Abstract
The vomeronasal organ (VNO) of terrestrial vertebrates plays a key role in the detection of pheromones, chemicals released by animals that elicit stereotyped sexual and aggressive behaviors among conspecifics. Sensory transduction in the VNO appears unrelated to that in the vertebrate olfactory and visual systems: the putative pheromone receptors of the VNO are evolutionarily independent from the odorant receptors and, in contrast to vertebrate visual and olfactory transduction, vomeronasal transduction is unlikely to be mediated by cyclic-nucleotide-gated channels. We hypothesized that sensory transduction in the VNO might instead involve an ion channel of the transient receptor potential (TRP) family, members of which mediate cyclic-nucleotide-independent sensory responses in Drosophila melanogaster and Caenorhabditis elegans and play unknown functions in mammals. We have isolated a cDNA (rTRP2) from rat VNO encoding a protein of 885 amino acids that is equally distant from vertebrate and invertebrate TRP channels (10–30% amino acid identity). rTRP2 mRNA is exclusively expressed in VNO neurons, and the protein is highly localized to VNO sensory microvilli, the proposed site of pheromone sensory transduction. The absence of Ca2+ stores in sensory microvilli suggests that, in contrast to a proposed mechanism of activation of mammalian TRP channels, but in accord with analysis of TRP function in Drosophila phototransduction, the gating of TRP2 is independent from the depletion of internal Ca2+ stores. Thus, TRP2 is likely to participate in vomeronasal sensory transduction, which may share additional similarities with light-induced signaling in the Drosophila eye.
Most terrestrial vertebrates have evolved two anatomically and functionally distinct olfactory organs, the main olfactory epithelium (MOE) and the vomeronasal organ (VNO). The VNO is a sensory structure adjacent to the ventral septum, which is thought to play a key role in the detection of pheromones (1). Pheromones are chemical signals released by animals that elicit stereotyped behavioral and neuroendocrine responses that ensure breeding and social hierarchy among conspecifics. In rodents these responses are dramatically impaired by surgical ablation of the VNO.
Molecular analysis has revealed that MOE and VNO sensory neurons use strikingly dissimilar mechanisms to transduce the olfactory information into changes in membrane potential. It has been shown that sensory transduction in the MOE involves the activation of a Gαs-like G protein by G-protein-coupled receptors, leading to the activation of adenylyl cyclase, the generation of cAMP, and ultimately the opening of cyclic-nucleotide-gated (CNG) ion channels (2). Accordingly, a large family of G-protein-coupled, seven-transmembrane-domain receptors has been identified that are likely to represent the mammalian odorant receptors (3, 4). In addition, genes encoding a specific Gαs-like G-protein subunit, Golf, a type III adenylyl cyclase, and an olfactory-specific CNG channel composed of at least two homologous subunits have been isolated (2, 5, 6). These components are highly enriched in the cilia of olfactory neurons, consistent with a role in odorant sensory transduction. Moreover, mouse knockout lines with targeted disruption of either the olfactory CNG channel or Golf display severe general anosmia, providing direct evidence that the excitatory olfactory signal transduction relies on G-protein-coupled, cyclic-nucleotide-dependent signals (7, 8).
The main elements of the MOE signal transduction apparatus, however, are not expressed in the VNO (9–12). Instead, two receptor gene families, Gαo-VN and Gαi2-VN, have been identified in rodent VNO; they are evolutionarily unrelated to the odorant receptor genes and are likely to encode distinct classes of pheromone receptors (9, 13–15). In addition, two G-protein α subunits, Gαo and Gαi2, cosegregate with these receptors in nonoverlapping subpopulations of VNO neurons and have been suggested to mediate pheromone receptor transduction (12, 32). The identity of downstream signaling components of the pheromone-evoked response has remained elusive. In particular, the search for functional subunits of CNG channels expressed in the VNO has been unsuccessful, and patch clamp analysis has revealed that, in contrast to vertebrate photoreceptors and MOE olfactory neurons, no CNG conductance can be elicited in VNO neurons (16). These results suggest the existence of a novel pathway for sensory transduction in the VNO, independent of CNG channels and distinct from that in the MOE.
Genetic analyses of phototransduction in Drosophila and chemosensation in Caenorhabditis elegans have recently revealed the essential role of members of the transient receptor potential (TRP) family of ion channels in mediating G-protein-regulated and cyclic-nucleotide-independent signaling pathways (17, 18). We reasoned that, similarly, a TRP homolog might be involved in vertebrate pheromone sensory transduction. Several vertebrate members of the TRP channel family have been cloned, at least in part, but their functions are not well understood (19–24). We report here the characterization of a member of the TRP family of ion channels (rTRP2) that is highly and specifically expressed in the rat VNO. The rTRP2 protein appears strictly localized to VNO sensory microvilli, where the pheromone-evoked response occurs, suggesting a direct role in pheromone signal transduction.
METHODS
Cloning of cDNA Encoding rTRP2.
