. We could not detect significant differences between the proliferation rate of normal mice and Gfi1-deficient animals in the CD4− CD8− (DN) and CD4+ CD8+ (DP) subsets, nor in the CD4lo CD8+ and CD8+ (SP) populations (Fig. 3 F and unpublished data). This excludes that the higher relative percentages of CD8+ cells that we see in Gfi1−/− mice are due to a higher proliferation rate. In contrast, we discovered that the CD4lo CD8lo and CD4+ CD8lo populations in Gfi1−/− mice had a significantly lower proliferative potential compared with the respective WT populations (Fig. 3 F). Because the CD4lo CD8lo and CD4+ CD8lo subsets still bear the potential to develop into both CD4 and CD8 Lins (42), we can again exclude that the higher percentages of CD8+ cells in the Gfi1−/− mice is due to a higher proliferation rate of these precursor cells.
Figure 4.
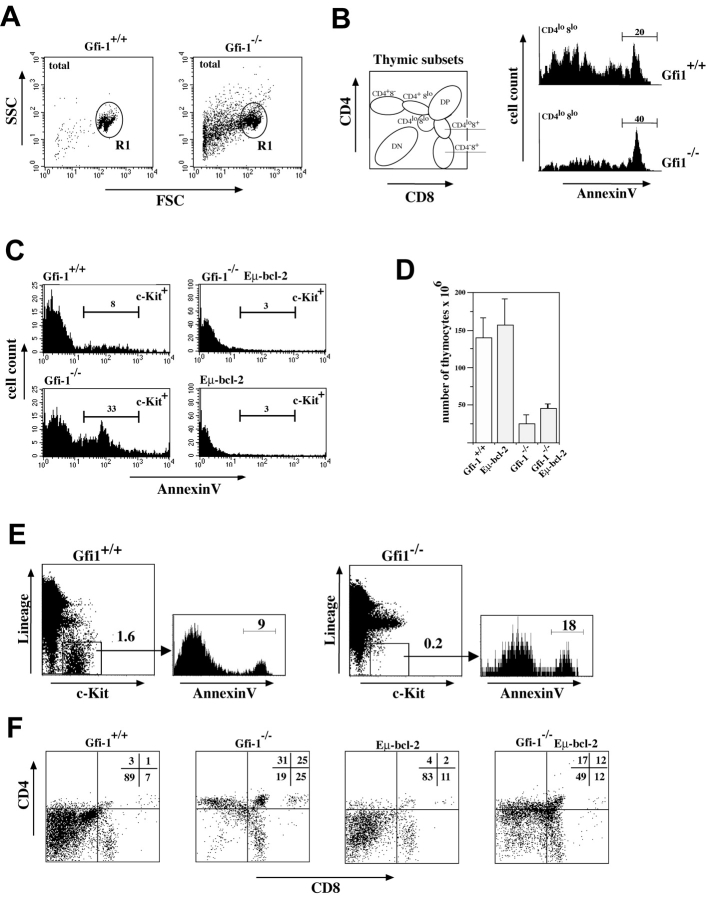
Cell survival and proliferation is disturbed in Gfi1−/− mice. (A) Thymocytes of Gfi1−/− mice (−/−) and WT controls (+/+) were analyzed on forward/sideward scatter for morphology and apparently intact cells were gated (R1). (B) Thymocytes of Gfi1−/− mice (−/−) and WT controls (+/+) were stained for CD4, CD8, and annexin V and gated according to Germain (reference 42) in seven subsets, which are schematically depicted. The subsets were electronically selected and analyzed for annexin V binding gated in forward/sideward scatter on R1. Histograms for the CD4lo CD8lo thymocyte population from WT (Gfi1+/+) or Gfi1-deficient (Gfi1−/−) mice are shown. (C) Thymocytes were stained with annexin V–FITC and c-Kit–PE and gated in forward/sideward scatter on R1. The percentages of c-Kit+ thymocytes that are able to bind annexin V–FITC for Gfi1−/− mice (−/−), WT controls (+/+), Eμ bcl-2 transgenic mice or double mutants lacking Gfi1 and expressing the Bcl-2 transgene (Gfi1−/−, Eμ bcl-2) are depicted. (D) Comparison of the total number of thymocytes of Gfi1+/+ (WT controls), Gfi1−/− mice, Eμ bcl-2 animals, and combinatorial mutant mice lacking Gfi1 and bearing the Eμ bcl-2 transgene (Gfi1−/−, Eμ bcl-2). (E) Thymocytes from WT (Gfi1+/+) or Gfi1−/− mice were stained with antibodies against Lin markers and for c-Kit expression and analyzed by three-color flow cytometry for annexin V binding (representative for n = 4 WT and n = 4 Gfi1−/− mice). (F) Thymocytes from WT (Gfi1+/+) or Gfi1−/− mice were stained for CD4, CD8, and c-Kit expression and were analyzed by three-color flow cytometry. The analysis shows CD4 and CD8 expression on electronically selected c-Kit+ cells. In particular, DN cells are depleted of c-Kit+ cells in the Gfi1−/− mice. All datasets are representative of four to five independent experiments with individual mice.
Gfi1− Mice Lose Cells of the Proliferating DN/c-Kit+ Subset Due to an Increased Cell Death Rate and a Lower Proliferative Capacity.
Another hallmark of Gfi1-deficient mice is their reduced thymic cellularity (Fig. 1 A). Because the expansion or deletion of particular T cell precursor subsets in the thymus relies on the regulation of cell proliferation and cell death, we analyzed forward/sideway scatter profiles of all thymic lymphoid cells. In Gfi1null mice a high percentage of cells with lower values of both parameters were detected suggesting significantly elevated numbers of dead cells (Fig. 4 A). To determine the extent of cell death initiated in Gfi1−/− mice, apparently intact cells were sorted (Fig. 4 A, R1) and analyzed for the presence of phosphatidylserine on the cell surface (by annexin V staining) and for propidium iodide uptake. The analysis of total thymocytes or the respective DN, DP, or SP subsets revealed no or only a moderate increase of dead or dying cells in Gfi1null mice compared with WT controls (unpublished data) excluding a general effect of Gfi1 on the survival of all thymocyte subsets. In addition, we found no altered susceptibility toward induced apoptosis using various stimuli in vitro (unpublished data). Next, we stained thymocytes for CD4 and CD8 expression to analyze different subsets and found that the CD4lo CD8lo subset in Gfi1null mice contained significantly higher percentages of annexin V+ cells than the corresponding population in WT controls (Fig. 4 B). The compiled data for four individual pairs of mice showed values of 26% (±5) for WT mice and 39% (±6) for Gfi1null mice. In addition, a significant increase of annexin V+ cells was observed when electronically gated c-Kit+ cells from Gfi1-deficient thymi were analyzed. The percentage of annexin V+ cells among the c-Kit+ population rose from 8% in WT mice to 33% in Gfi1-deficient animals (Fig. 4 C). Introduction of a Bcl-2 transgene that is active in the earliest DN T cell progenitors (Eμ-Bcl-2; reference 43) reduced the percentage of annexin V+ c-Kit+ cells back to 3%, similar to the levels found in single Eμ bcl-2 transgenic mice (Fig. 4 C). However, the overall thymic cellularity of Gfi1null mice could only marginally be elevated by the presence of Bcl-2 (Fig. 4 D).
