Abstract
We report the cloning and functional characterization in the mouse and the rat of a novel natural killer (NK) cell receptor termed KLRE1. The receptor is a type II transmembrane protein with a COOH-terminal lectin-like domain, and constitutes a novel KLR family. Rat Klre1 was mapped to the NK gene complex. By Northern blot and flow cytometry using newly generated monoclonal antibodies, KLRE1 was shown to be expressed by NK cells and a subpopulation of CD3+ cells, with pronounced interstrain variation. Western blot analysis indicated that KLRE1 can be expressed on the NK cell surface as a disulphide-linked dimer. The predicted proteins do not contain immunoreceptor tyrosine-based inhibitory motifs (ITIMs) or a positively charged amino acid in the transmembrane domain. However, in a redirected lysis assay, the presence of whole IgG, but not of F(ab′)2 fragments of a monoclonal anti-KLRE1 antibody inhibited lysis of Fc-receptor bearing tumor target cells. Moreover, the tyrosine phosphatase SHP-1 was coimmunoprecipitated with KLRE1 from pervanadate-treated interleukin 2–activated NK cells. Together, our results indicate that KLRE1 may form a functional heterodimer with an as yet unidentified ITIM-bearing partner that recruits SHP-1 to generate an inhibitory receptor complex.
Keywords: natural immunity, lymphocytes, immunological receptors, molecular sequence data, protein-tyrosine-phosphatase
Introduction
NK cells have the capacity to specifically recognize and kill certain tumor cells, virally infected cells and MHC-disparate normal hematopoietic cells (1–3). The target cell recognition depends upon an interplay between activating and inhibitory membrane receptors (4), recognizing a number of different ligands including MHC class I or class I–like molecules (5–7). The receptors belong to two different protein families, the killer cell lectin-like receptors (KLRs)* and receptors with immunoglobulin-like domains. Prominent among the latter are the killer cell immunoglobulin-like receptors (KIRs). Both contain inhibitory as well as activating members (8–10). As a rule, the inhibitory receptors have cytoplasmic tails with immunoreceptor tyrosine-based inhibitory motifs (ITIMs), with the consensus sequence I/V/L/SxYxxV/L (11).The activating receptors lack ITIM, but instead have a positively charged amino acid in the transmembrane (TM) domain, mediating association with adaptor molecules containing immunoreceptor tyrosine-based activating motifs (ITAM). After receptor engagement, tyrosine residues are phosphorylated, whereupon the ITIM and the ITAM recruit SH2 domain-containing tyrosine phosphatases and -kinases, respectively (4, 10–16). In particular, the protein tyrosine phosphatase SHP-1 has been demonstrated to associate with phosphorylated ITIMs, and to mediate inhibition of NK cell cytotoxicity (11–16). The KLRs are encoded by a distinct chromosomal region, the NK cell gene complex (NKC), which maps to homologous regions on mouse chromosome 6 (17), rat chromosome 4 (18), and human chromosome 12p13 (19). Several different NKC-encoded KLR families have so far been identified, typically expressed as homo- or heterodimers. The inhibitory members of the Ly-49 family are homodimers with one ITIM per molecule, yielding two ITIMs in close proximity. In contrast, the inhibitory heterodimer CD94/NKG2A recruits SHP-1 via the two ITIMs located in tandem within the cytoplasmic tail of one of the chains (NKG2A), whereas no direct signaling function has been described for the CD94 chain (20–22).
Here, we have cloned and characterized in the mouse and the rat a novel NK cell receptor termed KLRE1. The predicted proteins do not contain an ITIM in the cytoplasmic tail or a charged amino acid in the TM domain. We present evidence that despite the lack of ITIMs, KLRE1 forms part of an inhibitory receptor able to recruit the tyrosine phosphatase SHP-1.
Materials and Methods
Animals.
The inbred rat strains PVG, AUG, DA, AO, AGUS, LEW, DA.NKCb/b, and DA × (DA × PVG)F1 hybrids were reared under conventional conditions (routinely screened for pathogens) in Oslo, or obtained commercially (Harlan Olac). DA × (DA × PVG)F1 rats were generated from breeding pairs of male (DA × PVG)F1 and female DA rats. The NKC congenic rat strain DA.NKCb/b was generated by backcrossing (DA × PVG)F1 to DA for ten generations (selecting male offspring heterozygous at the NKC (NKCa/b) by RFLP analysis of the Nkrp1b locus) followed by inbreeding. The experimental protocol was approved by the institute veterinary surgeon and registered by the Experimental Animal Board under the Ministry of Agriculture of Norway. Inbred mice were purchased from The Jackson Laboratory.
Cloning of Mouse and Rat KLRE1 cDNA.
COS-7 cells were transfected by DEAE-dextran with a cDNA library prepared from IL-2 activated polyclonal mouse NK cells derived from CB.17 SCID mice in the pME18S vector (generously provided by Dr. Vinay Kumar, University of Texas, Dallas, Texas). After 72 h, the transfected COS-7 cells were stained with PE-conjugated DX6 mAb and the antigen-positive cells were isolated by fluorescence-activated cell sorting (Becton Dickinson). Antigen-positive cells were lysed in Hirt solution and plasmids were recovered by bacterial transformation. After three rounds of selection using transfected COS-7, individual plasmids were isolated and analyzed by transfection for DX6 antigen expression (23). A single plasmid was selected and confirmed to encode a protein reactive with the DX6 antibody.
The open reading frame (ORF) of the mouse DX6 antigen cDNA was used as a probe to screen a cDNA library made from IL-2 activated NK cells from F344 rats (provided by Dr. James C. Ryan, University of California, San Francisco, CA; reference 18). 2 × 106 plaques were lifted onto nylon membranes (Colony/Plaque Screen, NEN Life Science Products) and hybridized to radiolabeled probe (Megaprime DNA labeling system; Amersham Biosciences) under low stringency conditions, as described earlier (24). 10 independent clones were isolated and sequenced. Nested PCR was performed on genomic DNA extracted from DA and F344 liver cells (outer primer pair: 5′-TGCTATGAGGCTGTGAAGTGTGTT-3′ versus 5′-ATGTTTTTGGTATTTTTGCCTCTGCT-3′; inner primer pair: 5′-ACCCTGCTCCCAGTTCTGTCTTA-3′ versus 5′-TTGCCTCTGCTCAATGGAT-GACAAT-3′). To obtain cDNA corresponding to the two splice variants, forward primers made on the basis of genomic sequences (5′-CAGAATTTTTGTCTTTATA-3′ [rKLRE1 long] or 5′-CCTCCTTGCTTCAGAAGAGCAAA-3′ [rKLRE1 short]) were used together with a 3′UTR specific reverse primer (5′-ACCAAAAATTAAGACTTTA-3′) in reverse transcriptase PCR using total RNA from IL-2 activated DA NK cells (25). The PCR products were cloned into the pCR2.1-TOPO plasmid vector (Invitrogen), and three individual clones were completely sequenced on both strands as detailed previously (24).
Cell Preparations.
NK cells were purified from rat spleen by Isopaque-Ficoll density centrifugation (Lymphoprep; Nycomed) followed by passage through nylon wool (Leucopac; Fenwal Laboratories) and removal of T cells and B cells by negative selection with anti-CD3 mAb G4.18 (26), anti-TCRα/β mAb R73 (27), anti-CD5 mAb OX19 (28), anti-MHC class II mAb MRC OX6 (29), and immunomagnetic beads coated with sheep anti–mouse IgG (Dynal). Remaining cells were incubated at 37°C for 11 d in complete medium (RPMI 1640 [GIBCO BRL] containing 1 mM sodium pyruvate, 2 mM l-glutamine, 1% standard antibiotic/antimycotic solution and 10% FCS [GIBCO BRL]). 5 × 105 M 2-ME and rat rIL-2 (30) equivalent to 1,000 IU/ml of human IL-2 was added, yielding a >98% NKR-P1+ NK cell population as determined by flow cytometry using mAb 3.2.3 (31). Thoracic duct CD4+ T cells (>95% W3/25+ [32] by flow cytometry), CD8+ T cells (>91% OX8+ [33] by flow cytometry), and B cells were purified as described (24). The macrophage cell line R2 (34) and the mast cell line RBL-2H3 (35), and the F344-derived NK cell lines RNK-16 (36) and A181 (37) were cultured under standard conditions in complete medium. 2-ME was added to the NK cell medium. A stable, IL-2–dependent rat NK cell line termed RNKDA1 was generated from DA rat spleen NK cells by long term culture in complete medium with IL-2 and 2-ME as above. The cells were analyzed by flow cytometry and shown to be NKR-P1+/CD3−.
