Abstract
The PhoP/PhoQ two-component system controls the extracellular magnesium deprivation response in Salmonella enterica. In addition, several virulence-associated genes that are mainly required for intramacrophage survival during the infection process are under the control of its transcriptional regulation. Despite shared Mg2+ modulation of the expression of the PhoP-activated genes, no consensus sequence common to all of them could be detected in their promoter regions. We have investigated the transcriptional regulation and the interaction of the response regulator PhoP with the promoter regions of the PhoP-activated loci phoPQ, mgtA, slyB, pmrD, pcgL, phoN, pagC, and mgtCB. A direct repeat of the heptanucleotide sequence (G/T)GTTTA(A/T) was identified as the conserved motif recognized by PhoP to directly control the gene expression of the first five loci, among which the first four are ancestral to enterobacteria. On the other hand, no direct interaction of the response regulator with the promoter of phoN, pagC, or mgtCB was apparent by either in vitro or in vivo assays. These loci are Salmonella specific and were probably acquired by horizontal DNA transfer. Besides, sequence analysis of pag promoters revealed the presence of a conserved PhoP box in 6 out of the 12 genes analyzed. Our results strongly suggest that the expression of a set of Mg2+-controlled genes is driven by PhoP via unknown intermediate regulatory mechanisms that could also involve ancillary factors.
Salmonella enterica serovar Typhimurium responds to environmental magnesium deprivation by inducing the transcription of the PhoP-PhoQ regulon (10, 13, 42). Expression of this regulon is necessary for intramacrophage survival, resistance to acid pH and to antimicrobial peptides, modification of antigen presentation, formation of spacious vacuoles, and alteration of macrophage cell death (13). The coordination of this expression is governed by the activity of the PhoP/PhoQ two-component signal transduction system (14, 33). In this system extracellular Mg2+ interacts with the periplasmic domain of the sensor protein PhoQ, inducing a specific PhoP-phosphatase activity that controls the phosphorylation state of the transcriptional regulator PhoP (6, 7, 9, 47).
During the past years several PhoP-regulated loci have been discovered (3, 4, 18, 19, 23, 33, 42, 46). One group of PhoP-regulated loci appears to be ancestral among enterobacteria. It includes the phoPQ operon, which codes for the regulatory system, and the mgtA gene (10, 44), whose product is a P-type Mg2+ transporter, necessary for growth in low-Mg2+ environments. Other loci that form part of this group are slyB, previously reported as pcgH (42), which encodes a putative outer membrane lipoprotein; pagP (22), whose product is necessary for lipid A acylation and resistance to specific antimicrobial peptides; and pmrD (29), involved in the PhoP- and Mg2+-dependent activation of the PmrA-PmrB regulon, which controls modification of the overall negative charge of the lipopolysaccharide in response to ferric ions (20, 21, 38, 43, 48).
On the other hand, several genes discovered to be under PhoP regulation are Salmonella specific and were presumably incorporated into the Salmonella chromosome by horizontal DNA transfer. This group encompasses the Mg2+ transporter mgtCB operon (10, 40), which is part of Salmonella pathogenicity island 3 (SPI3); pagC and pagD (18); phoN (16), which encodes a periplasmic nonspecific acid phosphatase; pcgL (24), coding for a periplasmic d-alanine-d-alanine dipeptidase homologous to the VanX vancomycin resistance dipeptidase; pagL (45), whose product catalyzes a deacylation of lipid A precursors; the prophage-encoded pagJ, pagM, and pagK genes (19); and the host-induced virulence gene mig-14 (46). Other Salmonella-specific genes under the control of the magnesium starvation system are lpxO (11); spvB (23), located in the Salmonella virulence plasmid; pagN (8, 19), encoding a putative invasin located in Salmonella centisome 7; pqaA (2), which was found to be under PhoP/PhoQ control in Salmonella enterica serovar Typhi; and pgtE (17), an outer membrane protein posttranscriptionally regulated by the system.
PhoP belongs to the OmpR subfamily of response regulators (7, 28). Members of this subfamily recognize a direct repeat sequence in the promoter region of their target genes, and at least two Salmonella PhoP-regulated genes display a direct repeat motif 20 to 30 bases upstream of the site of transcriptional initiation (29, 41). Yamamoto et al. (49) described the presence of a similar repeat in the mgtA promoter region in Escherichia coli that is recognized by PhoP. Nevertheless, this motif cannot be detected in most of the described Salmonella PhoP-regulated genes by alignment of their promoter regions. This implies either the existence of a different recognition site for the response regulator or, alternatively, the presence of as yet unidentified factors that mediate a transcriptional cascade. In this report we characterize the interaction of PhoP with different PhoP-activated promoters. We show that the Salmonella response regulator recognizes and interacts with a direct-repeat sequence located in the promoter region of a set of pag genes. We also provide evidences that pag genes that do not harbor the PhoP box at their promoter regions are indirectly controlled by the magnesium-sensing system.
MATERIALS AND METHODS
Chemicals and reagents.
Nitrocellulose membranes were from Bio-Rad. [γ-32P]ATP was obtained from NEN Life Science Products. The oligonucleotides were purchased from Bio-Synthesis, Inc. (Lewisville, Tex.). Cell culture medium reagents were from Difco, and chemicals were from Sigma.