To identify the c-Kit+ subset that is affected by apoptosis in Gfi1-deficient mice, we stained thymocytes for Lin markers (CD3, CD4, CD8, Pan-NK, Gr-1, B220, Mac-1, and Ter-119) or only for CD4, CD8, and c-Kit expression. In a normal thymus (Gfi1+/+), 1.6% of the cells are Lin−/c-Kit+ whereas only 0.2% of Lin− cells express c-Kit in the Gfi1null mice (Fig. 4 E). As for the total c-Kit+ population, the percentage of annexin V+ cells among Lin− c-Kit+ cells is increased from 9% in the WT mouse to 18% in Gfi1null mice (Fig. 4 E). In addition, when the CD4/CD8 profile of c-Kit+ cells was analyzed we found that the c-Kit+ CD4− CD8− (DN) subpopulation is strongly decreased in Gfi1-deficient mice to 19 from 89% in the respective WT population (Fig. 4 F). This suggests that loss of Gfi1 leads to a strong depletion of c-Kit–expressing cells in the Lin− or CD4/CD8 DN subset. Coexpression of a bcl-2 transgene could rescue this depletion partially to 49% (Fig. 4 F).
Loss of Gfi1 Causes Defects in DN1 and DN2 Thymocyte Subsets That Affect Survival and Proliferation.
In the next step we wished to more closely analyze the effect of Gfi1 deficiency on the Lin− subpopulation. Lin− cells are also termed DN cells and form four subsets according to the relative expression of CD44 and CD25 surface markers designated DN1, DN2, DN3, and DN4 reflecting their developmental stage (refer to Introduction). When Lin− cells were analyzed for the presence of CD25 and CD44, we observed that in Gfi1−/− mice, the relative percentages of DN2, DN3, and DN4 cells were decreased compared with the respective WT subsets (Fig. 5 A) and, most strikingly, that a new population of CD44hi CD25int cells emerges in Gfi1−/− mice, which accounts for up to 24% of the DN compartment (Fig. 5 A, arrowhead). A similar accumulation of CD44hi CD25int cells was observed in d13 fetal thymocytes from Gfi1−/− animals after 5 d in culture (unpublished data). Cells of these new populations lack NK1.1, γ/δ TCR, Mac-1, Gr-1, CD19, or CD11c but expressed Thy 1.2 (unpublished data) and therefore are to be considered to belong to the T cell progenitor pool.
Figure 5.
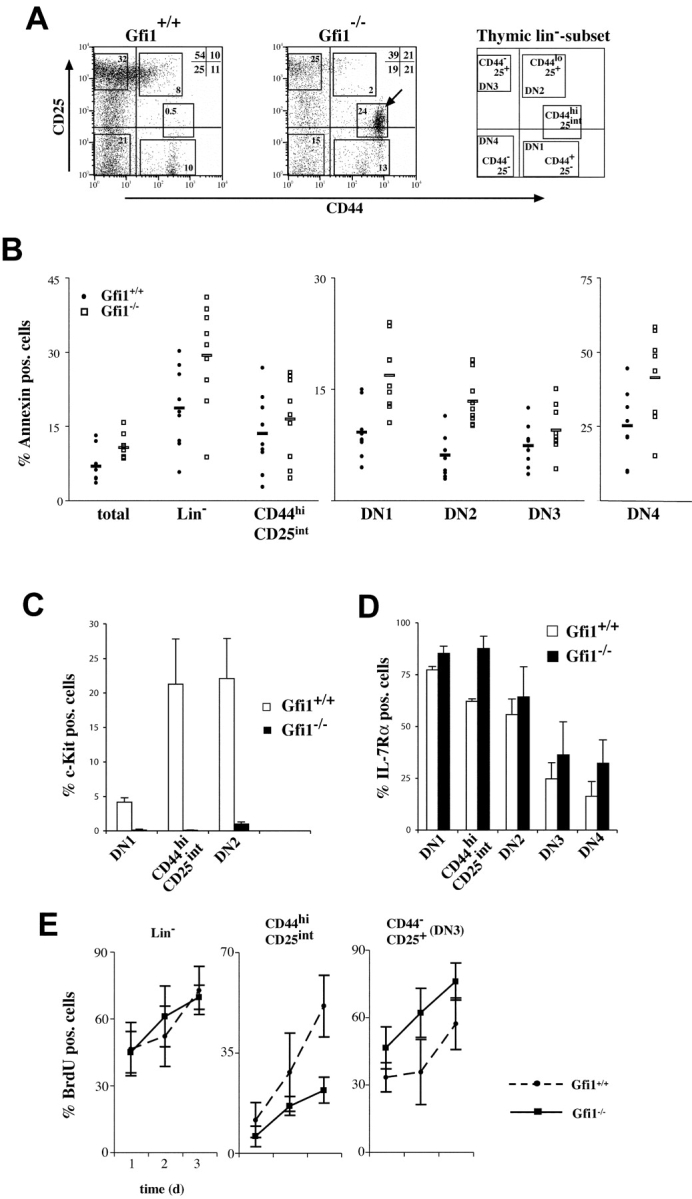
Depletion of c-Kit+ cells and alterations of DN subpopulations in Gfi1−/− mice. (A) Flow cytometric analysis of CD25 and CD44 expression among electronically gated Lin negative (Lin−) thymocytes. Relative percentages of DN2, DN3, and DN4 cells appear to be altered in Gfi1−/− mice compared with normal (Gfi1+/+) controls and the emergence of a new population is noted (arrowhead, CD44hi CD25int; n = 10 for each genotype). (B) Thymocytes from WT (Gfi1+/+) or Gfi1−/− animals were stained for expression of Lin markers CD44 and CD25. Lin− cells and the indicated subsets were analyzed for annexin V binding. The gates for the DN subsets were as indicated in A. The percentage of annexin V+ cells for the indicated subsets is given. The analysis reveals that only the DN1 and DN2 subsets of Gfi1null mice contain significantly more apoptotic cells (n = 9 for each genotype). The mean values for Gfi1-deficient and WT DN1 cells are 16.8 ± 4.8% and 9.1 ± 3.5%, respectively. This difference is significant at the 0.05 level (P = 0.004). The mean values for Gfi1-deficient and WT DN2 cells are 13.3 ± 3.3% and 6.0 ± 2.9%, respectively. This difference is significant at the 0.05 level (P = 0.001; Student's t test). (C) Lin− thymocytes from WT (Gfi1+/+) or Gfi1−/− mice were stained for CD44, CD25, and c-Kit expression. The percentage of c-Kit+ cells is given for the DN1, DN2, and CD25int CD44hi subset in WT and Gfi1null mice (n = 5 for each genotype). (D) Lin− thymocytes from WT (Gfi1+/+) or Gfi1−/− mice were stained for CD44, CD25, and IL-7Rα expression. The percentage of IL-7Rα+ cells is given for the DN1, DN2, DN3, DN4, and CD25int CD44hi subset in WT and Gfi1null mice (n = 4 for each genotype). (E) Three WT and three Gfi1null mice were analyzed for BrdU incorporation for each time point over a period of 3 d. The indicated cell subsets were analyzed by flow cytometric measurements and the percentages of BrdU+ cells were determined. Values are means with standard deviations and are plotted against the time in days (d).