Southern and Northern Blot Analysis.
DNA was extracted from rat liver, digested with restriction endonucleases (New England Biolabs, Inc.), subjected to horizontal agarose gel electrophoresis, and blotted onto nylon membranes (Biotrans membranes; ICN Biomedicals) by conventional methods. Extraction of total cellular RNA, formaldehyde agarose gel electrophoresis, and transfer to nylon membranes was performed by conventional methods. Equal loading of RNA in all lanes was verified by ethidium bromide staining. Hybridization was performed under conditions described earlier (18). All probes were radiolabeled with [32P] dCTP (Megaprime DNA labeling system; Amersham Biosciences).
Pulsed Field Gel Eectrophoresis.
Pulsed field gel electrophoresis (PFGE) was performed by standard methods, using single-cell suspensions from PVG lymph nodes embedded in agarose plugs. Horizontal agarose gels were run in a Rothaphor R22 electrophoresis chamber (Biometra), with microprocessor-controlled cooling, and ramping of voltage, switch time, and field angle. DNA was transferred to nylon membrane by exposing the gel to 302 nm UV light for 45 s, denaturing in 0.5 M NaOH, 1.5 M NaCl, neutralizing in 3 M sodium acetate pH 5.5, followed by capillary transfer in 20× SSC buffer for 24 h, and hybridized under conditions as described (18).
Transfections.
An expression construct of the short rat KLRE1 was generated as follows: Kozak sequence and ORF of KLRE1 (excluding stop codon) was amplified by PCR using gene specific primers containing XhoI and SalI restriction sites, respectively (5′-CTCGAGCCGATCCTGGAATTTCCTCCTT-3′/5′-GTCGACCTTCTTGCAAATGTACATTAGT-3′). The PCR product was cloned into pCR2.1-TOPO (Invitrogen) and the XhoI/SalI fragment inserted into pECHA (a bicistronic, stable expression vector with SRα promotor upstream of a cloning site followed by a HA tag sequence and a stop codon, and finally a ribosomal reentry site (IRES) from EMCV followed by the Neo resistance gene [unpublished data]). The resulting ORF contained the full KLRE1 with the amino acid sequence VDYPYDVPDYA added at the COOH terminus. 293T cells were transfected with the KLRE1-HA expression construct using Lipofectamine (GIBCO BRL): 10 μg plasmid DNA was preincubated with 60 μl lipofectamine for 15 min at room temperature in 1.4 ml OPTI-MEM (GIBCO BRL), mixed with 5.6 ml OPTI-MEM, and added to 20 × 106 293T cells in a 75 cm2 flask. After incubation for 5–6 h at 37°C, 7 ml of RPMI 1640/20% FCS was added, and the cells were further cultured at 37°C for 19 h. The transfection mixture was poured off, and the cells detached with 0.5 mM EDTA in PBS. Successful transfection was verified by flow cytometry analysis for surface staining with anti-HA and -KLRE1 antibodies. To stably transfect RNKDA1 cells with the KLRE1-HA expression construct, 3 × 106 cells in 0.4 ml complete medium with 2-ME and IL-2 were mixed with 20 μg linearized plasmid DNA at 4°C in a 2 mm cuvette and electroporated at 120 V, 960 uF (Gene-Pulser; Bio-Rad Laboratories). Stably transfected cell clones were selected by culture in 2 mg/ml geneticin (G-418 disulphate; Duchefa) and tested for surface expression of KLRE1-HA by flow cytometry with WEN27 and anti-HA mAbs.
Biochemistry.
DA NK cell and RNKDA1 cell membrane proteins were prepared by lysing cells in 1% Triton X-114 as described (38). The membrane protein fractions were analyzed on SDS-PAGE, transferred to PVDF membrane (Immobilon P; Millipore) and probed with 20 μg/ml WEN27 mAb, 1 μg/ml HA.11 mAb (MMS-101R; Covance), or 1 μg/ml rabbit anti-mouse SHP-1 antiserum (C-19; Santa Cruz Biotechnology, Inc.) followed by goat anti–mouse IgG HRP or goat anti–rabbit IgG HRP (Jackson ImmunoResearch Laboratories) and chemiluminescent detection (Super Signal Substrate system; Pierce Chemical Co./BioMaxMR film; Eastman Kodak Co.). For immunoprecipitations, cells were lysed in 1% digitonin buffer (25 mM Tris-HCl, 150 mM NaCl, pH 7.5, 1% digitonin, 1 mM NaF, 1 mM PMSF, 1 mM Na3Vo4, 10 μg/ml leupeptin, and 10 μg/ml aprotinin) for 30 min at 4°C, then centrifuged 15 min at 16,000 g in a microcentrifuge. The supernatant was collected, and precleared for 1 h with 20 μl protein A/G beads (Santa Cruz Biotechnology, Inc.), then centrifuged for 2 min at 2,300 g. The supernatant was collected and added specific antibody (6 μg rabbit anti–mouse SHP-1 antiserum or 3 μg rabbit HA probe [C-19 and Y-11, respectively; Santa Cruz Biotechnology, Inc.]) and incubated o/n at 4°C. 30 μl protein A/G beads (Santa Cruz Biotechnology, Inc.) were added and precipitation performed for 2 h at 4°C. The beads were washed three times with 0.5% digitonin lysis buffer and centrifugation for 3 min at 2,300 g, resuspended in SDS sample buffer, and boiled for 2 min. Samples were run on SDS-PAGE, transferred to PVDF membranes, and analyzed as described above. For sodium pervanadate stimulation, 100 × 106 cells in 0.5 ml PBS were preincubated at 37°C for 5 min, then added 5 μl of a 100× sodium pervanadate solution to 0.01% H2O2, 100 μM Na3VO4, and incubated for 5 min at 37°C. The stimulation was stopped by adding ice cold 2 × 1% digitonin lysis buffer, and the cells were lysed for 30 min at 4°C as described above.
Generation of Anti-mKLRE1 mAb DX6, Anti-rKLRE1 mAb WEN27, and WEN27 F(ab′)2 Fragments.
Spleen cells from outbred Lewis rats immunized with NK cells isolated from C57BL/6 mice were fused with the mouse SP2/0 myeloma cell line by conventional techniques. The DX6 hybridoma was selected by flow cytometry for mAbs reactive with splenic NK cells from C57BL/6 mice. The anti-rKLRE1 mAb WEN27 was obtained by immunization of BALB/c female mice with a soluble KLRE1/hFcγ1 fusion protein. This fusion protein was produced in 293T cells transiently transfected (Lipofectamine, see above) with an expression construct consisting of the human CD8α signal peptide coupled in-frame to the hinge, CH2 and CH3 regions of human IgG1, again coupled in-frame at its COOH-terminal end to the extracellular region of rKLRE1. WEN27 F(ab′)2 fragments were produced by digestion of the WEN27 mAb with 0.1 mg/ml pepsin (P7012; Sigma-Aldrich) in 0.2 M Na acetate buffer pH 4.0 for 3 h at 37°C. Digested immunoglobulin was dialyzed into PBS pH 8.0 after pepsin digestion, and integrity of the F(ab′)2 fragments was confirmed by gel electrophoresis and flow cytometry.
Flow Cytometry.