Bacterial strains, plasmids, and growth conditions.
Salmonella serovar Typhimurium 14028s and MS7953s (phoP::Tn10) (14) were used as the wild-type and phoP strains, respectively. Strains EG9523 (mgtA::MudJ phoP::Tn10), EG9313 (slyB::MudJ phoP::Tn10), EG9529 (mgtC::MudJ phoP::Tn10), EG9280 (pmrC::MudJ phoP::Tn10), and EG9332 (pcgL::MudJ phoP::Tn10) (10, 42, 43) were transformed with plasmid pEG9014 or with the pUHE21-2lacIq vector (41) and were used for PhoP-dependent pag expression analysis. E. coli PB1277 is strain BL21(DE3) carrying plasmid pPB1020 (7). pPB1020 harbors a phoP His tag fusion gene under the control of the T7 φ10 promoter of the pT7-7 plasmid. pPB1021 is a pMAL-c2 (New England Biolabs, Beverly, Mass.) derivative that encodes the maltose binding protein fused to the C-terminal cytoplasmic region of PhoQ (MBP-Qc) (7). For the β-galactosidase activity assay for which results are shown in Fig. 4A, bacteria were grown overnight with shaking at 37°C in Luria-Bertani medium with addition of different concentrations of isopropyl-β-d-thiogalactopyranoside (IPTG) as indicated. For the β-galactosidase activity assay for which results are shown in Fig. 4B, bacteria were grown overnight in N-minimal medium (pH 7.5) with 12 mM MgCl2 and diluted 1:20 into the same medium up to the exponential phase, washed three times in N-minimal medium (pH 7.5) with 8 μM MgCl2, resuspended in this medium, and grown with shaking at 37°C. Aliquots were withdrawn at the indicated time intervals. β-Galactosidase levels were determined as described previously (32). Plasmid DNA was introduced into bacterial strains by electroporation using a Bio-Rad apparatus as recommended by the manufacturer. Recombinant DNA techniques were performed according to standard protocols (39). The sequences of all DNA fragments were confirmed by DNA sequence analysis performed using the femtomole DNA sequencing system as recommended by the manufacturer (Promega). Ampicillin, kanamycin, and tetracycline were used at final concentrations of 50, 50, or 35 μg/ml, respectively.
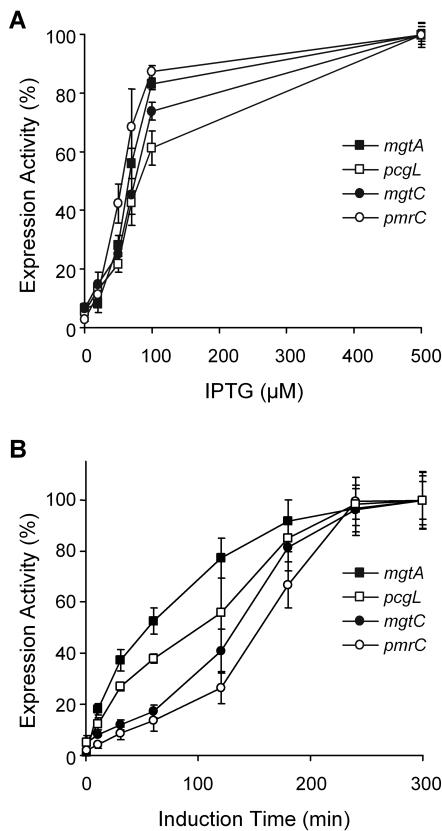
FIG. 4.
PhoP- and Mg2+-dependent expression of selected pag genes. Expression from the lacZ transcriptional fusions to mgtA, pcgL, mgtC, and the indirectly controlled gene pmrC was tested in a phoP::Tn10 background expressing PhoP from pEG9014 with addition of different concentrations of IPTG to the growth medium as indicated (A) and in a wild-type background (B), with aliquots withdrawn at different times as indicated, after transfer of the cells from a repressing (N-medium-12 mM MgCl2) to an inducing (N-medium-8 μM MgCl2) growth medium. β-Galactosidase activity from the corresponding lacZ transcriptional fusion was measured as described in Materials and Methods. Results are expressed as a percentage of the maximal expression achieved for each lacZ fusion and are averages from three independent assays performed in duplicate. Error bars, standard deviations.
RNA isolation and primer extension.
Overnight cultures of Salmonella serovar Typhimurium grown in N-minimal medium (pH 7.5) with 12 mM MgCl2 were washed three times in N-minimal medium (pH 7.5) without MgCl2 and diluted 1:50 into 10 ml of N-minimal medium (pH 7.5) containing either 8 μM or 12 mM MgCl2. Total RNA was extracted from mid exponential-phase cultures (A600, 0.3 to 0.5) as previously described (1). cDNA synthesis was performed using 2 pmol of 32P-end-labeled primers PROM MGTA-R (5′-GTAAATAATTTGCGCCGCGG-3′), PROM SLYB-R (5′-AAACGCTATTTCAGCATCCC-3′), PROM PCGL-R (5′-AGAGGATCCCATATGTTACACCTCG-3′), PROM MGTC-R (5′-AGG AATAGCGTGCGTTAACTT GCC-3′), PROM PAGC-R2, (5′-CTTAGCACGCTTTATTCCCG-3′), and PROM PHON-R (5′-TTAGCTAGCATCAGTGGTAG-3′), with 100 μg of total RNA and 1 U of SuperScript II RnaseH2 reverse transcriptase (Life Technologies, Inc.). The extension products were analyzed by electrophoresis on a 6% polyacrylamide-8 M urea gel and compared with sequence ladders initiated with the same 32P-labeled primer that was used for primer extension.