We have used four-color flow cytometry to analyze the percentage of apoptotic cells among the different Lin− populations and found that the DN1 and DN2 subsets in particular have significantly more cells that are annexin V+ in Gfi1null mice than in WT controls (Fig. 5 B). All other subsets such as the DN3, DN4, or the new CD44hi CD25int cells did not show significant differences with respect to annexin V staining whether Gfi1 was present or not (Fig. 5 B). Next, we stained Lin− thymocytes from both types of mice for CD25, CD44, and c-Kit expression and found that cells of the DN1 and DN2 subsets in Gfi1null mice lack c-Kit expression (Fig. 5 C). Also, the novel CD25int CD44+ population almost entirely lacked c-Kit+ cells (Fig. 5 C) but contained a significantly higher percentage of IL-7Rα+ cells (Fig. 5 D). The percentages of IL-7Rα+ cells was similar in WT and Gfi1−/− mice in all other DN subsets except for DN1 where a slight increase was observed in Gfi1null mice (Fig. 5 D).
To gain more insight in the effect of Gfi1 on the proliferative behavior of thymocytes, we have followed BrdU incorporation by flow cytometry over a period of 3 d in different Lin− subpopulations. For each time point, we have analyzed three individual control mice and three individual Gfi1null mice. Cell populations bearing the CD25 or CD44 markers were gated as depicted in Fig. 5 A. We found that the new intermediate cell population expressing CD44hi CD25int that appears in Gfi1−/− mice (Fig. 5 A) proliferates much less in the Gfi1 KO than the corresponding cells in WT mice (Fig. 5 E). The DN3 subset is slightly more proliferative in the Gfi1 Ko than in the WT control. The proliferation rate of all other Lin− subsets such as the DN1, DN2, or DN4 population do not differ significantly between Gfi1null and WT mice (unpublished data).
Gfi1 Affects Processes Associated with Pre-TCR Selection.
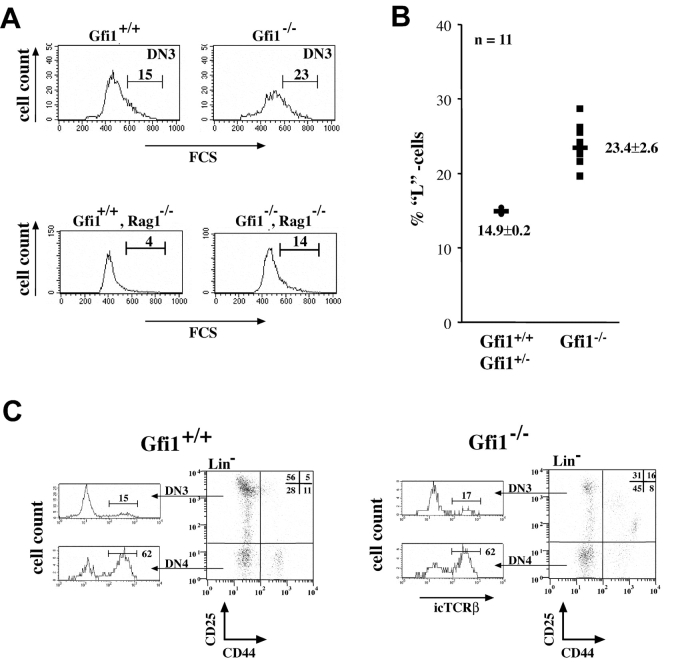
A critical step in the differentiation of DN cells is the pre-TCR selection that takes place in the DN3 subset (refer to Introduction). When DN3 cell were analyzed for their size distribution, we found that the relative frequency of larger L cells that have undergone pre-TCR selection are overrepresented in Gfi1−/− mice. In WT DN cells this population amounts to ∼14.9% (±0.2) but in Gfi1-deficient DN3 cells the L cell subset rises to 23.4% (±2.6). BrdU incorporation experiments demonstrated that the DN3 population of Gfi1−/− mice has a slightly higher proliferative capacity than the WT DN3 subset (Fig. 5 E). To test whether this effect led to uncontrolled formation of DP cells we analyzed DN3 cells in Gfi1−/−/RAG-1−/− mice. In normal RAG-1−/− with intact Gfi1, no DP cells are formed and the L subset percentage is limited to ∼3–4% of DN3 cells due to the lack of TCRβ chain expression (Fig. 6 A; reference 44). In contrast, Gfi1−/−/RAG-1−/− DN3 cells contained ∼14% L cells (Fig. 6 A), confirming the finding in Gfi1−/− mice. To test whether this observed effect on the E to L cell transition is associated with enhanced productive (in frame) rearrangements of the TCRβ locus, we measured the amount of intracellular TCRβ chains in Gfi1− DN3 cells. To this end, Lin− thymocytes were stained with antibodies against Lin markers and CD25 and CD44. The cells were then fixed and stained for cytoplasmic TCRβ according to described procedures (36). DN3 and DN4 cells from both WT and Gfi1−/− mice were electronically gated and showed very similar percentages of cells with identical cytoplasmic TCRβ expression levels (Fig. 6 C) suggesting that the lack of Gfi1 solely affects the regulation of cell size among the E and L type DN3 subsets or the expansion of the L subset but does not alter the efficiency of VDJ recombination nor the production of TCRβ chains from rearranged alleles.
Figure 6.