A 224-bp fragment was PCR amplified from rat VNO cDNA by using the degenerate primers GA(A/G) ACI TCI CA(A/G) TCI (C/T)TC TT(C/T) TGG and (C/T)TT IGT IC(G/T) IGC (A/G)AA (C/T)TT CCA (C/T)TC, which correspond to conserved sequences ETSQSLFW and EWKFARTK of TRP ion channels. The PCR product was subcloned into pBluescript KS (+) (Stratagene), and plasmid DNAs purified from isolated colonies were sequenced. 32P-labeled probes were generated from the plasmid inserts of mTRP2 and mTRP5 and hybridized at 65°C to duplicate filters (Hybond N+; Amersham) of 3 × 105 plaque-forming units from a rat VNO cDNA library (9). Phage inserts from 30 positive plaques were analyzed, which corresponded to independent isolates of a unique cDNA, likely to encode the rat ortholog of mTRP2 (rTRP2). Additional screening of 6 × 105 plaque-forming units from VNO cDNA libraries at a lower temperature of hybridization (55°C) failed to identify additional TRP homologs. DNA sequencing was performed on ABI 310 and 377 Genetic Analyzers (Perkin–Elmer).
Northern Blots.
Poly(A)+ RNAs were purified from rat VNO, olfactory epithelium, brain, eye, gut, kidney, lung, liver, muscle, testis, and spleen by using a poly(A)+ RNA isolation kit (Stratagene). VNO poly(A)+ RNA (0.5 μg) and poly(A)+ RNAs from all the other tissues (2 μg) were electrophoresed on a formaldehyde/agarose gel and transferred to nylon membrane (Hybond N+), and the blot was probed with 32P-labeled rTRP2 cDNA, then reprobed with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA as a control for loading.
In Situ Hybridization.
In situ hybridization was performed as described elsewhere (9). VNO, olfactory epithelium, retina, brain, utricle, testis, liver, kidney, and duodenum were dissected from adult (8- to 9-week-old) male and female rats. Whole heads were obtained from embryonic day 13 (E13), E15, E17, and E19 rat embryos. Tissues were embedded in Tissue-Tek OCT (Sakura). Antisense and sense digoxigenin-labeled probes were generated from the full-length rTRP2 cDNAs.
Production of an Anti-rTRP2 Polyclonal Antibody.
The carboxyl terminus of the rTRP2 gene (amino acids 814–885) was fused to glutathione S-transferase (GST) by using the pGEX 4T-3 vector (Pharmacia). The fusion protein was purified with glutathione-Sepharose-4b beads (Pharmacia), analyzed by PAGE, and used to inoculate three rabbits (Cocalico Biologicals, Reamstown, PA). Antibodies were affinity purified against a fusion protein of thioredoxin (Novagen) and the fragment of rTRP2 gene bound on a Sulfalink column (Pierce) as recommended by the manufacturer.
Western Blot.
HEK cells transfected (Lipofectamine; GIBCO/BRL) with rTRP2, untransfected HEK cells, rat brain, VNO, and olfactory epithelium were homogenized in RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) and spun at 14,000 × g to remove cellular debris. Protein was quantitated with a Bradford assay (Bio-Rad), and 20 μg from each tissue was separated by PAGE on a Tris⋅HCl 10% Ready Gel (Bio-Rad) and transferred to nitrocellulose. The protein was detected with enhanced chemiluminescence (ECL; Amersham).
Immunocytochemistry.
Tissue was fixed in 4% paraformaldehyde, equilibrated in sucrose, and embedded in Tissue-Tek OCT. Dissociated cells, prepared from rat as previously described (16), were fixed in paraformaldehyde and allowed to dry onto slides coated with Cell Tak (Collaborative Biomedical Products). Sections and cells permeabilized with 0.1% Triton X-100 were incubated with a 1:500 dilution of the primary antibody followed by a 1:200 dilution of Cy3- or Cy5-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch). Actin was visualized with Bodipy phalloidin Fl (Molecular Probes) at 1:500 dilution. Slides were mounted with Fluoromount G (Electron Microscopy Sciences). Images were obtained with an LSM 410 confocal microscope at 1024 × 1024 pixels, using an aperture of 60 (dissociated cells and low-magnification images of sections) or 26 (high-magnification images of sections). To calculate artifactual intensification of signal due to membrane area, we assumed 70–80 microvilli of 0.1-mm diameter on each knob of 3-mm diameter (25, 26).
Analysis of Human DNA.
Human genomic DNA was obtained from CLONTECH and hTRP2 sequences were amplified by using the following primers: GCACTGCGCCTCCTCCTGGCTGGGCTT and GAAGAGCACCTCAGCCAAGAACTGGG for Arg-496 and CTATGATCA CCAACTCCTTCCAGAAG and GGCAAATTCTCCTGAGAAGGTAGAAG for Arg-664. The PCR products were sequenced off both strands with the original primer pair or the internal primer CACACTGGAGAATCAAAGATGTGAGGAG.
RESULTS
A TRP Homolog Is Expressed in the Rodent VNO.