EMSA.
DNA fragments used for the electrophoretic gel mobility shift assays (EMSA) were amplified by PCR using Salmonella serovar Typhimurium chromosomal DNA as a template. Prior to the PCR, the primers that anneal to the coding strand of the promoters analyzed were labeled with T4 polynucleotide kinase and [γ-32P]ATP. The promoter regions of phoPQ, mgtA, slyB, pmrD, pcgL, mgtCB, pagC, and phoN were amplified using primers PROM PHOP-F (5′-GCAAATTATATCGGTCGCGC-3′), 369 (41), PROM MGTA-F (5′-TTGAATTCCCTACGCCGCTC-3′), PROM MGTA-R, PROM SLYB-F (5′-AGAGGATCCGCCTGTTGCGCAACCA-3′), PROM SLYB-R, PROM PMRD-F (5′-AGAGGATCCTCCCGTTGCGGTTGTG-3′), PROM PMRD-R (5′-AGAGGATCCCATAGCGCCCCCGTTT-3′), PROM PCGL-F (5′-AGAGGATCCGCAGTGGTCGTATAAG-3′), PROM PCGL-R, PROM PAGC-F (5′-AACGAATTCGTTAACCACTC-3′), PROM PAGC-R2, PROM PHON-F (5′-TGCGTGTCAGTCAGGCACG-3′), PROM PHON-R, PROM MGTC-F (5′-TACGGCGGCAATCAGGAC-3′), and PROM MGTC-R. PCR amplification rendered fragments of 242, 227, 349, 210, 290, 237, 280, and 348 bp, respectively. Approximately 6 fmol of labeled DNA in a 30-μl volume was incubated at room temperature for 30 min with the indicated amounts of purified unphosphorylated or phosphorylated PhoP-H6 protein. PhoP phosphorylation was performed by using MBP-Qc and ATP according to a protocol described previously (7). The binding buffer used for protein-DNA incubations contained 25 mM Tris-HCl (pH 8), 50 mM KCl, 0.5 mM EDTA, and 10% glycerol. Samples were run on a 5% nondenaturing Tris-glycine polyacrylamide gel at room temperature. After electrophoresis the gel was dried and autoradiographed.
Protein purification protocols.
The His-tagged fusion protein PhoP-H6 was purified from strain PB1277. PhoP-H6 was expressed by addition of 0.7 mM IPTG to induce the DE3-encoded T7 RNA polymerase (7). The MBP-Qc fusion protein was purified from the E. coli strain TB1 (obtained from the pMAL Protein Fusion and Purification system [NEB, Inc.]) transformed with plasmid pPB1021. MBP-Qc was expressed as described previously (7). All procedures were carried out at 4°C. The protein profile of the purified proteins was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
DNase I footprinting assay.
DNase I protection assays were done for both DNA strands using the appropriate labeled primer (1). PhoP-H6 protein was incubated with MBP-Qc, with or without the addition of ATP for 20 min at 37°C as described previously (7). Binding reactions with different amounts of PhoP-H6 protein and 6 fmol of labeled DNA were performed as described for the gel mobility shift assay. DNase I (Life Technologies, Inc.) (0.05 U) was added and incubated for 2 min at room temperature in a final volume of 70 μl. The reaction was stopped by adding 90 μl of 20 mM EDTA (pH 8), 200 mM NaCl, and 100 μg of tRNA/ml. DNA fragments were purified by phenol-chloroform extraction and resuspended in 7 μl of H2O. Samples (3 μl) were analyzed by denaturing polyacrylamide (6%) gel electrophoresis by comparison with a DNA sequence ladder generated with the appropriate primer.
RESULTS
Mapping the transcription start sites of different PhoP-activated promoters.