Gfi1 deficiency disturbs passage through pre-TCR selection but does not affect VDJ recombination. (A) Analysis of the DN3 subset of mice with the indicated genotypes for cell size distribution by forward angle light scattering (FCS). The boundary between E and L cells was set according to Hoffman et al. (reference 26) and as previously described (12). The bracket indicates the percentages of L cells that emerge after pre-TCR selection. Analyses are representative for three individual sets of animals for each genotype. (B) Compiled data from 11 WT or heterozygous (Gfi1+/−) and 11 Gfi1-deficient mice indicating L cell percentages. A mean value and a standard deviation are given. (C) Intracellular TCRβ chain expression levels in DN3 and DN4 subsets. Lin− thymocytes were analyzed for expression of CD25 and CD44. The DN3 and DN4 subsets were electronically gated to detect intracellular levels of TCRβ. No difference could be found between normal thymocytes and those negative for Gfi1.
Disruption of Thymocyte Development in Gfi1-deficient Mice Is Associated with Deregulated Expression of Potential Gfi1 Effector Genes.
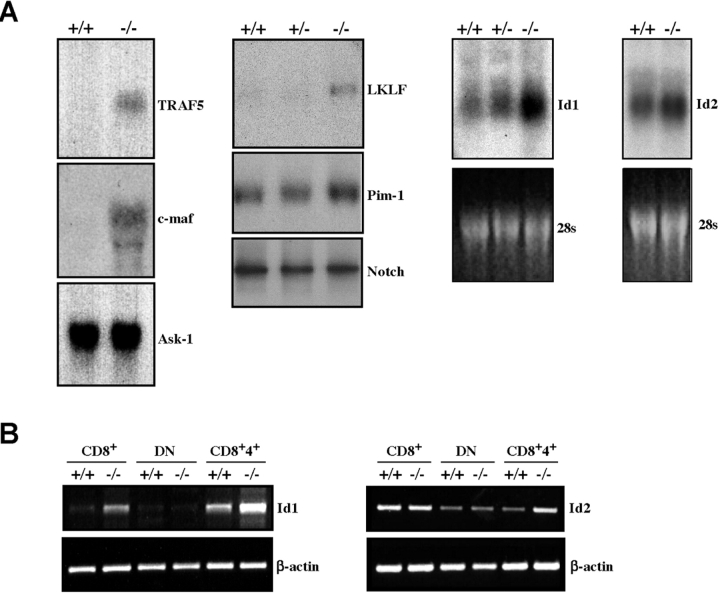
To identify potential downstream effector genes of Gfi1, which could shed light on the role of Gfi1 in T cell development, we used high density DNA microarrays to compare 11,000 murine genes for their expression level in normal and Gfi1−/− mice. Two independent experiments with RNA targets from pooled Gfi1−/− and WT thymi and two independent experiments with FACS®-sorted CD4+/CD8+ DP cells from WT and Gfi1−/− mice were performed. Evaluation of the array analysis and verification by Northern blot and RT-PCR analysis led to the identification of four genes that most prominently showed up-regulated expression in Gfi1−/− total thymocytes or Gfi1−/− DP cells compared with their WT counterparts (Table I and Fig. 7 A). We found that the genes for the bZIP transcription factor (c-Maf), the lung Krüppel-type factor (LKLF), the adaptor protein TNFR-associated factor (TRAF)5, and for the helix-loop-helix proteins Id1 and Id2 were among those up-regulated in total Gfi1−/− thymocytes (Table I and Fig. 7 A). Id3 expression appeared to be unaffected by Gfi1 (Table I). TRAF5, LKLF, and the genes for the helix-loop helix factors Id1 and Id2 were also up-regulated in Gfi1−/− DP thymocytes (Table I and Fig. 7 B). The expression of the vast majority of other genes including Pim-1, Notch, Ask-1 (Fig. 7 A), or members of the Bcl-2 family (unpublished data) were unaffected by the loss of Gfi1. Moreover, the experiments showed that Id1 expression is also up-regulated in Gfi1−/− CD8 SP cells, however, the expression of both Id1 and Id2 appeared to be unaffected in DN cells (Fig. 7 B). This suggests that Gfi1 negatively regulates these genes in particular thymocytes subsets and that their expression is relieved from this repression when Gfi1 is absent.
Table I.
Genes with Altered Expression Levels in Gfi-1−/− Thymocytes
| Total thymocytesMurine 11K array
|
Sorted CD4+/CD8+ (DP) cellsMurine U74A array
|
|||
|---|---|---|---|---|
| Up-regulated inGfi-1−/− thymocytes | Fold change(experiment 1) | Fold change(experiment 2) | Fold change(experiment 1) | Fold change(experiment 2) |
| c-Maf | 10.3 | 30.6 | 1.3 | 1.4 |
| TRAF5 | 6.1 | 2.4 | 9.7 | 8.4 |
| LKLF | 3.6 | 2.9 | 8.3 | 10.2 |
| Id1 | absent | absent | 7.4 | 6 |
| Id2 | 1.3 | n.t. | 4.2 | 3.5 |
| Id3 | absent | absent | 1.3 | 1.5 |
Total RNA was isolated from total thymocytes or from FACS®-sorted CD4+/CD8+ cells of Gfi1−/− (Gfi1−/−) or WT animals and was used to synthesize target cRNAs for the hybridization of four separate murine 11K DNA probe arrays and four separate U74A arrays (Affymetrix). The fold change in expression levels in Gfi1−/− thymocytes or DP cells in relation to WT thymocytes or WT DP cells of two independent experiments for each array set is shown. An average fold change of >2.5 was considered significant.
Figure 7.
Expression of potential downstream effector genes of Gfi1 in thymic T cells. (A) RNA from Gfi1−/− (−/−), Gfi1 heterozygous (+/−), or WT (+/+) animals was analyzed after electrophoretic separation for the expression of TRAF5, c-Maf, Ask-1, LKLF, Pim-1, Notch, as well as Id1 and Id2. The autoradiograms of a representative analysis are depicted. The level of TRAF5, c-Maf, LKLF, and Id1-specific transcripts appears to be strongly up-regulated in Gfi1−/− mice and Id2 transcripts appear to be weakly up-regulated in cells lacking Gfi1. Expression levels of Ask-1, Pim-1, or Notch 1 were not detectably altered in Gfi1-deficient thymocytes compared with WT or heterozygous controls. (B) Expression of Id1 and Id2 in three different subsets of WT (+/+) or Gfi1-deficient (−/−) mice. CD8+ and DN cells were isolated by magnetic beads and the DP cells were obtained by FACS® sorting. Total RNA was isolated from the indicated thymic subsets and was used for an RT-PCR reaction.