For single-color analysis, 50 μl of cells (107 cells/ml) were incubated with 50 μl of Ab for 30 min on ice (anti-rKLRE1 mAb WEN27, anti-mKLRE1 mAb DX6, and anti-HA mAb HA.11). After three washes, labeled cells were incubated with F(ab′)2 of FITC conjugated sheep anti–mouse or goat anti–rat Ig (Sigma-Aldrich). For two-color analysis of rat cells from 2-mo-old individuals, mesenterial lymph nodes were disrupted in PBS and filtered (CellStrainer; Becton Dickinson). Spleen cells were obtained by density centrifugation (Lymphoprep; Nycomed) followed by passage through nylon wool (Leucopac; Fenwal Laboratories). Primary labeling was with WEN27, followed by FITC conjugated F(ab′)2 of sheep anti–mouse IgG (F-2266; Sigma-Aldrich), mouse IgG, and either PE-conjugated anti-CD3 (PE-G4.18; BD Biosciences), biotin-labeled anti-Ig kappa light chain (OX12 [39]), or biotin-labeled anti-NKR-P1 (3.2.3 [31]). Biotinylated mAbs were detected using PE-conjugated streptavidin (Jackson ImmunoResearch Laboratories). C57BL/6 spleen cells were analyzed using PE-anti-NK1.1, PE-DX5 (40), PE-anti-CD19, Cychrome-anti-CD3, biotin-labeled DX6, and PE-and biotin-labeled isotype control antibodies (BD Biosciences). Biotin-labeled antibodies were detected using Alexa-488 conjugated streptavidin (Molecular Probes). Stained cells were analyzed by flow cytometry (FACScan™; Becton Dickinson).
Redirected Lysis Assay.
Standard 4 h in vitro cytotoxicity assays were performed as described (41). Exponentially growing YAC-1, P388D1, and P815 cells were harvested and labeled for 1 h at 37°C with 100 μCi 51CrNa2O4. Labeled target cells were washed and resuspended at 105 cells/ml and 100 μl of this cell suspension was added to each assay well. IL-2–stimulated DA NK effector cells were harvested and resuspended in growth medium at densities indicated in the figure legends. 15-min preincubations with WEN27, WEN27 F(ab′)2, and control IgG were performed where indicated. 100 μl of effector cells were added to each assay well. Plates were centrifuged for 1 min at 130 g and incubated at 37°C for 4 h. The supernatant was harvested and counted in a gamma counter. The results are presented as median values from triplicates for each effector/target cell ratio. Spontaneous release of 51Cr was less than 10%.
Results
Cloning of Mouse and Rat KLRE1.
A rat monoclonal antibody toward mouse NK cells, named DX6, was obtained by immunizing rats with mouse NK cells, and selecting by flow cytometry for mAbs reactive with splenic NK cells from C57BL/6 mice. A mouse cDNA clone encoding the DX6 antigen was isolated by expression cloning, using COS-7 cells transfected with a cDNA expression library prepared from IL-2–stimulated polyclonal mouse NK cells derived from CB.17 SCID mice. The transfected cells were stained with DX6 and the antigen positive cells isolated by cell sorting. After three rounds of selection, individual plasmids were isolated and analyzed for DX6 antigen expression after transfection. A single plasmid was selected for sequence analysis, and contained a cDNA insert of 1324 nucleotides, with an ORF predicting a polypeptide of 226 amino acids, with a relative molecular mass of 26.2 kD and two potential sites for N-linked glycosylation (GenBank/EMBL/DDBJ accession no. AY100458).
Using the ORF fragment of the mouse cDNA clone to screen an F344 rat NK cell cDNA library, a rat cDNA clone of 940 nucleotides was isolated, containing an ORF of 202 amino acids, 82.2/88.3% identical to the mouse clone at the amino acid/nucleotide levels (GenBank/EMBL/DDBJ accession no. AF86187). Five additional rat clones were completely or partially sequenced, all apparently identical. No additional clones were obtained by rehybridizing the filters with a rat probe. The predicted mouse and rat polypeptides are type II integral membrane proteins with a COOH-terminal C-type lectin-like domain (Fig. 1 A). Sequence comparisons showed that they belong to the KLRs, but constitute the first members of a separate, novel family (family E) and have thus been termed mouse and rat KLRE1 (Fig. 1 B).
Figure 1.
(A) Alignment of the amino acid sequences of rat and mouse KLRE1. Identical amino acids are indicated by dashes, and the transmembrane region of the KLRE1 polypeptide has been underlined. Cysteines forming intra-chain disulphide bridges are marked with asterisks, and a free cysteine possibly involved in interchain bonding is marked with a diamond. No ITIM motif is present in the cytoplasmic tails, and no charged amino acid in the transmembrane domains. The five intron sites within the coding region are marked with dots (with phases 1, 1, 1, 0, and 2, in that order). (B) Dendrogram displaying total amino acid identity among members of the different KLR families yet identified. Rat and mouse KLRE1 constitute a separate branch of the KLRs, most closely related to CD94. The GenBank/EMBL/DDBJ accession nos. of the analyzed sequences are: M25812, U10304 (mLy-49A and B, respectively), U56863, U56822, (rLy-49.9 and 0.12, respectively), AF416564 (rKLRH1), AF350409, AF350410 (mCLR-B and -F, respectively), AF321552 (rOCIL), X96719 (hAICL), L23638 (mCD69), NM010652, NM010653 (mNKG2A and C, respectively), AF021350, AF021349 (rNKG2A and C, respectively), AF030313 (mNKG2D), AF009511 (rNKR-P2), AY100458 (mKLRE1), AF486186 (rKLRE1), AF030311 (mCD94), AF009133 (rCD94), AF200949 (hCLEC1), AF201457 (mCLEC2), AJ010751, X79812 (mouse and rat MAFA, respectively), M62891, U56936 (rNKR-P1A and B, respectively), M77676, M77677 (mNKR-P1A and B, respectively), AF139769 (mMDL1), and NM016523 (hKLRF1).
Compared with mKLRE1, the cloned rKLRE1 cDNA lacked 70 nucleotides, including the mKLRE1 start codon, predicting a protein with a shorter cytoplasmic domain. Genomic sequencing of rat liver DNA revealed an intron at position –24 relative to the rKLRE1 start codon. The 3′-terminal part of the intron showed high sequence similarity with the missing 70 nucleotides and with a consensus 3′ splice site immediately upstream, suggesting the possibility of alternative splicing. A new set of clones was obtained by RT-PCR of mRNA from IL-2 stimulated DA NK cells with a 3′-UTR–specific reverse primer and a 5′-UTR–specific forward primer spanning the expected alternative splice site. Three individual clones were sequenced. The 760 nt consensus sequence contained an ORF of 226 amino acids, similar in length to mKLRE1, showing 80.5/87.8% identity at the amino acid/nucleotide levels (GenBank/EMBL/DDBJ accession no. AF486186; Fig. 1 A). The presence of a short KLRE1 variant in DA NK cells, identical to the F344 KLRE1, was confirmed by cloning using a 5′ forward primer specific for the short rKLRE1, followed by sequence analysis of three individual RT-PCR clones (GenBank/EMBL/DDBJ accession no. AF86185).
Southern blot analysis of rat genomic DNA digested with different restriction enzymes showed a single band when hybridized to a probe derived from the middle exon of the lectin-like domain, indicating a single gene without close relatives (unpublished data). The short and long rKLRE1 sequences are therefore not products of separate genes, but result from alternative splicing of a 5′UTR intron, where a single donor site is spliced to two different acceptor sites. In the rat, the two splice sites are each followed by in-frame start codons. In the mouse, however, only the first of the corresponding two codons was ATG, whereas the second was ATT (isoleucine). Searching the NCBI rat Trace archive using the rat KLRE1 sequence yielded 19 genomic trace sequences covering all six translated exons. The deduced intron sites and phases were typical for KLR genes (Fig. 1 A). rKLRE1 has two putative sites for N-linked glycosylation, and the unglycosylated forms have a predicted relative molecular mass of 23.6 (short) and 26.2 (long) kD, respectively. The cytoplasmic domains of both mouse and rat KLRE1 lack ITIMs, and the transmembrane domains do not contain charged amino acids (Fig. 1 A).
The Klre1 Locus Maps to the NK Cell Gene Complex.