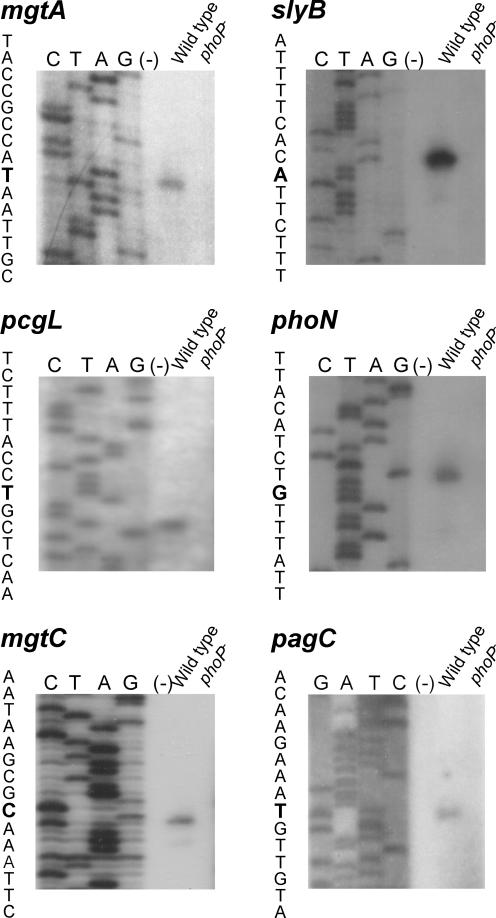
To characterize the DNA motif required for PhoP to recognize its target genes in the regulon, we mapped the transcription start sites of six PhoP-activated loci. We included mgtA and slyB, two genes also present in other enterobacterial species, and four Salmonella-specific loci, mgtCB, pagC, phoN, and pcgL (31). We performed primer extension using RNAs isolated from late-exponential-phase cultures of the wild-type Salmonella strain ATCC 14028s and the phoP strain MS7953s grown in N-minimal medium with 8 μM Mg2+. A single PhoP-dependent primer extension product was detected for each pag gene (Fig. 1) (the transcription start site and the putative promoter region for each gene studied in this report are shown in Fig. 2). For pagC, the transcription start site corresponded to a T residue located 559 bp upstream of the pagC start codon, which correlated to the size of the mRNA detected by Pulkkinen and Miller (36). For mgtA and mgtCB, 263- and 284-nucleotide (nt) 5′ untranslated leader mRNAs were identified, respectively (40, 44). The position of the transcription start site for mgtA corresponded to a T residue, which coincided with the previously identified transcription start site of the E. coli mgtA gene (27), although these two loci are only 65% identical at the promoter region. Single transcription start sites were also detected for the phoN, pcgL, and slyB genes, corresponding to a G, a C, and an A residue located 31, 43, and 99 bp upstream of their translation start codons, respectively (16, 24, 30). On the other hand, no primer extension products were detected for any of the six loci with RNA isolated from the phoP strain (Fig. 1), corroborating their transcriptional dependence on PhoP.
FIG. 1.
Mapping the transcription start sites of the selected Salmonella serovar Typhimurium pag genes. Primer extension products were generated after reverse transcription of total RNA isolated from mid-exponential-phase 14028s (wild-type) or MS7953s (phoP) cells grown in N-minimal medium, pH 7.5, with 8 μM MgCl2. As negative control, a primer extension reaction was carried out without addition of RNA (−). The primer extension products were run on a 6% polyacrylamide sequencing gel against dideoxy sequencing reactions primed with the same primer. The sequences spanning the transcription start sites are shown, and the transcription start sites are boldfaced.
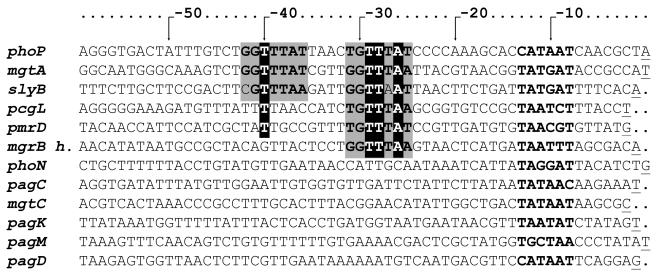
FIG. 2.
Alignment of the promoter regions of all identified Salmonella serovar Typhimurium pag promoters. Sequences from the pag promoter regions either previously described (18, 19, 26, 29, 41) or identified in this work were piled up. The (G/T)GTTTA(A/T) direct repeats are boxed, and the proposed −10 regions are boldfaced. The T and A residues essential for expression of the E. coli mgtA gene are highlighted. The transcription start sites are underlined. The putative promoter region of the mgrH homologue was included, based on the transcription start site determined in E. coli (27).
Analysis of PhoP-promoter DNA interaction by EMSA.
Figure 2 shows the alignment of the promoter region of the twelve pag genes from which the transcription start sites were identified, including those identified in the present analysis (18, 19, 29, 41). The presence of the putative PhoP box, defined in E. coli as the tandem direct repeat of the heptanucleotide (T)G(T)TT(AA) (49), can be distinguished only in the promoter regions of phoP, mgtA, and slyB, whereas a single heptanucleotide sequence can be detected in the promoters of pcgL and pmrD. The absence of a common motif among all pag promoters could imply that, in addition to the described motif, Salmonella PhoP is able to recognize a different sequence than the response regulator from E. coli, despite the 93% identity of these response regulators (15). Alternatively, it is possible that the Mg2+ regulation of the loci that do not harbor the repeat is indirect, driven by a yet unidentified regulatory cascade, as has been described for the PmrA-regulated genes (29).
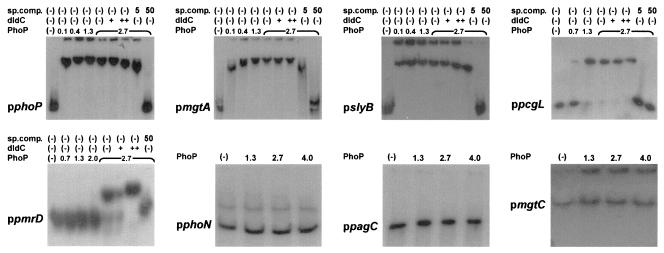
EMSA was performed using PCR-amplified fragments derived from the promoter regions of all genes investigated. Each PCR product encompasses the transcriptional start site of the downstream PhoP-regulated gene. The PCR fragments extend 135, 172, 272, 115, 258, 178, 237, and 272 bp upstream of the transcriptional start sites of phoPQ, mgtA, slyB, pcgL, pmrD, phoN, pagC, and mgtCB, respectively. Purified PhoP-H6 fusion protein was used for these assays. A plasmid harboring the gene coding for the His-tagged PhoP (pPB1020) complemented a phoP mutant strain for the Mg2+-controlled activation of the regulon in a PhoQ-dependent manner (7).