Discussion
The transcriptional repressor Gfi1 is expressed at very high levels in thymic lymphoid cells compared with other organs and cell Lins (3, 7, 12, 34, 45). Previous analyses demonstrated that Gfi1 is present in DN and DP thymocytes but is either absent or expressed at low levels in the CD4 or CD8 SP cells (12), which suggested a role of Gfi1 in early T cell development. Gfi1-deficient mice were generated to shed more light on the role of this transcription factor in these processes and pin down the developmental stages where Gfi1 comes into action. The two most prominent features of Gfi1−/− mice are a drastically reduced thymic cellularity and a significant bias toward the production or selection of CD8 SP cells. This already demonstrated that Gfi1 is generally required for a normal production of T cell progenitors and their differentiation.
The most striking hallmark of Gfi1−/− animals, which is certainly detrimental for T cell development, is the decreased thymic cellularity. The results from our analysis of the early DN thymocyte subsets suggest that the decreased number of cells in Gfi1−/− thymi is the consequence of ongoing apoptosis in c-Kit+ cells and in particular in the DN1 and DN2 subpopulations that we could detect by annexin V staining of the respective subpopulations. This correlates not only with generally reduced DN cell numbers but also with a depletion of c-Kit+ cells in the DN (or Lin−) subset. Hence, we propose that Gfi1 is responsible for the survival or expansion of c-Kit+ cells in the DN1 and DN2 subpopulations. Further, our data suggest that the lack of Gfi1 blocks differentiation from DN1 to DN2 cells and therefore aggravates the depletion of the DN2 and following subsets even more. This view is supported by the appearance of a novel, very prominent, distinct population in Gfi1−/− mice, which is Lin− and CD44hi CD25int. It is conceivable that this novel population represents an accumulation of intermediate cells, which are blocked on their way from the DN1 to the DN2 stage. These cells are c-Kit−, Thy1.2+, and also appear in fetal thymi of Gfi1−/− mice except when they represent other, mature B or T cells immigrating from the periphery. Because in the WT thymus cells of the DN2 population are proliferating and significantly contribute to thymus size and cell numbers, their depletion or reduction in the absence of Gfi1 due to apoptosis may well explain the smaller thymus size and the low thymic cellularity in Gfi1-deficient mice. In addition, the newly arising CD44hi CD25int population is only slowly proliferating. Provided that this population is a transitory subset, this may present an additional reason for a delayed development of DN2 from DN1 cells.
Our data support a role of Gfi1 in the same uncommitted DN T cell progenitors (i.e., DN1 and DN2) that critically depend on SCF and IL-7 (Fig. 5). Both SCF as well as IL-7 are expressed in Gfi1−/− mice (unpublished data) excluding a lack of ligands as a cause for the observed perturbations in the DN/c-Kit+ population. The Gfi1−/− phenotype shares some strikingly similar aspects with the phenotype of c-Kit– (18) or IL-7Rα–deficient mice (21, 20, 46). The strong depletion of c-Kit+ cells or the down-regulation of c-Kit expression that is observed in DN1 and DN2 cells of Gfi1−/− mice suggests that the observed phenotype in Gfi1null mice is the consequence of a c-Kit deficiency. However, it cannot be decided at present whether the loss of Gfi1 directly affects either c-Kit or IL-7R pathways or parallel or downstream signaling events. It is not unlikely that Gfi1 is one endpoint of c-Kit signaling and is required to translate the physiological effects of c-Kit, namely survival and proliferation of T cell progenitors, or alternatively, that lack of Gfi1 interferes with a correct c-Kit expression. Our finding that Bcl-2 was able to restore the loss of the c-Kit+ cells but failed to reconstitute a normal thymic cellularity in Gfi1−/− mice suggests that Gfi1 is involved in both survival and proliferation of c-Kit+ T cells.
Our finding that the TRAF5 adaptor molecule is strongly up-regulated in Gfi1-deficient thymocytes points to the alternative possibility that costimulatory signals provided by members of the TNF family are affected by the loss of Gfi1. It has been previously shown that TRAF5 is involved in CD27, CD30, CD40, lymphotoxin β receptor, and OX40 signaling and that overexpression of TRAF5 alone is sufficient for the subsequent activation of nuclear factor κB and Jun kinase (47–49). The importance of these pathways during the early T cell development is poorly understood but there is growing evidence for a function of TNFR superfamily members in regulating apoptosis and proliferation among DN thymocytes and during negative selection (50, 51). The importance of death receptor signaling in DN thymocyte development has been demonstrated recently by constructing transgenic mice that express a FADD dominant-negative mutant (52). It is possible that the observed cell death phenotype of Gfi1−/− mice results from interference with this type of signal.
We have previously suggested a role for Gfi1 in DN3 cells that was supported by earlier findings with transgenic mice that express constitutively high levels of Gfi1 in pre-T cells. In these animals a block at the E to L transition exactly where pre-TCR selection occurs and an accumulation of DN3 cells (12) is observed. In the Gfi1 KO mouse we observe the opposite phenotype i.e., a higher proportion of L cells than in WT controls. It is unclear how Gfi1 affects the pathways and regulatory mechanisms that control transition from DN3 to DN4 or from E to L cells within DN3. Both Gfi1 overexpressing transgenic mice and Gfi1−/− mice display a complete TCR repertoire (unpublished data) and in both types of mice an influence on TCRβ chain locus rearrangement could be excluded. In particular, the lack of Gfi1 did not affect the production of cytoplasmic or membrane-bound TCRβ chains, suggesting that Gfi1 is instead associated with processes that regulate the cell size distribution among E and L cell subsets or the expansion or survival of L cells than with the efficiency of the VDJ recombination process (Fig. 6).
Another characteristic of Gfi1-deficient mice is the preponderance of CD8 SP cells with respect to CD4 SP cells. We could exclude that the higher relative percentages of CD8+ cells in Gfi1 KO mice are due to an overrepresentation of CD8 ISP cells or their delayed development or are the result of an increased proliferation of mature selected CD8+ cells. We suggest that Gfi1 plays a role in whether DP cells differentiate into the CD4 or CD8 Lin and thereby in the process of positive selection of DP cells. This is supported by several findings in Gfi1−/− mice: first, the general higher relative percentages of CD8+ cells and lower percentages of CD4+ cells; second, the similar altered ratio of CD4/CD8 percentages in CD69+ and IL-7R+ thymocytes; and third, an apparently increased positive selection of MHC I–restricted HY-TCR–bearing CD8+ T cells.