Southern blot analysis of rat and mouse genomic DNA was consistent with a single KLRE1 gene. Southern blot of liver DNA from PVG and DA rats digested with BstXI, gave different restriction patterns when hybridized to a full-length KLRE1 probe. This polymorphism was used to analyze a panel of 223 DA × (PVG × DA)F1 rats previously involved in mapping of several other KLR genes. The Cd94, Nkrp2, and Nkg2 genes have previously been mapped to the middle part of the NKC, flanked proximally by the Nkrp1 and distally by the Ly-49 gene clusters (18, 24–25, 42). Klre1 was found to cosegregate with Nkrp1b, Cd94, Nkrp2, Nkg2, Klrh1 (43) and proximal Ly49 genes in all 223 rats, thus mapping the Klre1 locus to the NKC on rat chromosome 4. For more precise mapping, genomic PFGE analysis was performed. The full-length KLRE1 probe hybridized to a Pme I fragment of ∼115 kb containing the members of the Nkg2 gene family, the Nkrp2 and Cd94 loci. The mapping was confirmed by data recently released by the Rat Genome Sequencing Consortium (www.ncbi.nlm.nih.gov/genome/guide/rat) where the rat KLRE1 sequence was positioned immediately proximal to rat CD94 (distance ∼12 kb). Analysis of the genomic sequence released by the Mouse Genome Sequencing Consortium (www.ncbi.nlm.nih.gov/genome/guide/mouse), showed conservation of localization, orientation, and distance to CD94, with the mKLRE1 sequence located ∼15 kb proximal to mouse CD94 in the middle part of the mouse NKC on chromosome 6.
KLRE1 Is Expressed by NK Cells and a Subset of CD3+ Cells.
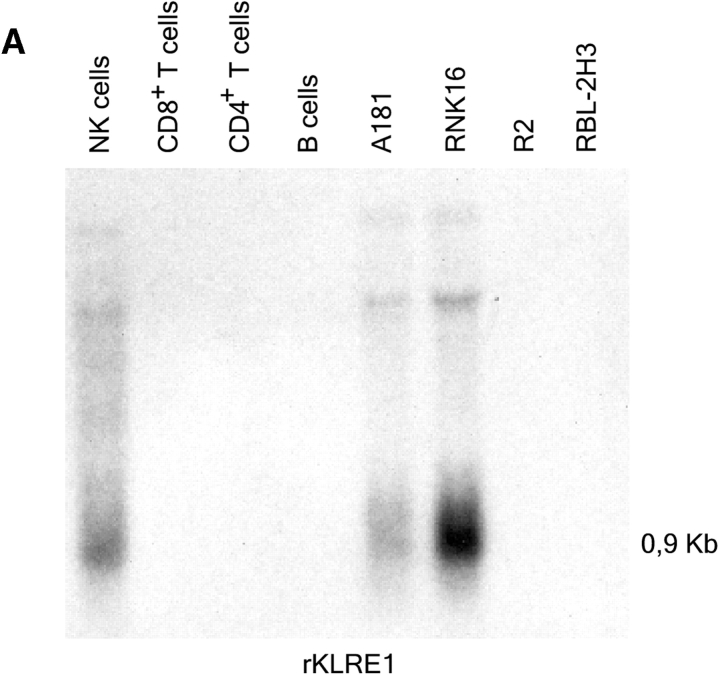
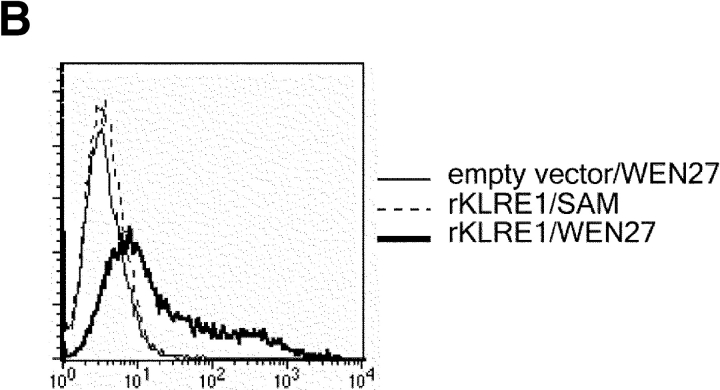
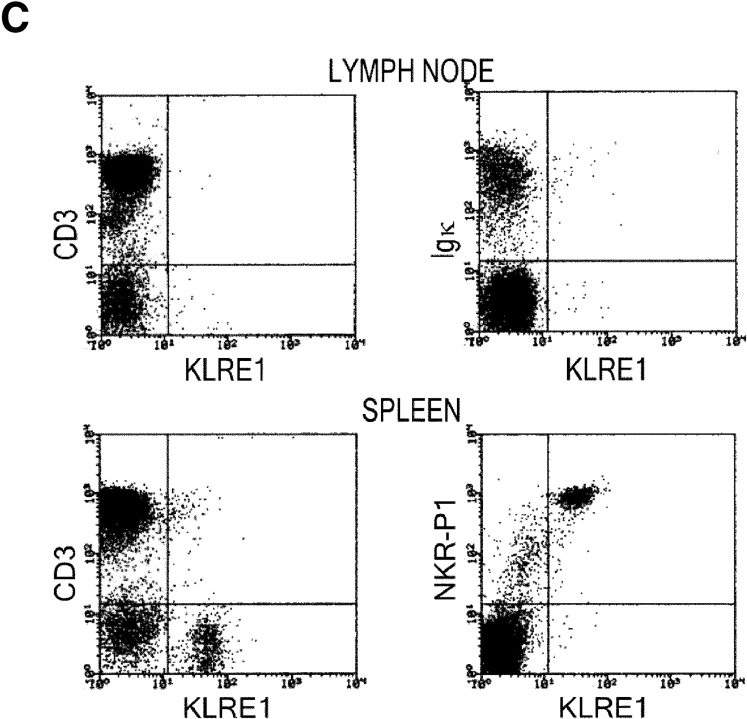
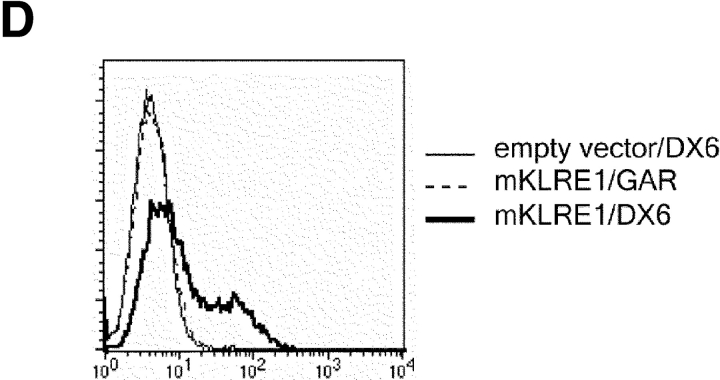
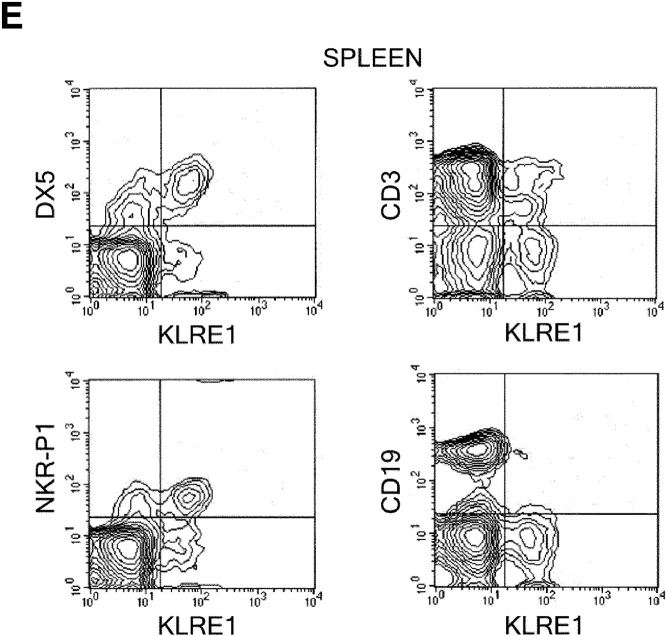
In the rat, Northern blot analysis using the full-length rat KLRE1 probe showed KLRE1 transcripts in IL-2–activated DA NK cells and the NK cell lines RNK16 and A181, but not in T cells, B cells, the macrophage cell line R2 or the mast cell line RBL-2H3 (Fig. 2 A). To investigate surface level expression, a monoclonal antibody toward rKLRE1, termed WEN27 (Fig. 2 B) was generated by immunizing mice with a soluble fusion protein of the extracellular region of rKLRE1 coupled to the Fc region of human IgG1. Flow cytometry with WEN27 showed that all NKR-P1bright spleen cells in DA rats expressed KLRE1. A small population (∼1.5%) of the CD3+ cells from spleen showed weak KLRE1 expression, whereas all CD3+ cells from lymph node were KLRE1−. B cells from lymph node or spleen (Ig kappa light chain positive) did not express KLRE1 (Fig. 2 C). Similarly, flow cytometry analysis of mouse spleen cells with the anti-KLRE1 antibody DX6 showed that mKLRE1 was expressed by most NK cells (stained by either DX5 or NK1.1), and in addition by a subset of the CD3+ cells (Fig. 2, D and E). The fraction of NK cells that was stained with KLRE1 was strain dependent (see below).