Band retardation of the promoter fragments of phoP, mgtA, and slyB was detected when they were incubated with 0.1 μM or higher concentrations of the purified response regulator (Fig. 3). A single retarded band was detected with these three promoter fragments, even with the highest concentration of PhoP-H6 tested, suggesting that a single PhoP-binding site was present in each fragment. A 1,000-fold excess of competing poly(dI-dC) did not affect the interaction, while no shifted radioactive band was detected when a 50-fold excess of unlabeled promoter fragment was included in the mixture, indicating that the interaction of the response regulator with each promoter fragment was specific. In the case of the promoter regions of pcgL and pmrD, band retardation was observed by using at least 1.3 and 2.7 μM PhoP, respectively. Footprinting analysis of these two promoters could not be successfully performed even when high PhoP concentrations were used (data not shown). Incubation of the promoter fragment of phoN, pagC, or mgtCB with as much as 4.0 μM PhoP-H6 did not affect the electrophoretic migration of these fragments under the conditions tested. Although phosphorylation of PhoP lowered the concentration required for the shift in the first six promoters 2- to 10-fold, no interaction was observed for the promoter fragments of phoN, pagC, or mgtCB when higher concentrations of PhoP (4 μM) or phospho-PhoP were used (Fig. 3 and data not shown). In these experiments, PhoP concentrations above 4 μM could not be used because of protein aggregation.
FIG. 3.
PhoP binds to the promoter regions of the phoP, mgtA, slyB, pcgL, and pmrC genes but not to the promoter region of phoN, pagC, or mgtC. EMSA was performed using the 32P 3′-end-labeled PCR fragment of the promoter region of phoP, mgtA, slyB, pcgL, pmrC phoN, pagC, or mgtC, respectively, incubated with different amounts of purified PhoP-H6, in the absence or presence of different amounts of either poly(dI-dC) as a nonspecific competitor (dIdC) or the corresponding unlabeled PCR fragment (sp.comp.).
There are two conceivable explanations for the lack of PhoP interaction with the promoter fragments of phoN, pagC, and mgtCB: either (i) the PhoP control of these loci is indirect or (ii) due to a lower affinity of PhoP for the putative binding site in these promoters, a higher protein concentration or activation status would be necessary for the interaction to be detected in the in vitro assay. To distinguish between these alternatives, we first analyzed the expression of mgtA, pcgL, mgtC, and pmrC as an indirectly PhoP-regulated gene (21, 43) in a phoP::Tn10 strain, expressing PhoP from the IPTG-regulated plasmid pEG9014. As previously reported, this Tn10 insertion in phoP is nonpolar and could be complemented by expressing PhoP from pEG9014 (42). This assay showed that pcgL required higher levels of IPTG, and thus a higher intracellular concentration of activated PhoP, to achieve the same levels of induction as the other three loci analyzed. This was consistent with the results obtained in the DNA mobility shift assays (see Fig. 3). On the other hand, both pmrC and mgtC showed an expression pattern similar to that of the directly regulated gene mgtA, suggesting that mgtC does not harbor a low-affinity site for PhoP. We then analyzed the time course of expression of the four genes in a wild-type background when the cells were transferred from repressing to inducing conditions. Figure 4B shows that there was a delay in the onset of expression not only for pmrC but also for mgtC compared to mgtA or pcgL. This retardation strongly suggests that, in analogy to what happens with pmrC, an induction cascade could be required to activate mgtC.
Determination of the PhoP-binding site in the promoter region of phoPQ.
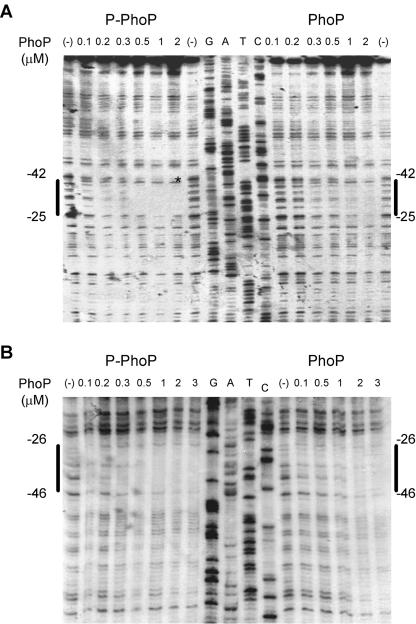
We then examined the DNA sequence recognized by PhoP in the promoter region of the phoPQ operon as a representative of a directly PhoP controlled promoter (Fig. 5). DNase I footprinting analysis was performed on both the coding and noncoding strands of the promoter fragment by using either phosphorylated or unphosphorylated PhoP-H6 protein.
FIG. 5.