Degenerate oligonucleotides were designed, based on conserved sequences of the putative pore and of the carboxy-terminal domain of known Drosophila and mammalian TRP genes (19–24). PCR amplification with cDNA prepared from VNO, but not from MOE, generated an abundant PCR product of the expected size (224 bp), suggesting the existence of one or several VNO-specific TRP ion channels. The screening of a rat VNO cDNA library with 32P-labeled probes generated from the subcloned PCR products led to the identification of a cDNA present at a frequency of 1 in 10,000. This cDNA is nearly identical to a 405-bp fragment of a mouse TRP homolog (mTRP2, 94% identity at the nucleic acid level) identified by PCR amplification from liver and brain RNA, and to a human expressed sequence tag (EST) (hTRP2) identified in a fetal cDNA library (19–21). The cDNA isolated from the VNO is therefore likely to encode the rat ortholog of TRP2 (rTRP2). Although the initial amplification of VNO cDNA had also generated a minor product likely to represent the rat ortholog of TRP5, extensive screening of the VNO cDNA library failed to confirm the presence of TRP5 in the VNO. Moreover, subsequent screening of the VNO cDNA library with the initial PCR probes as well as with the 3.4-kb full-length rTRP2 cDNA under high- and low-stringency conditions of hybridization and washing failed to reveal any additional TRP homologs.
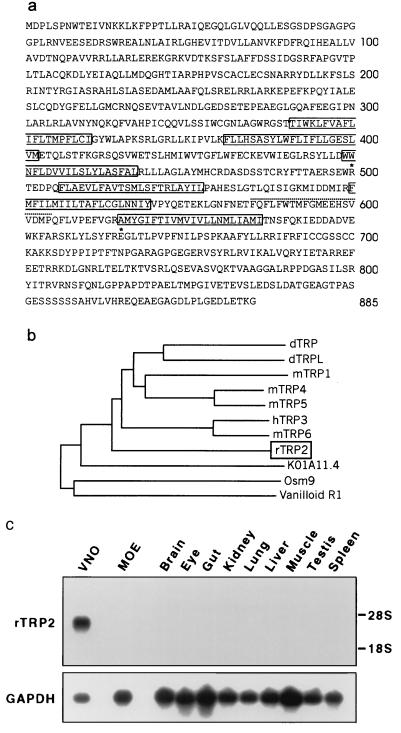
The cDNA of rTRP2 encodes a predicted protein of 885 amino acids (Fig. 1a) which contains six hydrophobic regions predicted to span the membrane, and a putative pore domain located between the fifth and six transmembrane domains. The amino terminus of rTRP2 displays an ankyrin-repeat consensus sequence (amino acids 147–179) (27) and the carboxyl terminus contains a stretch of amino acids (744–785) predicted to form a coiled-coil domain. Phylogenetic analysis, based on sequence alignment within the TRP family of ion channels, indicates that rTRP2 is equally distant from Drosophila and from other mammalian TRPs (Fig. 1b), with which it shares between 25% and 30% sequence identity. It is more distantly related to the nematode sequences OSM9 (18) and K01A11.4 and to the vertebrate vanilloid receptor (28) (respectively 21%, 18%, and 12% sequence identity).
Figure 1.
rTRP2 is a VNO-specific ion channel of the TRP family. (a) Predicted amino acid sequence encoded by rTRP2 cDNA, showing the position of six putative transmembrane domains (solid outlines), and pore region (dotted underline). Stars indicate the positions of two mutations introducing stop codons within the ORF of the corresponding human sequence. (b) Phylogenetic tree of members of the TRP family in Drosophila (dTRP and dTRPL), mouse (mTRP1, -4, -5, and -6), human (hTRP3 and vanilloid R1), and C. elegans (OSM9 and KO1A11.4). Full-length protein sequences were aligned in CLUSTAL_X (42), and phylogeny was calculated by the neighbor-joining method, after removal of gapped regions, with 1,000 bootstrap trials. GenBank accession numbers are P19334 (dTRP), M88185 (dTRPL), U73625 (mTRP1), U50922 (mTRP4), AF029983 (mTRPS), N13758 (hTRP3), U49069 (mTRP6), Z66514 (K01A11.4), AF0115639 (OSM9), and AF029310 (VR1). (c) Northern blot analysis shows that rTRP2 transcripts are confined to the VNO.
Expression Pattern of rTRP2.
The distribution of rTRP2 was investigated by Northern blot and in situ hybridization experiments. In Northern blots (Fig. 1c) 32P-labeled rTRP2 probe hybridized strongly to RNA purified from the VNO. A very faint signal could be detected in RNA from the MOE after longer exposure of the blot (data not shown), but no signal was detected in other tissues, even after extensive exposure of the blot.