According to a currently favored model, the efficient recruitment of the tyrosine kinase Lck to the TCR and a stronger Lck-mediated signal, resulting from TCR–MHC class II interaction, promotes commitment to the CD4 Lin whereas weaker Lck signals that are initiated after the TCR interacts with MHC class I leads to development into CD8 Lin cells (53–55, for review see 42). Recently, an alternative so-called “kinetic signaling” model for CD4/CD8 Lin determination and positive selection has been put forward (56). Here, the Lin decision depends on the persistence and not the strength of the Lck signal and CD4 cells are selected from the DP pool upon a persistent TCR/MHC-mediated signal (56). CD8+ SP cells develop from a CD4+CD8lo cell pool by a process termed coreceptor reversal upon cessation of the TCR signal (54). Interestingly, in the kinetic signaling model, the coreceptor reversal and the development toward the CD8 Lin is mediated by IL-7R signaling (56). Because Gfi1−/− mice clearly have higher relative numbers of DP/IL-7Rα+ cells than WT mice, this could explain the enhanced positive selection of CD8+ cells and the bias toward an overproduction of CD8+ cells in mice lacking Gfi1. However, we also found that in Gfi1-deficient mice the CD4lo CD8lo cell pool that is under selection contains more apoptotic cells than its WT counterpart. The possibility exists that this alleviates development of CD8+ cells because positive selection of CD8+ cells might be more efficient when selectable thymocytes are limiting.
Based on these models, we propose that Gfi1 might be involved in the maintenance of a strong or persistent TCR/MHC-mediated signal and suggest that the higher relative percentages of CD8 SP cells and increased positive selection in Gfi1-deficient mice is a consequence of a weakened or shortened Lck signaling or a premature cessation of the Lck-mediated signals. We have observed that during selection of DP cells, the relative percentage of CD69+ TCRβhi cells is decreased in Gfi1−/− mice compared with WT mice, although the same relative percentage of the preceding CD69+ TCRβlo population is not reduced. This argues for a signaling defect in Gfi1−/− mice because up-regulation of TCRβ during selection depends on efficient TCR/MHC signal transduction (36). In addition, we found that CD4lo CD8lo and CD4+ CD8lo populations in Gfi1−/− had a significantly lower proliferative potential in the Gfi1null mice than in WT animals. These subsets contain precisely those cells that are in the process of positive/negative selection and Lin decision. Their lower proliferation in the absence of Gfi1 may also reflect a weaker signaling during selection with the result of a preferred development of CD8+ cells. Another argument for a signaling defect in Gfi1−/− DP cells are their high levels of LKLF. In WT cells, expression of the LKLF gene is very efficiently down-regulated in cells that receive signals from TCR–MHC interactions (57). By contrast, LKLF expression rises only in postselection SP cells after that TCR/MHC signal has been completely shut off (57). Hence, a premature appearance of LKLF in DP thymocytes as is observed here is consistent with a lower or absent TCR signaling.
How Gfi1 affects the strength or the duration of TCR/MHC signaling remains to be clarified. We have previously shown that the Gfi1 gene is an effector of TCR signaling and is up-regulated shortly after stimulation of the TCR with anti-CD3 antibodies in peripheral T cells (45). We hypothesize that downstream effectors of Gfi1 might be critical to maintain the signal to regulate the selection process. Because one of the potential effectors of Gfi1, LKLF, maintains the quiescent state of SP T cells and is quickly down-regulated upon antigenic stimulation (57), it is conceivable that constitutive high levels of LKLF, as seen in Gfi1−/− cells, would counteract TCR-mediated signaling events and as a consequence negatively affect selection and Lin determination. More intriguingly, however, our finding that the expression of the helix-loop-helix transcription factors Id1 and Id2 is up-regulated in Gfi1−/− DP thymocytes may offer an alternative explanation. Both molecules are inhibitors of the E-box proteins E2A, E47, and HEB (58). In particular, E47 and E1A deficiency has been shown to accelerate positive selection and increase the percentage of CD8 SP cells (59). All of these features are also observed in Gfi1−/− mice, strongly suggesting that Gfi1 exerts these functions by acting as an upstream negative regulator of Id1 and Id2 expression and subsequently of E-protein dosage. From this it appears likely that deficiency of Gfi1 results in the relief of the repression of both Id proteins and therefore to the inhibition of E-box protein function, among them possibly E2A and E47.
Acknowledgments
The authors thank Anne-Odile Hueber for the introduction to FTOC. We also thank Andreas Strasser for Eμ bcl-2 animals, Pierre Ferrier for RAG-1–deficient mice, and Philippe Poussier for the T3.70 antibody. We are indebted to Nadine Pieda, Adriane Parchatka, Inge Spratte, Eva Gau, and Klaus Lennartz for excellent technical assistance and Petra Plessow and Tomas Civela for excellent animal care.
This work was supported by the Deutsche Forschungsgemeinschaft, DFG, (grant Mo 435/10-3, 10-4), the “Fonds der chemischen Industrie,” by the European Community Framework 5 Program, and the “IFORES Program” of the University of Essen Medical School.
R. Yücel and H. Karsunky contributed equally to this work.
H. Karsunky's present address is Department of Pathology, Stanford University, B259 Beckman Center, Stanford, CA 94305.
Footnotes
Abbreviations used in this paper: BrdU, bromo-deoxy-uridine; c-Maf, bZIP transcription factor; DN, double negative; DP, double positive; FTOC, fetal thymic organ culture; Id, inhibitor of DNA binding; ISP, immature single positive; Lin, lineage; LKLF, lung Krüppel-like factor; PIAS, protein inhibitor of activated signal transducer and activator of transcription; SCF, stem cell factor; SP, single positive; STAT, signal transducer and activator of transcription; TRAF, TNFR-associated factor.