DNA footprinting analysis of the phoP promoter. Footprinting analysis of the promoter region of phoP was performed on both end-labeled coding (A) and noncoding (B) strands. The concentration of unphosphorylated (PhoP) or phosphorylated (P-PhoP) PhoP-H6 protein added to the DNA fragments is given above the gels. Phosphorylation of PhoP was achieved by incubation with MBP-Qc and ATP as described in Materials and Methods. Solid lines indicate the PhoP-protected region. The position of the area of protection was determined by comparison with sequence ladders, obtained by using the same labeled primer as was used for the probe. The hypersensitive DNase I site is indicated by an asterisk.
The phosphorylated PhoP-H6 protein protected nt −25 to nt −42 relative to the transcription start site of promoter pphoP1 in the coding strand and nt −26 to nt −46 in the noncoding strand of phoP (Fig. 5). Thus, there was an overlap of 17 bp between the two strands protected by the PhoP protein. The protected region encompassed the direct repeat (G/T)GTTTAT. The C residue at position −23 relative to the transcription start site was observed to be hypersensitive to DNase I. Protection with unphosphorylated PhoP was observed by using 2 μM response regulator. On the other hand, the same protected region was detected by using 0.5 μM phosphorylated PhoP-H6.
DISCUSSION
In Salmonella enterica the two-component system PhoP/PhoQ controls the transcription of at least 30 different loci (13). Among these, two groups could be distinguished according to the presence of orthologous sequences in related bacteria. A set of PhoP-regulated loci, including the phoPQ operon, can be found in other enterobacteria such as E. coli, while other pag genes are Salmonella specific (31). The incorporation of the latter genes into the bacterial chromosome was suggested to be driven by horizontal transfer after divergence from E. coli some 100 million years ago. In this report we provide evidence that PhoP/PhoQ directly controls the expression of the majority of the ancestral genes, while it indirectly exerts Mg2+ regulation of most of the horizontally acquired pag genes.
We found that PhoP recognizes the sequence (G/T)GTTTA(A/T) in the promoter regions of five of the investigated pag promoters. This sequence is arranged as a direct repeat motif between 25 and 42 bp upstream from the transcriptional start site of the pag loci phoPQ, mgtA, and slyB. This PhoP box is essentially in agreement with the (T/G)GTTTA sequence proposed in E. coli by Minagawa et al. (34) during the preparation of this report. In spite of the fact that divergence from this sequence found in different E. coli pag promoters was suggested to be responsible for distinct binding affinities, a direct PhoP interaction was detected in all cases analyzed. In contrast, we could detect only one copy of the repeat in the Salmonella PhoP directly controlled pmrD and pcgL promoters, which required higher concentrations of the response regulator for the in vitro interaction and, in the case of pcgL, for maximal in vivo expression. It is worthwhile to point out here that both promoters harbor a T residue at position −38 (corresponding to position −40 in the consensus pag promoter), shown to be essential for expression of the E. coli mgtA gene (49). Although we could not successfully carry out footprinting analysis of these two promoters, Kato et al. (26) recently showed that the pmrD promoter was protected by PhoP in a region that encompasses 42 to 9 bp upstream of the pmrD transcription start site. Collectively, these results indicate that PhoP is able to recognize and interact with these two promoters even when they do not display an intact tandem repeat. Nevertheless, further experiments are being conducted to discern which residues in the pag promoters are essential for direct PhoP-induced expression.
Taking into account that the physiological function of the PhoP-PhoQ regulon in enteric bacteria is to cope with low-magnesium environments by both increasing the uptake of the divalent cation and reducing its envelope requirement (13), most of the ancestral enteric gene products are expected to have a role in this process. Indeed, six loci were described to belong to this group of PhoP-activated genes. The phoPQ operon was previously demonstrated to be autoregulated (41). It was recently postulated that the autoregulation of the signal transduction components might be associated with a “learning behavior” that, after sensing an inducing environment, will prompt the cells to mount a faster and larger response in a subsequent event (25). Autoregulation helps to amplify and modulate the transduction process signaled by Mg2+ starvation. The fact that mgtA was PhoP controlled led us to define the inducing signal of the regulon and to understand its physiological role (10). Accordingly, mutants in this locus showed impaired growth in low-magnesium environments (42). The role of pagP in Mg2+ homeostasis was recently assessed in Legionella pneumophila, where the growth of an rcp (the pagP-like gene) mutant in low-magnesium media was affected (37). The pagP gene codes for an enzyme that catalyzes the incorporation of palmitate into lipopolysaccharide (LPS), increasing bacterial resistance to some cationic antimicrobial peptides (22). slyB encodes a putative outer membrane lipoprotein. As a homologue of Pal lipoproteins (5), it could participate in stabilizing the outer membrane by reducing the requirement of Mg2+ as an outer membrane counterion. An in silico analysis of the Salmonella genome shows a homologue of E. coli mgrB (27) with an identity of 80% (31). It encodes a putative 47-amino-acid inner membrane protein for which no function has been described to date. Finally, pmrD encodes a protein that postranscriptionally mediates PhoP induction of the PmrA-PmrB regulon during magnesium limitation (29). This regulon participates in the modulation of the overall charge and the Mg2+ content of LPS, allowing a reduction in the requirements for the bacterial envelope Mg2+ content in favor of its cytoplasmic uptake in low-magnesium environments (13, 29, 43, 48).