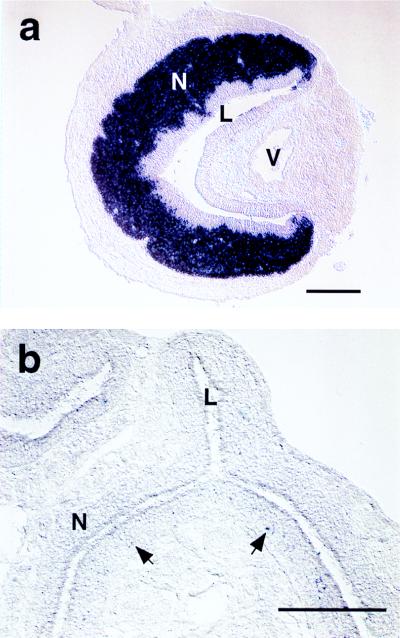
In situ hybridization with a digoxigenin-labeled rTRP2 antisense RNA probe further confirmed the striking specificity of rTRP2 expression. An intense hybridization signal was detected in the VNO neuroepithelium, restricted to sensory neurons (Fig. 2a). The rTRP2 probe equally labeled both apical and basal subpopulations of VNO neurons which are otherwise distinguished by expression of distinct Gα subunits and pheromone receptor families. No signal was observed when we examined sections from an exhaustive series of other sensory and nonsensory tissues (see Methods). A very faint signal was detected in a small population (less than 1%) of basally located cells in the adult MOE (Fig. 2b), which are likely to account for the barely detectable signal in Northern blots. The location of the labeled cells in the olfactory epithelium is consistent with that of immature sensory neurons. Accordingly, scattered labeled cells were also observed at higher density in MOE from late embryonic stages (E14 to E19; data not shown). However, no protein was detected in adult and embryonic MOEs (see below and Fig. 3).
Figure 2.
rTRP2 is strongly expressed in adult rat VNO neurons. (a) In VNO cross sections hybridized to digoxigenin probe for rTRP2, the VNO neuroepithelium (N) appears strongly and specifically labeled. L, VNO lumen; V, VNO vein. (b) In the MOE, some sparse cells are faintly labeled (arrows) that are mostly located in the basal half of the neuroepithelium. N, neuroepithelium; L, lumen.
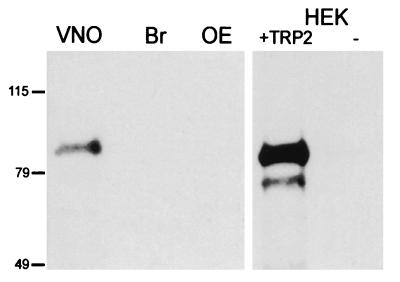
Figure 3.
Detection of rTRP2 protein in immunoblots. Western blots were probed with an antibody raised against the carboxyl terminus of rTRP2. Lanes contain total protein (20 μg per lane) from VNO, brain (Br), and olfactory epithelium (OE), and from HEK cells transfected with TRP2 or untransfected (−).
Our data suggest that, in contrast to other TRP channels characterized so far in mammals, rTRP2 is specifically expressed in mature VNO neurons and is virtually absent from any other neuronal and nonneuronal tissues. Previous amplification of TRP2 from mouse liver and human embryonic brain and cloning of TRP2 fragments from bovine testis (19–21) might therefore result from minute expression of TRP2 in these tissues that contrasts with the high level of rTRP2 expression detected in VNO neurons (0.1% of the transcripts in cDNA libraries prepared from individual VNO sensory neurons, data not shown). Alternatively, TRP2 tissue distribution might differ in different species.
rTRP2 Is Localized to Microvilli of VNO Sensory Neurons.
In the MOE, olfactory sensory transduction occurs in specialized cilia extended by olfactory neurons into the nasal cavity (2). Similarly, pheromone signal transduction is likely to occur in the sensory microvilli that protrude from the dendrites of VNO sensory neurons and that line the VNO lumen. Restricted localization in VNO microvilli would therefore distinguish components of the pheromone sensory transduction from those involved in more general signaling processes. Antisera were raised against the carboxyl terminus of the rTRP2 protein and were affinity-purified. On immunoblots, the anti-rTRP2 antisera recognized a band of about 90 kDa in VNO, but not in brain or in MOE (Fig. 3). A band of the same molecular mass was recognized in HEK cells transfected with rTRP2 but not in untransfected cells.
Immunolabeling of VNO sections showed intense staining of the luminal surface of the VNO but faint staining of neuronal cell bodies (Fig. 4a–d). In contrast, but consistent with the distribution of rTRP2 message, no staining was observed in sections from MOE (not shown). At high magnification, the TRP2 protein appeared strictly localized to the microvillar zone of the adult VNO neuroepithelium immediately distal to the actin-rich terminal web of supporting cells.