References
- 1.Tong, B., H.L. Grimes, T.Y. Yang, S.E. Bear, Z. Qin, K. Du, W.S. El-Deiry, and P.N. Tsichlis. 1998. The Gfi-1B proto-oncoprotein represses p21WAF1 and inhibits myeloid cell differentiation. Mol. Cell. Biol. 18:2462–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rödel, B., T. Wagner, M. Zörnig, J. Niessing, and T. Möröy. 1998. The human homologue (GFI1B) of the chicken GFI gene maps to chromosome 9q34.13-A locus frequently altered in hematopoietic diseases. Genomics. 54:580–582. [DOI] [PubMed] [Google Scholar]
- 3.Grimes, H.L., T.O. Chan, P.A. Zweidler-McKay, B. Tong, and P.N. Tsichlis. 1996. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. 16:6263–6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zweidler-Mckay, P.A., H.L. Grimes, M.M. Flubacher, and P.N. Tsichlis. 1996. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol. 16:4024–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rödel, B., K. Tavassoli, H. Karsunky, T. Schmidt, M. Bachmann, F. Schaper, P. Heinrich, K. Shuai, H.P. Elsasser, and T. Möröy. 2000. The zinc finger protein Gfi-1 can enhance STAT3 signaling by interacting with the STAT3 inhibitor PIAS3. EMBO J. 19:5845–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, C.D., J. Liao, B. Liu, X. Rao, P. Jay, P. Berta, and K. Shuai. 1997. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 278:1803–1805. [DOI] [PubMed] [Google Scholar]
- 7.Gilks, C.B., S.E. Bear, H.L. Grimes, and P.N. Tsichlis. 1993. Progression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol. Cell. Biol. 13:1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zörnig, M., T. Schmidt, H. Karsunky, A. Grzeschiczek, and T. Möröy. 1996. Zinc finger protein GFI-1 cooperates with myc and pim-1 in T-cell lymphomagenesis by reducing the requirements for IL-2. Oncogene. 12:1789–1801. [PubMed] [Google Scholar]
- 9.Scheijen, B., J. Jonkers, D. Acton, and A. Berns. 1997. Characterization of pal-1, a common proviral insertion site in murine leukemia virus-induced lymphomas of c-myc and Pim-1 transgenic mice. J. Virol. 71:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt, T., M. Zörnig, R. Beneke, and T. Möröy. 1996. MoMuLV proviral integrations identified by Sup-F selection in tumors from infected myc/pim bitransgenic mice correlate with activation of the gfi-1 gene. Nucleic Acids Res. 24:2528–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt, T., H. Karsunky, E. Gau, B. Zevnik, H.P. Elsasser, and T. Möröy. 1998. Zinc finger protein GFI-1 has low oncogenic potential but cooperates strongly with pim and myc genes in T-cell lymphomagenesis. Oncogene. 17:2661–2667. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt, T., H. Karsunky, B. Rödel, B. Zevnik, H.P. Elsasser, and T. Möröy. 1998. b. Evidence implicating Gfi-1 and Pim-1 in pre-T-cell differentiation steps associated with beta-selection. EMBO J. 17:5349–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godfrey, D.I., J. Kennedy, T. Suda, and A. Zlotnik. 1993. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 150:4244–4252. [PubMed] [Google Scholar]
- 14.Murray, R., T. Suda, N. Wrighton, F. Lee, and A. Zlotnik. 1989. IL-7 is a growth and maintenance factor for mature and immature thymocyte subsets. Int. Immunol. 1:526–531. [DOI] [PubMed] [Google Scholar]
- 15.Baird, A.M., R.M. Gerstein, and L.J. Berg. 1999. The role of cytokine receptor signaling in lymphocyte development. Curr. Opin. Immunol. 11:157–166. [DOI] [PubMed] [Google Scholar]
- 16.Fehling, H.J., and H. von Boehmer. 1997. Early αβ T cell development in the thymus of normal and genetically altered mice. Curr. Opin. Immunol. 9:263–275. [DOI] [PubMed] [Google Scholar]
- 17.Rodewald, H.R., K. Kretzschmar, W. Swat, and S. Takeda. 1995. Intrathymically expressed c-kit ligand (stem cell factor) is a major factor driving expansion of very immature thymocytes in vivo. Immunity. 3:313–319. [DOI] [PubMed] [Google Scholar]
- 18.Rodewald, H.R., M. Ogawa, C. Haller, C. Waskow, and J.P. DiSanto. 1997. Pro-thymocyte expansion by c-kit and the common cytokine receptor gamma chain is essential for repertoire formation. Immunity. 6:265–272. [DOI] [PubMed] [Google Scholar]
- 19.Fehling, H.J., S. Gilfillan, and R. Ceredig. 1999. αβ/γδ lineage commitment in the thymus of normal and genetically manipulated mice. Adv. Immunol. 71:1–76. [PubMed] [Google Scholar]
- 20.Peschon, J.J., P.J. Morrissey, K.H. Grabstein, F.J. Ramsdell, E. Maraskovsky, B.C. Gliniak, L.S. Park, S.F. Ziegler, D.E. Williams, C.B. Ware, et al. 1994. Early lymphocyte expansion is severely impaired in interleukin 7 receptor–deficient mice. J. Exp. Med. 180:1955–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Freeden-Jeffry, U., P. Vieira, L.A. Lucian, T. McNeil, S.E. Burdach, and R. Murray. 1995. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 181:1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrie, H.T., F. Livak, D. Burtrum, and S. Mazel. 1995. T cell receptor gene recombination patterns and mechanisms: cell death, rescue, and T cell production. J. Exp. Med. 182:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudley, E.C., H.T. Petrie, L.M. Shah, M.J. Owen, and A.C. Hayday. 1994. T cell receptor beta chain gene rearrangement and selection during thymocyte development in adult mice. Immunity. 1:83–93. [DOI] [PubMed] [Google Scholar]
- 24.Godfrey, D.I., and A. Zlotnik. 1993. b. Control points in early T-cell development. Immunol. Today. 14:547–553. [DOI] [PubMed] [Google Scholar]
- 25.Godfrey, D.I., J. Kennedy, P. Mombaerts, S. Tonegawa, and A. Zlotnik. 1994. Onset of TCR-beta gene rearrangement and role of TCR-beta expression during CD3-CD4-CD8- thymocyte differentiation. J. Immunol. 152:4783–4792. [PubMed] [Google Scholar]
- 26.Hoffman, E.S., L. Passoni, T. Crompton, T.M. Leu, D.G. Schatz, A. Koff, M.J. Owen, and A.C. Hayday. 1996. Productive T-cell receptor β-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 10:948–962. [DOI] [PubMed] [Google Scholar]
- 27.Anderson, S.J., and R.M. Perlmutter. 1995. A signaling pathway governing early thymocyte maturation. Immunol. Today. 16:99–105. [DOI] [PubMed] [Google Scholar]
- 28.Penit, C., B. Lucas, and F. Vasseur. 1995. Cell expansion and growth arrest phases during the transition from precursor (CD4−8−) to immature (CD4+8+) thymocytes in normal and genetically modified mice. J. Immunol. 154:5103–5113. [PubMed] [Google Scholar]