Figure 2.
KLRE1 is expressed by NK cells and a subset of T cells. (A) Northern blot analysis of total RNA from different cell types, hybridized to a full-length KLRE1 probe. A181 and RNK16 are rat NK cell lines, R2 is a rat macrophage line, and RBL-2H3 is a rat mast cell line. (B) The newly generated mAb WEN27 specifically recognizes rKLRE1, demonstrated by flow cytometry analysis of 293T cells transfected with empty vector or with a rKLRE1 expression construct. Staining of transfected cells with secondary antibody (sheep anti–mouse) alone is shown for comparison. (C) Flow cytometry analysis of double stained lymph node cells and spleen cells from DA rats with anti-KLRE1 mAb WEN27, and mAbs specific for rat CD3, Ig kappa light chain and NKR-P1. (D) The DX6 mAb specifically recognizes mKLRE1. Surface expression of a mKLRE1 expression construct in transfected 293T cells. Thin line: empty vector/DX6, dotted line: mKLRE1/goat anti–rat-FITC, thick line: mKLRE1/DX6. (E) Flow cytometry analysis of mouse spleen cells with the anti-mKLRE1 monoclonal antibody DX6. C57BL/6 spleen (adult) was stained with PE-anti-NKR-P1, PE-DX5, PE-anti-CD19, Cychrome-anti-CD3, biotin-labeled DX6, and PE-and biotin-labeled isotype control antibodies.
KLRE1 Expression Varies Between Strains of Mice and Rats.
In the mouse, KLRE1 expression levels were tested by triple staining flow cytometry on DX5 positive cells from 24 different inbred strains. NK cells from all the strains expressed KLRE1, although mostly at low levels (KLRE-1dim cells; Table I). Only five of the strains, SWR/J (H-2a), MA/MyJ, C58/J, and C3H/HEN (H-2k) and SJL/J (H-2s) harbored KLRE1bright cells. In seven of the strains tested only fractions of the NK cells were stained by DX6. In the remaining strains, all NK cells were KLRE1+ (Table I). All the strains tested bearing the H-2b and H-2d haplotypes were KLRE1dim. However, NK cells from only one of three strains with the H-2a haplotype and three of eleven with the H-2k haplotypes exhibited strong KLRE1 expression (Table I). The data therefore do not provide sufficient evidence to suggest MHC-induced down-regulation of KLRE1 expression in H-2b and H-2d haplotypes.
Table I.
KLRE1 Expression in Inbred Mouse Strains
| H-2 haplotypes
|
|||||
|---|---|---|---|---|---|
| KLRE1a | a | b | d | k | z/s/v |
| Bright | Ma/MyJ 100 | ||||
| C58/J 50 | |||||
| SWR/J 75 b | C3H/HEN 75 | (s) SJL/J 50 | |||
| Dim | CE/J 75 | ||||
| C57BR/cdJ 100 | |||||
| ST/bj 100 | |||||
| AKR/J 50 | |||||
| C57BL/6J 100 | C3H/HeJ 100 | ||||
| C57BL/10J 100 | NZB/BINJ 25 | C3HeB/FeJ 100 | |||
| A/J 100 | C57L/J 100 | BALB/c 100 | CBA/CaJ 100 | (z) NZW/LacJ 100 | |
| DBA/1J 100 | LP/J 100 | DBA-2J 100 | CBA/J 100 | (v) SM/J 100 | |
The table shows flow cytometric analyses of KLRE1 expression levels in DX5-positive spleen cells from 24 different inbred mouse strains, grouped according to H-2 haplotypes.
KLRE1 staining intensity.
Percentage of KLRE1-positive NK.
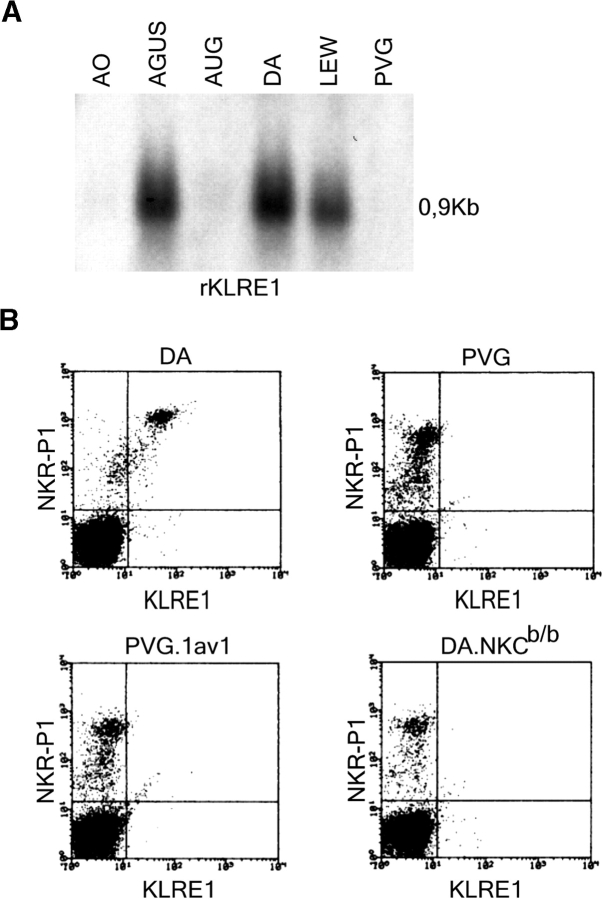
Strain-dependent KLRE1 expression was also demonstrated in the rat, where Northern blot analysis of IL-2 activated NK cells demonstrated markedly higher levels of transcription in NK cells from AGUS, LEW and DA rats than from PVG, AO and AUG rats (Fig. 3 A). Flow cytometry analysis of KLRE1 surface expression on NK cells stimulated with IL-2 for 14 d showed bright staining of all DA NK cells, but hardly detectable staining of PVG NK cells. KLRE1 was not expressed on NK cells from the MHC congenic rat strain PVG.1AV1, or on NK cells from a congenic DA rat strain carrying the NKC of the PVG haplotype (DA.NKCb/b), indicating that the difference in expression levels between these two particular strains is not controlled by the MHC, but by factors intrinsic to the NKC (Fig. 3 B). By Southern blot analysis, the PVG allele of Klre1 did not appear to contain large deletions (unpublished data). The difference in expression between these two strains applied both to freshly isolated and IL-2 stimulated NK cells (unpublished data).
Figure 3.
KLRE1 expression is strain dependent. (A) Northern blot analysis of total RNA from IL-2 activated NK cells from different inbred rat strains, hybridized to a full length KLRE1 probe. (B) Flow cytometry analysis of double stained spleen cells from the DA, PVG, PVG.1AV1, and DA.NKCb/b rat strains.
KLRE1 Can Be Expressed As a Disulphide-linked Dimer.