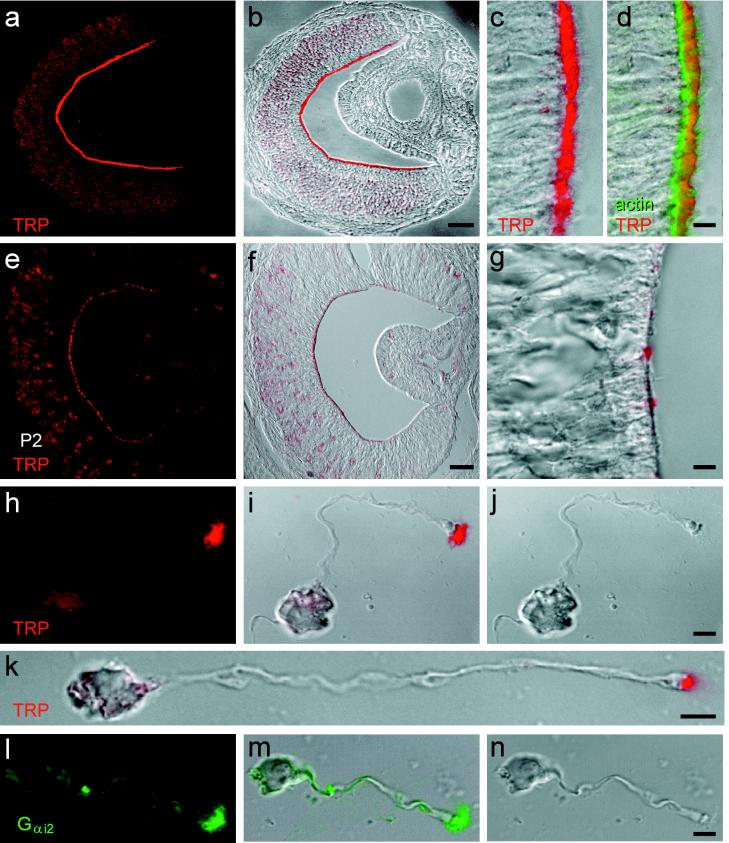
Figure 4.
Localization of rTRP2 to the sensory microvilli of vomeronasal neurons. (a and b) Adult VNO labeled with affinity-purified anti-rTRP2 antibody (red) and shown as the fluorescence signal (a) or a digital addition of the fluorescence and bright-field images (b). rTRP2 is located at the luminal surface of the sensory epithelium. (c and d) Higher magnification of the VNO sensory epithelium labeled with the anti-rTRP2 antibody (red) and Bodipy phalloidin to label actin (green). Label is shown as the red fluorescence channel combined digitally with the differential interference contrast (DIC) image (c), or both red and green channels combined with DIC. TRP2 is located mostly distal to the actin-rich terminal web. (e–g) Postnatal-day-2 VNO, labeled with anti-rTRP2 and shown as in a–c. Luminal labeling is present but more punctate than in adult. (h–j) Localization of rTRP2 in microvilli, in a dissociated rat VNO neuron. Images are fluorescence (h), combined fluorescence and DIC (i), or DIC alone (j). Label is restricted to the microvilli and not apparent in the dendritic knob. (k) Another VNO neuron labeled as in i. (l–n) Localization of Gi2–3 immunoreactivity in the microvilli of a dissociated VNO neuron. (Scale bars: a and b, 100 μm; e and f, 50 μm; c, d, g, and h–n, 5 μm.)
Even more striking results were obtained by immunolabeling of dissociated sensory neurons. As shown in Fig. 4 h–k, the expression of rTRP2 is remarkably confined to sensory microvilli, whereas the neuronal cell body and dendritic knob show a much fainter staining. Because the microvilli have more membrane area than other regions of VNO neurons, label on a uniformly distributed membrane protein might appear more intense in this region of the cell. On the basis of the number and size of VNO neuronal microvilli and the focal depth of the confocal microscope, we calculate that such an effect might artifactually enhance fluorescent signal from the microvilli by 2.5-fold. In fact, measurements from confocal images showed that intensity in the microvilli was increased by a factor of 40 ± 9 (mean ± SD; n = 3) over adjacent regions of the dendritic knob or cell body.
TRP2 protein was detected in only a few scattered VNO neurons in late-stage embryos (E19 and E20) but was readily detectable in newborn animals (postnatal day 2; see Fig. 4 e–g). At this stage the TRP2 protein showed a more patchy distribution at the luminal surface and a relatively stronger distribution in neuronal cell bodies. The adult distribution of rTRP2 is achieved at postnatal day 21 (not shown).
The selective enrichment of the rTRP2 protein in the VNO microvilli is highly suggestive of a direct role of rTRP2 in the pheromone-induced sensory cascade. Similarly, the G protein Gαi2, which is expressed by a subset of VNO neurons, with high concentration at the lumen, has been suggested to participate in the pheromone transduction pathway (12). Using an antibody to Gαi2, we observed that Gαi2 is also restricted to the microvilli of dissociated VNO neurons, in a pattern identical to that of TRP2 (Fig. 4 l–n).
rTRP2 in Humans.
Curiously, the human ortholog of rTRP2 is considered to be a pseudogene, as several independent ESTs indicate the presence of deleterious mutations introducing stop codons within the ORF (19). We confirmed the presence of two of these mutations (see Fig. 1a) by partial sequencing of hTRP2 obtained from human genomic DNA. The existence of a functional vomeronasal system in Old World primates, including apes and humans, is debated (29). It is thus possible that, in higher primates in which VNO function may not be essential for breeding, the selection pressure to retain functional VNO transduction components is much reduced.