In this work, direct PhoP regulation was demonstrated for the ancestral genes phoPQ, mgtA, slyB, and pmrD. Moreover, a putative PhoP-binding site was found to be located in the promoter region of the mgrB homologue in Salmonella serovar Typhimurium (Fig. 2), and a sequence harboring a putative PhoP box (AGATTATN4TGTTTAT) could also be detected in the pagP promoter region.
On the other hand, the (T/G)GTTTA(A/T) conserved motif could not be detected in the promoter regions of 6 of the 12 Salmonella serovar Typhimurium genes analyzed. Most of these genes were found to be required for virulence and intramacrophage survival, and their role in magnesium homeostasis remains unclear. One possible explanation is that most of these genes evolved to be under PhoP/PhoQ control in order to guarantee a coordinated expression pattern inside the host cell. Then, while most of the ancestral genes were directly modulated by the Mg2+-regulatory system, the Salmonella-specific genes were recruited under PhoP control, probably through a regulatory cascade. This point is emphasized by the observation that, while we have not been able to observe PhoP binding in vitro, these genes are activated in vivo at phospho-PhoP levels equivalent to those for the directly activated genes (Fig. 4A). This result indicates that lack of interaction was not due to the presence of a low-affinity recognition site. Moreover, the delay in the onset of expression shown for mgtC compared to mgtA and pcgL (Fig. 4B) strongly contributes to the suggested indirect induction process. Lending support to this concept is the fact that three transcription factors, and thus putative intermediaries for induction cascades, SlyA, Mig-14, and RstA, have recently been described to be under PhoP transcriptional control (34, 35, 46). Besides, we cannot eliminate the potential involvement of a PhoP-dependent anti-terminator mechanism (12). In this regard, it is worth mentioning that long noncoding leaders were found both in directly and indirectly regulated pag genes. Further work has to be carried out to fully understand the complexity of the Mg2+ stimulon signaling network.
In conclusion, we can postulate that the set of genes acquired horizontally, necessary for Salmonella to succeed throughout all steps of infection, have been enrolled under the indirect control of the ancestral PhoP/PhoQ system and in this way are coordinately expressed under a meaningful host signal.
Acknowledgments
We are grateful to the anonymous reviewers for helpful suggestions.
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (Argentina), Fundación Antorchas, Third World Academy of Sciences (Trieste, Italy), CONICET, and Ministerio de Salud de la República Argentina. E.G.V. is a career investigator of the National Research Council (CONICET, Argentina), and S.L. and A.A. are fellows of the same institution. F.C.S. is a member of the Rosario National University Research Council (CIUNR) and CONICET, and he is also an International Research Scholar of the Howard Hughes Medical Institute.
REFERENCES
- 1.Aguirre, A., S. Lejona, E. García Véscovi, and F. C. Soncini. 2000. Phosphorylated PmrA interacts with the promoter region of ugd in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:3874-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, S. J., C. Daniels, and R. Morona. 1997. PhoP/Q regulated genes in Salmonella typhi: identification of melittin sensitive mutants. Microb. Pathog. 22:165-179. [DOI] [PubMed] [Google Scholar]
- 3.Behlau, I., and S. I. Miller. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 175:4475-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belden, W. J., and S. I. Miller. 1994. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect. Immun. 62:5095-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cascales, E., A. Bernadac, M. Gavioli, J.-C. Lazzaroni, and R. Lloubes. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castelli, M. E., A. Cauerhff, M. Amongero, F. C. Soncini, and E. García Véscovi. 2003. The H box-harboring domain is key to the function of the Salmonella enterica PhoQ Mg2+-sensor in the recognition of its partner PhoP. J. Biol. Chem. 278:23579-23585. [DOI] [PubMed] [Google Scholar]
- 7.Castelli, M. E., E. García Véscovi, and F. C. Soncini. 2000. The phosphatase activity is the target for Mg2+ regulation of the sensor protein PhoQ in Salmonella. J. Biol. Chem. 275:22948-22954. [DOI] [PubMed] [Google Scholar]
- 8.Folkesson, A., A. Advani, S. Sukupolvi, J. D. Pfeifer, S. Normark, and S. Lofdahl. 1999. Multiple insertions of fimbrial operons correlate with the evolution of Salmonella serovars responsible for human disease. Mol. Microbiol. 33:612-622. [DOI] [PubMed] [Google Scholar]
- 9.García Véscovi, E., Y. M. Ayala, E. Di Cera, and E. A. Groisman. 1997. Characterization of the bacterial sensor protein PhoQ. Evidence for distinct binding sites for Mg2+ and Ca2+. J. Biol. Chem. 272:1440-1443. [DOI] [PubMed] [Google Scholar]
- 10.García Véscovi, E., F. C. Soncini, and E. A. Groisman. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165-174. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons, H. S., S. Lin, R. J. Cotter, and C. R. H. Raetz. 2000. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, a new Fe2+/alpha ketoglutarate dependent dioxygenase homologue. J. Biol. Chem. 275:32940-32949. [DOI] [PubMed] [Google Scholar]
- 12.Gollnick, P., and P. Babitzke. 2002. Transcription attenuation. Biochim. Biophys. Acta 1577:240-250. [DOI] [PubMed] [Google Scholar]