- 29.Saito, T., and N. Watanabe. 1998. Positive and negative thymocyte selection. Crit. Rev. Immunol. 18:359–370. [DOI] [PubMed] [Google Scholar]
- 30.Nossal, G.J. 1994. Negative selection of lymphocytes. Cell. 76:229–239. [DOI] [PubMed] [Google Scholar]
- 31.Kisielow, P., and H. von Boehmer. 1995. Development and selection of T cells: facts and puzzles. Adv. Immunol. 58:87–209. [DOI] [PubMed] [Google Scholar]
- 32.Guidos, C.J. 1996. Positive selection of CD4+ and CD8+ T cells. Curr. Opin. Immunol. 8:225–232. [DOI] [PubMed] [Google Scholar]
- 33.Engel, I., and C. Murre. 2001. The function of E- and Id proteins in lymphocyte development. Nat. Rev. Immunol. 1:193–199. [DOI] [PubMed] [Google Scholar]
- 34.Karsunky, H., H. Zeng, T. Schmidt, B. Zevnik, R. Kluge, K.W. Schmid, U. Duhrsen, and T. Möröy. 2002. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 30:295–300. [DOI] [PubMed] [Google Scholar]
- 35.Möröy, T., A. Grzeschiczek, S. Petzold, and K.U. Hartmann. 1993. Expression of a Pim-1 transgene accelerates lymphoproliferation and inhibits apoptosis in lpr/lpr mice. Proc. Natl. Acad. Sci. USA. 90:10734–10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfer, A., T. Bakker, A. Wilson, M. Nicolas, V. Ioannidis, D.R. Littman, P.P. Lee, C.B. Wilson, W. Held, H.R. MacDonald, et al. 2001. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat. Immunol. 2:235–241. [DOI] [PubMed] [Google Scholar]
- 37.Lipshutz, R.J., S.P. Fodor, T.R. Gingeras, and D.J. Lockhart. 1999. High density synthetic oligonucleotide arrays. Nat. Genet. 21:20–24. [DOI] [PubMed] [Google Scholar]
- 38.Ikawa, T., S. Fujimoto, H. Kawamoto, Y. Katsura, and Y. Yokota. 2001. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc. Natl. Acad. Sci. USA. 98:5164–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucas, B., and R.N. Germain. 1996. Unexpectedly complex regulation of CD4/CD8 coreceptor expression supports a revised model for CD4+CD8+ thymocyte differentiation. Immunity. 5:461–477. [DOI] [PubMed] [Google Scholar]
- 40.Barthlott, T., H. Kohler, and K. Eichmann. 1997. Asynchronous coreceptor downregulation after positive thymic selection: prolonged maintenance of the double positive state in CD8 lineage differentiation due to sustained biosynthesis of the CD4 coreceptor. J. Exp. Med. 185:357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kisielow, P., H. Bluthmann, U.D. Staerz, M. Steinmetz, and H. von Boehmer. 1988. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 333:742–746. [DOI] [PubMed] [Google Scholar]
- 42.Germain, R.N. 2002. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol. 2:309–322. [DOI] [PubMed] [Google Scholar]
- 43.Strasser, A., A.W. Harris, and S. Cory. 1991. bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell. 67:889–899. [DOI] [PubMed] [Google Scholar]
- 44.Mombaerts, P., J. Iacomini, R.S. Johnson, K. Herrup, S. Tonegawa, and V.E. Papaioannou. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 68:869–877. [DOI] [PubMed] [Google Scholar]
- 45.Karsunky, H., I. Mende, T. Schmidt, and T. Möröy. 2002. High levels of the onco-protein Gfi-1 accelerate T-cell proliferation and inhibit activation induced T-cell death in Jurkat T-cells. Oncogene. 21:1571–1579. [DOI] [PubMed] [Google Scholar]
- 46.von Freeden-Jeffry, U., N. Solvason, M. Howard, and R. Murray. 1997. The earliest T lineage-committed cells depend on IL-7 for Bcl-2 expression and normal cell cycle progression. Immunity. 7:147–154. [DOI] [PubMed] [Google Scholar]
- 47.Arch, R.H., and C.B. Thompson. 1998. 4-1BB and Ox40 are members of a tumor necrosis factor (TNF)-nerve growth factor receptor subfamily that bind TNF receptor-associated factors and activate nuclear factor kappaB. Mol. Cell. Biol. 18:558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakano, H., H. Oshima, W. Chung, L. Williams-Abbott, C.F. Ware, H. Yagita, and K. Okumura. 1996. TRAF5, an activator of NF-kappaB and putative signal transducer for the lymphotoxin-beta receptor. J. Biol. Chem. 271:14661–14664. [DOI] [PubMed] [Google Scholar]
- 49.Akiba, H., H. Nakano, S. Nishinaka, M. Shindo, T. Kobata, M. Atsuta, C. Morimoto, C.F. Ware, N.L. Malinin, D. Wallach, et al. 1998. CD27, a member of the tumor necrosis factor receptor superfamily, activates NF-kappaB and stress-activated protein kinase/c-Jun N-terminal kinase via TRAF2, TRAF5, and NF-kappaB-inducing kinase. J. Biol. Chem. 273:13353–13358. [DOI] [PubMed] [Google Scholar]
- 50.Baseta, J.G., and O. Stutman. 2000. TNF regulates thymocyte production by apoptosis and proliferation of the triple negative (CD3−CD4−CD8−) subset. J. Immunol. 165:5621–5630. [DOI] [PubMed] [Google Scholar]
- 51.Li, R., and D.M. Page. 2001. Requirement for a complex array of costimulators in the negative selection of autoreactive thymocytes in vivo. J. Immunol. 166:6050–6056. [DOI] [PubMed] [Google Scholar]
- 52.Newton, K., A.W. Harris, and A. Strasser. 2000. FADD/MORT1 regulates the pre-TCR checkpoint and can function as a tumour suppressor. EMBO J. 19:931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hogquist, K.A. 2001. Signal strength in thymic selection and lineage commitment. Curr. Opin. Immunol. 13:225–231. [DOI] [PubMed] [Google Scholar]
- 54.Matechak, E.O., N. Killeen, S.M. Hedrick, and B.J. Fowlkes. 1996. MHC class II-specific T cells can develop in the CD8 lineage when CD4 is absent. Immunity. 4:337–347. [DOI] [PubMed] [Google Scholar]
- 55.Itano, A., P. Salmon, D. Kioussis, M. Tolaini, P. Corbella, and E. Robey. 1996. The cytoplasmic domain of CD4 promotes the development of CD4 lineage T cells. J. Exp. Med. 183:731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bosselut, R., L. Feigenbaum, S.O. Sharrow, and A. Singer. 2001. Strength of signaling by CD4 and CD8 coreceptor tails determines the number but not the lineage direction of positively selected thymocytes. Immunity. 14:483–494. [DOI] [PubMed] [Google Scholar]
- 57.Kuo, C.T., M.L. Veselits, and J.M. Leiden. 1997. LKLF: a transcriptional regulator of single-positive T cell quiescence and survival. Science. 277:1986–1990. [DOI] [PubMed] [Google Scholar]
- 58.Quong, M.W., W.J. Romanow, and C. Murre. 2002. E protein function in lymphocyte development. Annu. Rev. Immunol. 20:301–322. [DOI] [PubMed] [Google Scholar]
- 59.Bain, G., M.W. Quong, R.S. Soloff, S.M. Hedrick, and C. Murre. 1999. Thymocyte maturation is regulated by the activity of the helix-loop-helix protein, E47. J. Exp. Med. 190:1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]