DISCUSSION
The transduction of pheromone signals by VNO sensory neurons represents the initial step in a cascade of molecular events that ultimately leads to specific arrays of behavioral and neuroendocrine changes characterizing the mammalian pheromone response. We have identified a member of the TRP family of ion channels, rTRP2, that is likely to participate in the pheromone signaling cascade. Our data suggest molecular similarities in the sensory signaling pathways of mammalian VNO sensory neurons and Drosophila photoreceptor cells and point to pheromone transduction as an experimental system with which to study TRP channel function. The initial observation, confirmed here, that TRP2 is a pseudogene in humans (19), suggests that responses to pheromones in humans (30) might result from detection by MOE neurons. This idea is in accord with data obtained in several animal species showing that the VNO may not be the only site for pheromone detection, and that some pheromonal cues are processed by the main olfactory system (1).
Genetic studies in invertebrates have demonstrated that members of TRP family are essential for G-protein-regulated and cyclic-nucleotide-independent sensory pathways. In C. elegans the OSM9 channel is required for the sensory response of AWA olfactory neurons to the activation of the odorant receptor ODR10 (18) and mediates mechanosensation and olfactory adaptation in other sensory neurons. In Drosophila photoreceptors, the activation of rhodopsin triggers a light-induced conductance formed by the TRP and TRP-like channels (7, 31). Similarly, rodent VNO neurons appear insensitive to cyclic nucleotides (16) and display high and specific expression of rTRP2, a mammalian homolog of the TRP channels. The exclusive expression of the rTRP2 protein by VNO neurons, its striking localization in VNO sensory microvilli, together with its similarity to other channels involved in G-protein-regulated and cyclic-nucleotide-independent sensory pathways, is consistent with a direct role of rTRP2 in the pheromone evoked-response. The rTRP2 channel might represent the primary conductance activated by the pheromone signal, or it could mediate a secondary amplification or modification of the sensory response. Direct evidence that rTRP2 indeed represents the primary component of the pheromone-induced conductance will require the demonstration that the rTRP2 channel is activated by pheromone signals. These experiments are not possible at the moment, because of the absence of well characterized mammalian pheromones and of specific activators or inhibitors of TRP channels.
A Model for Pheromone-Induced Signaling in the VNO.
Molecular and functional properties shared between Drosophila photoreceptors and mammalian VNO sensory neurons suggest a common model of signal transduction in which TRP channels represent the primary conductance. In the Drosophila eye, photoisomerization of rhodopsin activates a Gα protein of the Gq class that triggers a phosphatidylinositol signaling cascade, leading, by a mechanism still to be defined, to the opening of the cation-selective dTRP and dTRPL channels (17, 31).
In rodents, two families of receptor genes expressed exclusively by VNO neurons have been identified, each thought to contain about 50–100 genes. A novel family of putative seven-transmembrane-domain receptors is expressed in the most apical domain of the VNO, and a second gene family related to the metabotropic neurotransmitter receptors is found in a more basal zone of the neuroepithelium (9, 13–15). It is therefore likely that VNO signal transduction involves G-protein signaling. Immunocytochemical analysis in opossum and rodents (12, 32) showed high expression of the two G-protein subunits Gαo and Gαi2 in nonoverlapping populations of VNO neurons. Using high-resolution confocal microscopy on isolated VNO sensory neurons, we confirmed the specific cellular localization of Gαi2 in VNO microvilli, consistent with potential role in signal transduction of pheromones. In addition, a high level of adenylyl cyclase type II (ACII) was found in all VNO neurons (12). In contrast to ACIII found in MOE, ACII can be directly stimulated by G-protein subunits βγ in the presence of low levels of Gαs (33) leading to the production of cAMP and potential activation of a CNG channel. However, only the β subunit of the CNG channel (roCNC2) is expressed in the VNO (5, 6, 10), and because it does not form a detectable CNG conductance when expressed alone in an heterologous expression system, it is unlikely to be functional by itself in the VNO. Moreover, and in contrast to results obtained with MOE neurons, rodent VNO neurons appear insensitive to cyclic nucleotides (16), making this pathway unlikely.
On the basis of identification of rTRP2 as a candidate transduction channel in the VNO we propose a model according to which the activation of Gαo or Gαi2 by pheromone receptors and the subsequent release of βγ [or alternatively, the activation of Gαq also identified in the VNO (12)] would stimulate a phospholipase C (34), leading to the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) into inositol trisphosphate (IP3) and diacylglycerol (DAG). Elevation of IP3 or DAG would then lead to the opening of the TRP2 channel and alteration in membrane potential. This model would be consistent with the increase in IP3 reported in snake and hamster VNO neurons in response to pheromones (35–37).
Activation of TRP Channels.