- 13.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groisman, E. A., E. Chiao, C. J. Lipps, and F. Heffron. 1989. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc. Natl. Acad. Sci. USA 86:7077-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groisman, E. A., F. Heffron, and F. Solomon. 1992. Molecular genetic analysis of the Escherichia coli phoP locus. J. Bacteriol. 174:486-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groisman, E. A., M. H. Saier, Jr., and H. Ochman. 1992. Horizontal transfer of a phosphatase gene as evidence for mosaic structure of the Salmonella genome. EMBO J. 11:1309-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guina, T., E. C. Yi, H. Wang, M. Hackett, and S. I. Miller. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 182:4077-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunn, J. S., C. M. Alpuche-Aranda, W. P. Loomis, W. J. Belden, and S. I. Miller. 1995. Characterization of the Salmonella typhimurium pagC/pagD chromosomal region. J. Bacteriol. 177:5040-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunn, J. S., W. J. Belden, and S. I. Miller. 1998. Identification of PhoP-PhoQ activated genes within a duplicated region of the Salmonella typhimurium chromosome. Microb. Pathog. 25:77-90. [DOI] [PubMed] [Google Scholar]
- 20.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 21.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189-198. [DOI] [PubMed] [Google Scholar]
- 23.Heithoff, D. M., C. P. Conner, U. Hentschel, F. Govantes, P. C. Hanna, and M. J. Mahan. 1999. Coordinate intracellular expression of Salmonella genes induced during infection. J. Bacteriol. 181:799-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hilbert, F., F. García del Portillo, and E. A. Groisman. 1999. A periplasmic d-alanyl-d-alanine dipeptidase in the gram-negative bacterium Salmonella enterica. J. Bacteriol. 181:2158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffer, S. M., H. V. Westerhoff, K. J. Hellingwerf, P. W. Postma, and J. Tommassen. 2001. Autoamplification of a two-component regulatory system results in “learning” behavior. J. Bacteriol. 183:4914-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato, A., T. Latifi, and E. A. Groisman. 2003. Closing the loop: the PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. Proc. Natl. Acad. Sci. USA 100:4706-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato, A., H. Tanabe, and R. Utsumi. 1999. Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J. Bacteriol. 181:5516-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenney, L. J. 2002. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 5:135-141. [DOI] [PubMed] [Google Scholar]
- 29.Kox, L. F., M. M. Wosten, and E. A. Groisman. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig, A., C. Tengel, A. Bauer, R. Benz, H. J. Mollenkopf, and W. Goebel. 1995. SlyA, a regulatory protein from Salmonella typhimurium, induces a haemolytic and pore-forming protein in Escherichia coli. Mol. Gen. Genet. 249:474-486. [DOI] [PubMed] [Google Scholar]
- 31.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minagawa, S., H. Ogasawara, A. Kato, K. Yamamoto, Y. Eguchi, T. Oshima, H. Mori, A. Ishihama, and R. Utsumi. 2003. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J. Bacteriol. 185:3696-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norte, V. A., M. R. Stapleton, and J. Green. 2003. PhoP-responsive expression of the Salmonella enterica serovar Typhimurium slyA gene. J. Bacteriol. 185:3508-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pulkkinen, W. S., and S. I. Miller. 1991. A Salmonella typhimurium virulence protein is similar to a Yersinia enterocolitica invasion protein and a bacteriophage lambda outer membrane protein. J. Bacteriol. 173:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robey, M., W. O′Connell, and N. P. Cianciotto. 2001. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect. Immun. 69:4276-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roland, K. L., L. E. Martin, C. R. Esther, and J. K. Spitznagel. 1993. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J. Bacteriol. 175:4154-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Snavely, M. D., C. G. Miller, and M. E. Maguire. 1991. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J. Biol. Chem. 266:815-823. [PubMed] [Google Scholar]
- 41.Soncini, F. C., E. García Véscovi, and E. A. Groisman. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177:4364-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soncini, F. C., E. García Véscovi, F. Solomon, and E. A. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soncini, F. C., and E. A. Groisman. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 178:6796-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao, T., M. D. Snavely, S. G. Farr, and M. E. Maguire. 1995. Magnesium transport in Salmonella typhimurium: mgtA encodes a P-type ATPase and is regulated by Mg2+ in a manner similar to that of the mgtB P-type ATPase. J. Bacteriol. 177:2654-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trent, M. S., W. Pabich, C. R. H. Raetz, and S. I. Miller. 2001. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J. Biol. Chem. 276:9083-9092. [DOI] [PubMed] [Google Scholar]
- 46.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 47.Waldburger, C. D., and R. T. Sauer. 1996. Signal detection by the PhoQ sensor-transmitter. Characterization of the sensor domain and a response-impaired mutant that identifies ligand-binding determinants. J. Biol. Chem. 271:26630-26636. [DOI] [PubMed] [Google Scholar]
- 48.Wosten, M. M., L. F. Kox, S. Chamnongpol, F. C. Soncini, and E. A. Groisman. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113-125. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto, K., H. Ogasawara, N. Fujita, R. Utsumi, and A. Ishihama. 2002. Novel mode of transcription regulation of divergently overlapping promoters by PhoP, the regulator of a two-component system sensing external magnesium availability. Mol. Microbiol. 45:423-438. [DOI] [PubMed] [Google Scholar]