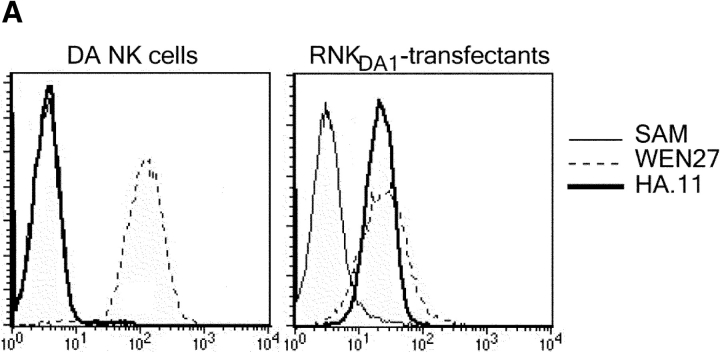
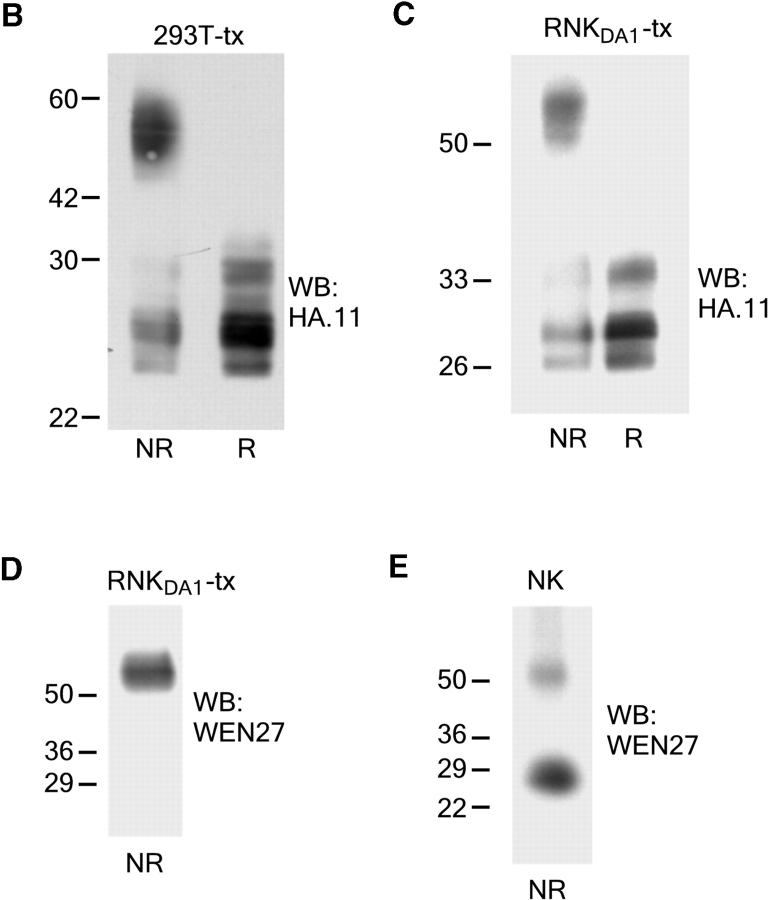
A rat NK cell line termed RNKDA1 was generated by isolation of NK cells from the spleen of DA rats. The NK cells were kept growing in media containing IL-2 and shown by flow cytometry to be NKR-P1+/CD3−. The RNKDA1 cell line was stably transfected with a rKLRE1-HA tag expression construct (Fig. 4 A) and Western blot analyses performed on cell lysates of these cells as well as of 293T cells transiently transfected with the same expression construct. Using anti-HA antibodies or the WEN27 mAb a ∼54 kD band was found under nonreducing conditions and a ∼27 kD band under reducing conditions, demonstrating that after transfection KLRE1 formed disulphide linked dimers (Fig. 4, B–D). Unexpectedly, in IL-2–stimulated DA NK cells and in the nontransfected RNKDA1 cell line (unpublished data), the 27 kD band, probably corresponding to a monomeric form of KLRE1, was more prominent than the ∼54 kD band under nonreducing conditions (Fig. 4 E).
Figure 4.
KLRE1 can be expressed as a dimer in the NK cell membrane. The IL-2–dependent RNKDA1 cell line was stably transfected with a KLRE1-HA expression construct. (A) RNKDA1–tx cells express KLRE1-HA in the cell surface as demonstrated by flow cytometry. Untransfected cells are shown for comparison. (B) Whole cell lysate of 293T cells transiently transfected with the KLRE1-HA expression construct (293T-tx) was separated by SDS-PAGE under nonreducing (NR) and reducing (R) conditions and analyzed by Western blotting using anti-HA mAb. (C) The RNKDA1 NK cell line was stably transfected with a KLRE1-HA expression construct (RNKDA1-tx), lysed in Triton X-114 buffer, and the membrane protein fraction was separated by SDS-PAGE, blotted, and probed with anti-HA mAb or (D) the anti-KLRE1 mAb WEN27. (E) Triton X-114 lysate of IL-2 activated DA NK cells was separated by SDS-PAGE and analyzed by Western blotting using the WEN27 mAb.
KLRE1 Inhibits NK Cell Cytotoxicity.
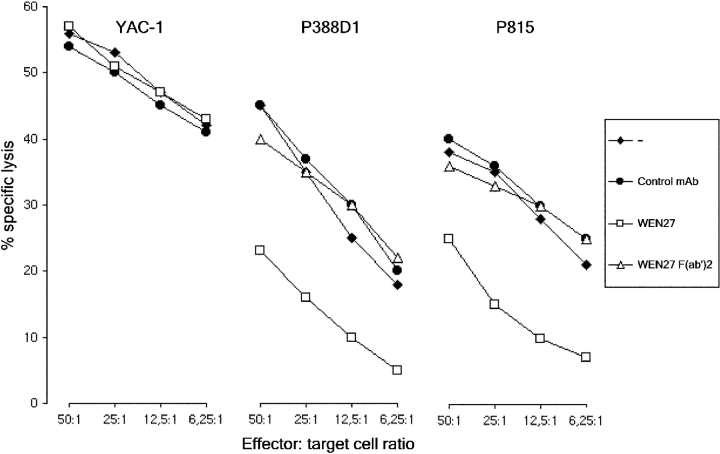
The functional activity of KLRE1 was assessed by a redirected lysis assay, using IL-2–stimulated DA NK cells as effector cells and the Fc-receptor bearing (FcR+) tumor cell-lines P388D1 and P815 and the FcR− line YAC-1 as targets. In the absence of WEN27 antibody, all target cell types were efficiently lysed. Killing of the FcR+ targets was inhibited when the effector cells had been preincubated with WEN27 antibody (whole IgG molecules). Incubation with WEN27 F(ab′)2 fragments did not affect killing, demonstrating that the inhibition observed with intact mAb was not a result of blocking of an activating receptor. Treatment with WEN27 mAb did not influence lysis of YAC-1 cells. Pre-treating the effector cells with an isotype matched mAb had no effect on the level of cytotoxicity against P815 and P388D1, whereas pretreatment with an antibody against the activating receptor NKp46 augmented killing (Berg, S.F., I.H. Westgaard, personal communication). The results show that cross-linking of KLRE1 inhibits NK cell mediated cytotoxicity (Fig. 5) .
Figure 5.
KLRE1 inhibits NK cytotoxicity. 4 h chromium release redirected lysis assay with IL-2–activated DA NK cells as effector cells. Labeled target cells are denoted above each graph, and effector:target cell ratio denoted below the graphs. The effector cells were preincubated with control mAb, WEN27 whole Ig or WEN27 F(ab′)2 as indicated.
KLRE1 in Dimeric Form Associates with SHP-1.