The mechanisms by which any of the TRP channels are gated are still largely mysterious and the object of controversy (38, 39). Mammalian homologs of dTRP have been identified that show predominant expression in the brain and in a variety of nonexcitable tissues, such as adrenal gland, lung, testis, and ovary (19–24). The subcellular localization of these channels has not yet been described, and their function is unknown. On the basis of in vitro studies, it has been suggested that the mammalian homologs of dTRP are involved in the refilling of Ca2+ stores induced by receptor-mediated store depletion and that, by analogy, dTRPs might also be store-operated (20, 40). However, (i) the localization of dTRP in photoreceptor microvilli distant from internal Ca2+ stores, (ii) the absence of activation of dTRP channels in vivo after release of Ca2+ stores induced by thapsigargin, and (iii) the activation of the light-induced conductance in mutants lacking functional InsP3 receptor strongly challenge this hypothesis (17, 31, 41). Similarly, the highly restricted localization of rTRP2 to sensory microvilli that do not have a vesicular compartment representing a potential Ca2+ store (25, 26) argues against a store-associated activation. Moreover, in preliminary experiments, we have been unable to activate rTRP2 by agents that deplete Ca2+ stores. Thus, our results suggest that TRP2 and probably other mammalian TRP channels are activated, as is dTRP, by second-messenger signaling pathways that are store-independent.
Taken together, our data underline the importance of TRP channel-mediated transduction in a variety of sensory systems across evolution, including Drosophila phototransduction, C. elegans olfaction, and mammalian VNO pheromone transduction, and provide additional support for a mode of activation of TRP channels in sensory neurons that is not directly mediated by intracellular store depletion.
Acknowledgments
We thank Fay Wang and Ljiljianna D. Jelaska for excellent technical assistance, Josephine Austrie for help in preparing the manuscript, and members of the Dulac, Corey, Peralta, and Sheng labs and D. Arnold for helpful discussions. This work was supported by the Howard Hughes Medical Institute (D.P.C. and C.D.), Harvard University (C.D.), and Grants DC3903-01 and DC02889 (E.R.L.) from the National Institute on Deafness and Other Communication Disorders. E.R.L. was an Associate, D.P.C. is an Investigator, and C.D. is an Assistant Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- MOE
main olfactory epithelium
- VNO
vomeronasal organ
- CNG
cyclic-nucleotide-gated
- TRP
transient receptor potential
- En
embryonic day n
Note Added in Proof
While this paper was in press, articles were published describing the activation of dTRP and dTRPL by polyunsaturated fatty acids (43) and of hTRP6 and hTRPC3 by diacylglycerol (44).
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF136401).
References
- 1.Halpern M. Annu Rev Neurosci. 1987;10:325–362. doi: 10.1146/annurev.ne.10.030187.001545. [DOI] [PubMed] [Google Scholar]
- 2.Schild D, Restrepo D. Physiol Rev. 1998;78:429–466. doi: 10.1152/physrev.1998.78.2.429. [DOI] [PubMed] [Google Scholar]
- 3.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 4.Zhao H, Ivic L, Otaki J M, Hashimoto M, Mikoshiba K, Firestein S. Science. 1998;279:237–242. doi: 10.1126/science.279.5348.237. [DOI] [PubMed] [Google Scholar]
- 5.Bradley J, Li J, Davidson N, Lester H, Zinn K. Proc Natl Acad Sci USA. 1994;91:8890–8894. doi: 10.1073/pnas.91.19.8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liman E R, Buck L B. Neuron. 1994;13:611–621. doi: 10.1016/0896-6273(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 7.Brunet L J, Gold G H, Ngai J. Neuron. 1996;17:681–693. doi: 10.1016/s0896-6273(00)80200-7. [DOI] [PubMed] [Google Scholar]
- 8.Belluscio L, Gold G H, Nemes A, Axel R. Neuron. 1998;20:69–81. doi: 10.1016/s0896-6273(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 9.Dulac C, Axel R. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 10.Berghard A, Buck L B, Liman E R. Proc Natl Acad Sci USA. 1996;93:2365–2369. doi: 10.1073/pnas.93.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y, Trindelli R, Ryba N J P. Biochem Biophys Res Commun. 1996;220:900–904. doi: 10.1006/bbrc.1996.0503. [DOI] [PubMed] [Google Scholar]
- 12.Berghard A, Buck L B. J Neurosci. 1996;16:909–918. doi: 10.1523/JNEUROSCI.16-03-00909.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrada G, Dulac C. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- 14.Matsunami H, Buck L B. Cell. 1997;90:775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- 15.Ryba N J, Tirindelli R. Neuron. 1997;19:371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- 16.Liman E R, Corey D P. J Neurosci. 1996;16:4625–4637. doi: 10.1523/JNEUROSCI.16-15-04625.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niemeyer B A, Suzuki E, Scott K, Jalink K, Zuker C S. Cell. 1996;85:651–659. doi: 10.1016/s0092-8674(00)81232-5. [DOI] [PubMed] [Google Scholar]
- 18.Colbert H A, Smith T L, Bargmann C I. J Neurosci. 1997;17:8259–8269. doi: 10.1523/JNEUROSCI.17-21-08259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wes P D, Chevesich J, Jeromin A, Rosenberg C, Stetten G, Montell G. Proc Natl Acad Sci USA. 1995;92:9652–9656. doi: 10.1073/pnas.92.21.9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu X, Jiang M, Peyton M, Boulay G, Hurst R, Stefani E, Birnbaumer L. Cell. 1996;85:661–671. doi: 10.1016/s0092-8674(00)81233-7. [DOI] [PubMed] [Google Scholar]
- 21.Wissenbach U, Schroth G, Philipp S, Flockerzi V. FEBS Lett. 1998;429:61–66. doi: 10.1016/s0014-5793(98)00561-4. [DOI] [PubMed] [Google Scholar]
- 22.Boulay G, Zhu X, Peyton M, Jiang M, Hurst R, Stefani E, Birnbaumer L. J Biol Chem. 1997;272:29672–29680. doi: 10.1074/jbc.272.47.29672. [DOI] [PubMed] [Google Scholar]
- 23.Okada T, Shimizu S, Wakamori M, Maeda A, Kurosaki T, Takada N, Imoto K, Mori Y. J Biol Chem. 1998;273:10279–10287. doi: 10.1074/jbc.273.17.10279. [DOI] [PubMed] [Google Scholar]
- 24.Philipp S, Cavalie A, Freichel M, Wissenbach U, Zimmer S, Trost C, Marquart A, Murakami M, Flockerzi V. EMBO J. 1996;15:6166–6171. [PMC free article] [PubMed] [Google Scholar]
- 25.Adams D. Microsc Res Technique. 1992;23:86–97. doi: 10.1002/jemt.1070230108. [DOI] [PubMed] [Google Scholar]
- 26.Vaccarezz O L, Sepich L N, Tramezzani J H. J Anat. 1981;132:167–185. [PMC free article] [PubMed] [Google Scholar]
- 27.Lux S E, John K M, Bennett V. Nature (London) 1990;344:36–43. doi: 10.1038/344036a0. [DOI] [PubMed] [Google Scholar]
- 28.Caterina M J, Schumacher M A, Tominaga M, Rosen T A, Levine J D, Julius D. Nature (London) 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 29.Wisocky C J. Neurosci Biobehav Rev. 1979;3:301–341. doi: 10.1016/0149-7634(79)90015-0. [DOI] [PubMed] [Google Scholar]
- 30.Stern K, McClintock M R. Nature (London) 1998;392:177–179. doi: 10.1038/32408. [DOI] [PubMed] [Google Scholar]
- 31.Ranganathan R, Malicki D M, Zuker C S. Annu Rev Neurosci. 1995;18:283–317. doi: 10.1146/annurev.ne.18.030195.001435. [DOI] [PubMed] [Google Scholar]
- 32.Halpern M, Shapiro L W S, Jia C. Brain Res. 1995;677:157–161. doi: 10.1016/0006-8993(95)00159-n. [DOI] [PubMed] [Google Scholar]
- 33.Tang W J, Gilman A G. Science. 1991;254:1500–1503. doi: 10.1126/science.1962211. [DOI] [PubMed] [Google Scholar]
- 34.Sternweiss P C. Curr Opin Cell Biol. 1994;6:198–203. doi: 10.1016/0955-0674(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 35.Luo Y, Lu S, Chen P, Wang D, Halpern M. J Biol Chem. 1994;269:16867–16877. [PubMed] [Google Scholar]
- 36.Kroner C, Breer H, Singer A G, O’Connell R J. NeuroReport. 1996;7:2989–2992. doi: 10.1097/00001756-199611250-00038. [DOI] [PubMed] [Google Scholar]
- 37.Wekesa K S, Anholt R R. Endocrinology. 1997;138:3497–3504. doi: 10.1210/endo.138.8.5338. [DOI] [PubMed] [Google Scholar]
- 38.Clapham D E. Neuron. 1996;16:1069–1072. doi: 10.1016/s0896-6273(00)80132-4. [DOI] [PubMed] [Google Scholar]
- 39.Scott K, Zuker C. Curr Opin Neurobiol. 1998;8:383–388. doi: 10.1016/s0959-4388(98)80065-2. [DOI] [PubMed] [Google Scholar]
- 40.Minke B, Selinger Z. Curr Opin Neurobiol. 1996;6:459–466. doi: 10.1016/s0959-4388(96)80050-x. [DOI] [PubMed] [Google Scholar]
- 41.Acharya J K, Jalink K, Hardy R, Hartenstein V, Zuker C S. Neuron. 1997;18:881–887. doi: 10.1016/s0896-6273(00)80328-1. [DOI] [PubMed] [Google Scholar]
- 42.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgings D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chyb S, Raghu P, Hardie R C. Nature (London) 1999;397:255–259. doi: 10.1038/16703. [DOI] [PubMed] [Google Scholar]
- 44.Hofmann T, Obukhov A G, Schaefer M, Harteneck C, Gudermann T, Schultz G. Nature (London) 1999;397:259–263. doi: 10.1038/16711. [DOI] [PubMed] [Google Scholar]