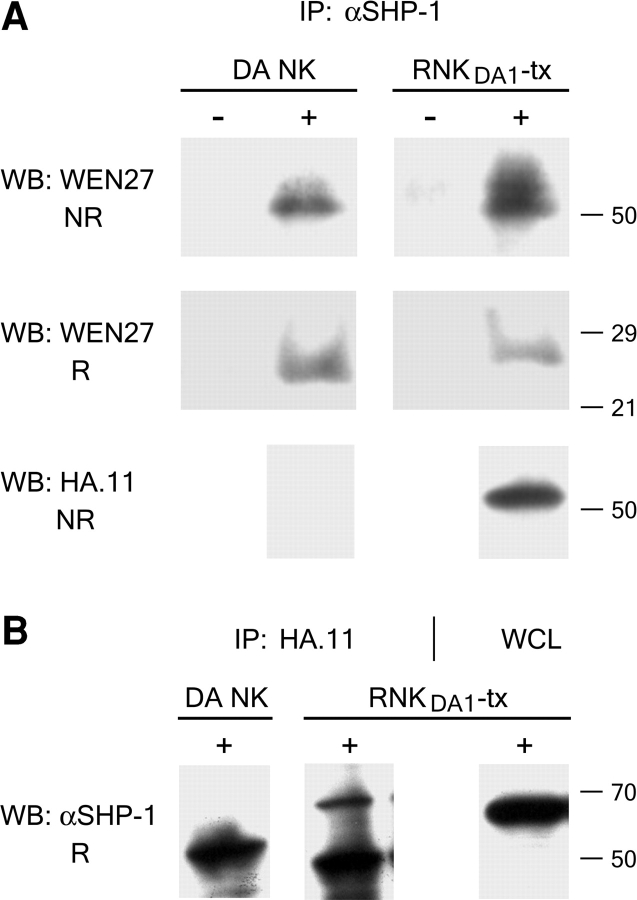
As a rule, the inhibitory NK receptors transmit their signals via protein tyrosine phosphatases, in particular SHP-1, docking to the phosphorylated ITIMs after receptor engagement (11–16, 21). To examine whether SHP-1 plays a role in signal transduction via KLRE1, cells from the KLRE1-HA transfected, IL-2–dependent DA NK cell line (RNKDA1-tx) were pretreated with pervanadate, and digitonin lysates of the cells were precipitated with anti-HA or anti-SHP-1 antibodies. By Western blotting, anti-SHP-1 was found to precipitate KLRE1 and vice versa (Fig. 6, A and B) . Whereas the most prominent band detected on Western blots from nontransfected DA NK cells was ∼27 kD, the major anti-KLRE1 immunoreactivity following immunoprecipitation with anti-SHP-1 was found at a ∼54 kD band (Fig. 6 A), indicating covalent dimerization with a partner of ∼27 kD. To investigate whether KLRE1 interacts with SHP-1 directly, 293T cells were cotransfected with KLRE1-HA and rat SHP-1 expression constructs. After pervanadate stimulation, whole cell lysates were immunoprecipitated with anti–SHP-1 or with anti-HA antibodies. In this situation, KLRE1-HA could not be coprecipitated with SHP-1, or vice versa (unpublished data), suggesting that the SHP-1 binding observed with the dimeric form of KLRE1 is mediated by an as yet unidentified partner chain expressed in IL-2–activated NK cells, but lacking in 293T cells.
Figure 6.
KLRE1 is associated with SHP-1. Freshly isolated NK cells from DA rats (DA NK) and RNKDA1 cells transfected with a KLRE1-HA expression construct (RNKDA1-tx) were analyzed by immunoprecipitation with anti-SHP-1 (αSHP-1) (A) or anti-HA antibody (HA.11) (B). (A) Pervanadate stimulated (5 min at 37°C) (+) and unstimulated (−) cells were lysed in 1% digitonin buffer and immunoprecipitated with αSHP-1. The immune complexes were separated by SDS-PAGE under nonreducing (NR) and reducing (R) conditions respectively, blotted onto a membrane, and probed with WEN27 or anti-HA antiserum. Equal amounts of immunoprecipitated SHP-1 were verified by Western blotting using anti–SHP-1 antibodies (unpublished data). (B) Western blots of HA.11 immunoprecipitates from pervanadate stimulated cells probed with αSHP-1. The right lane with whole cell lysate (WCL) shows the position of SHP-1 (∼70 kD). The ∼50 kD band in the HA.11 immunoprecipitate lanes represents the Ig heavy chain of the anti-HA antibodies (SDS-PAGE performed under reducing conditions).
Discussion
The sequence similarities, selective transcription by NK cells, genetic localization to the NKC, and a simple pattern on genomic Southern blots constitute evidence that the cloned cDNA molecules belong to a novel single-gene KLR family, and represent orthologs in the mouse and the rat. Attempts to clone a human ortholog by low stringency hybridization to a human NK cell cDNA library failed, and searching the genome, EST and htgs databases revealed no human counterpart. Two splice variants differing in position of the cytoplasmic, NH2-terminal start codon were found in the rat. In the mouse, the short splice variant was not detected, probably reflecting a T→G transversion at position –10 in the upstream pyrimidine tract of the putative second splice site, consistent with the lack of an in-frame ATG following this site.
As the protein lacked cytoplasmic ITIMs as well as a charged amino acid in the TM domain, the deduced amino acid sequence gave no indication as to whether KLRE1 would transmit activating or inhibitory signals. Two observations indicate an inhibitory role. First, in the redirected lysis assay, adding whole Ig WEN27 mAb to the culture medium inhibited lysis of NK sensitive Fc receptor positive tumor target cells, whereas addition of WEN27 F(ab′)2 fragments did not. Second, SHP-1 could be precipitated with KLRE1 from lysates of pervanadate treated DA NK cells. So far SHP-1 is only known to associate with KLRs via the binding of its tandem SH2 domains to phosphorylated ITIMs (11–16, 21). Although mouse and rat KLRE1 lack ITIMs, the KLRE1 polypeptide contains a tyrosine residue in the intracellular region that could be subject to phosphorylation, and might form part of an as yet unidentified motif involved in binding SHP-1. However, KLRE1 could not be coimmunoprecipitated with SHP-1 from lysates of pervanadate treated 293T cells cotransfected with rat SHP-1 and KLRE1.
At the start of the lectin domain most KLR molecules contain two conserved cysteines that have been shown to form a disulphide bridge, creating a loop that probably functions to stabilize the base of the lectin domain (44–47). These cysteines are also present in KLRE1 (Cys113 and Cys 124 in the mouse and the long rat splice variant), probably contributing to loop-formation. Within this presumed loop both rat and mouse KLRE1 contain an extra cysteine (Cys 112, marked with a diamond in Fig. 1 A). As there are no other membrane external cysteines that could participate in intrachain disulphide bonding, Cys112 represents a candidate for inter-chain disulphide bonding. The fact that KLRE1 can form covalently linked dimers was shown by transient transfection of 293T cells and by stable transfection of a DA NK cell line (RNKDA1) with the KLRE1-HA expression construct. In both systems Western blot analysis performed under nonreducing conditions showed a band corresponding to ∼54 kD, using anti-HA antibodies, WEN27, or a polyclonal antipeptide antiserum. We would therefore expect that native KLRE1 also formed homo- or heterodimers in untransfected cells, as is the case for the other KLRs investigated. Surprisingly, Western blot analysis of nontransfected DA NK cells showed strong staining of a ∼27 kD band, both under nonreducing and reducing conditions, plus a weaker band corresponding to the expected dimer under nonreducing conditions. The nature of the 27 kD band has not been investigated, but may represent a monomeric or noncovalently linked heterodimeric form of KLRE1, not necessarily expressed on the cell surface. Nevertheless, when KLRE1 was coimmunoprecipitated with antibody against SHP-1 from digitonin lysates of pervanadate-treated DA NK cells, a strong ∼54 kD KLRE1 band was obtained, suggesting that the dimeric form of KLRE1 is the functionally active, inhibitory form of the receptor. An analogous type of receptor is the CD94/NKG2A heterodimer, that recruits SHP-1 through two tandem ITIM motifs in the cytoplasmic region of NKG2A, whereas the CD94 chain appears to lack a direct signaling function (19–21). Sequence comparison showed that KLRE1 is more closely related to CD94 than to other KLR families. After cotransfection of 293T cells with rKLRE1-HA and rNKG2A-FLAG expression constructs, the two chains could not be coprecipitated with either anti-HA or anti-FLAG antibodies (unpublished data). Together, the presented data indicate that KLRE1 may form a functional heterodimer with an as yet unidentified ITIM-bearing partner that recruits SHP-1 to generate an inhibitory receptor complex.
These studies indicate the existence of a novel NKC-encoded C-type lectin-like NK cell receptor in rodents that may inhibit immune function. Efforts to identify the putative heterodimeric partner chain are currently proceeding.
Acknowledgments
We thank W. Jensen, M. Lauritzen, and J. Norrström for technical assistance, T. Vang for advice, and J.C. Ryan, V. Kumar, A.N. Barclay, T. Hünig, J.G.M.C. Damoiseaux, and J.C. Hiserodt for sharing reagents.
This work was supported by the Norwegian Research Council, the Norwegian Cancer Society, Odd Fellow Medical Research Fund, and Bergljot and Sigurd Skaugen's Fund (I.H. Westgaard, E. Dissen, K.M. Torgersen, and S. Fossum); and by National Institutes of Health grant CA89294 (L.L. Lanier). L.L. Lanier is an American Cancer Society Research Professor.
Note added in proof: After the resubmittance of this manuscript the mouse KLRE1 cDNA sequence was described. Reference: Wilhelm, B.T., and D.L. Mager. 2003. Identification of a new murine lectin-like gene in close proximity to CD94. Immunogenetics. 55:53–56.
Footnotes
Abbreviations used in this paper: ITIM, immunoreceptor tyrosine-based inhibitory motif; KLR, killer cell lectin-like receptor; NKC, NK cell gene complex; ORF, open reading frame; PFGE, pulsed field gel electrophoresis; TM, transmembrane.
References
- 1.Biron, C.A., K.B. Nguyen, G.C. Pien, L.P. Cousens, and T.P. Salazar-Mather. 1999. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu. Rev. Immunol. 17:189–220. [DOI] [PubMed] [Google Scholar]
- 2.Cerwenka, A., and L.L. Lanier. 2001. Natural killer cells, viruses and cancer. Nat. Rev. Immunol. 1:41–49. [DOI] [PubMed] [Google Scholar]
- 3.Rolstad, B., J.T. Vaage, C. Naper, D. Lambracht, K. Wonigeit, E. Joly, and G.W. Butcher. 1997. Positive and negative MHC class I recognition by rat NK cells. Immunol. Rev. 155:91–104. [DOI] [PubMed] [Google Scholar]
- 4.Lanier, L.L. 1998. NK cell receptors. Annu. Rev. Immunol. 16:359–393. [DOI] [PubMed] [Google Scholar]
- 5.Diefenbach, A., and D.H. Raulet. 2001. Strategies for target cell recognition by natural killer cells. Immunol. Rev. 181:170–184. [DOI] [PubMed] [Google Scholar]
- 6.Karlhofer, F.M., R.K. Ribaudo, and W.M. Yokoyama. 1992. MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature. 358:66–70. [DOI] [PubMed] [Google Scholar]
- 7.Arase, H., E.S. Mocarski, A.E. Campbell, A.B. Hill, and L.L. Lanier. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 296:1323–1326. [DOI] [PubMed] [Google Scholar]
- 8.Moretta, A., S. Sivori, M. Vitale, D. Pende, L. Morelli, R. Augugliaro, C. Bottino, and L. Moretta. 1995. Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J. Exp. Med. 182:875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mason, L.H., S.K. Anderson, W.M. Yokoyama, H.R. Smith, R. Winkler-Pickett, and J.R. Ortaldo. 1996. The Ly-49D receptor activates murine natural killer cells. J. Exp. Med. 184:2119–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanier, L.L., B. Corliss, J. Wu, and J.H. Phillips. 1998. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 8:693–701. [DOI] [PubMed] [Google Scholar]
- 11.Bruhns, P., P. Marchetti, W.H. Fridman, E. Vivier, and M. Daeron. 1999. Differential roles of N- and C-terminal immunoreceptor tyrosine-based inhibition motifs during inhibition of cell activation by killer cell inhibitory receptors. J. Immunol. 162:3168–3175. [PubMed] [Google Scholar]
- 12.Burshtyn, D.N., A.M. Scharenberg, N. Wagtmann, S. Rajagopalan, K. Berrada, T. Yi, J.P. Kinet, and E.O. Long. 1996. Recruitment of tyrosine phosphatase HCP by the killer cell inhibitor receptor. Immunity. 4:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olcese, L., P. Lang, F. Vely, A. Cambiaggi, D. Marguet, M. Blery, K.L. Hippen, R. Biassoni, A. Moretta, L. Moretta, et al. 1996. Human and mouse killer-cell inhibitory receptors recruit PTP1C and PTP1D protein tyrosine phosphatases. J. Immunol. 156:4531–4534. [PubMed] [Google Scholar]
- 14.Fry, A.M., L.L. Lanier, and A. Weiss. 1996. Phosphotyrosines in the killer cell inhibitory receptor motif of NKB1 are required for negative signaling and for association with protein tyrosine phosphatase 1C. J. Exp. Med. 184:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell, K.S., M. Dessing, M. Lopez-Botet, M. Cella, and M. Colonna. 1996. Tyrosine phosphorylation of a human killer inhibitory receptor recruits protein tyrosine phosphatase 1C. J. Exp. Med. 184:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura, M.C., E.C. Niemi, M.J. Fisher, L.D. Shultz, W.E. Seaman, and J.C. Ryan. 1997. Mouse Ly-49A interrupts early signaling events in natural killer cell cytotoxicity and functionally associates with the SHP-1 tyrosine phosphatase. J. Exp. Med. 185:673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown, M.G., S. Fulmek, K. Matsumoto, R. Cho, P.A. Lyons, E.R. Levy, A.A. Scalzo, and W.M. Yokoyama. 1997. A 2-Mb YAC contig and physical map of the natural killer gene complex on mouse chromosome 6. Genomics. 42:16–25. [DOI] [PubMed] [Google Scholar]
- 18.Dissen, E., J.C. Ryan, W.E. Seaman, and S. Fossum. 1996. An autosomal dominant locus, Nka, mapping to the Ly-49 region of a rat natural killer (NK) gene complex, controls NK cell lysis of allogeneic lymphocytes. J. Exp. Med. 183:2197–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renedo, M., I. Arce, K. Montgomery, P. Roda-Navarro, E. Lee, R. Kucherlapati, and E. Fernandez-Ruiz. 2000. A sequence-ready physical map of the region containing the human natural killer gene complex on chromosome 12p12.3-p13.2. Genomics. 65:129–136. [DOI] [PubMed] [Google Scholar]
- 20.Lazetic, S., C. Chang, J.P. Houchins, L.L. Lanier, and J.H. Phillips. 1996. Human natural killer cell receptors involved in MHC class I recognition are disulfide-linked heterodimers of CD94 and NKG2 subunits. J. Immunol. 157:4741–4745. [PubMed] [Google Scholar]
- 21.Le Drean, E., F. Vely, L. Olcese, A. Cambiaggi, S. Guia, G. Krystal, N. Gervois, A. Moretta, F. Jotereau, and E. Vivier. 1998. Inhibition of antigen-induced T cell response and antibody-induced NK cell cytotoxicity by NKG2A: association of NKG2A with SHP-1 and SHP-2 protein-tyrosine phosphatases. Eur. J. Immunol. 28:264–276. [DOI] [PubMed] [Google Scholar]
- 22.Carretero, M., G. Palmieri, M. Llano, V. Tullio, A. Santoni, D.E. Geraghty, and M. Lopez-Botet. 1998. Specific engagement of the CD94/NKG2-A killer inhibitory receptor by the HLA-E class Ib molecule induces SHP-1 phosphatase recruitment to tyrosine-phosphorylated NKG2-A: evidence for receptor function in heterologous transfectants. Eur. J. Immunol. 28:1280–1291. [DOI] [PubMed] [Google Scholar]
- 23.Chang, C., and L.L. Lanier. 2000. Use of cDNA library expression cloning to identify components of heterodimeric receptor complexes. Methods Mol. Biol. 121:273–281. [DOI] [PubMed] [Google Scholar]
- 24.Dissen, E., S.F. Berg, I.H. Westgaard, and S. Fossum. 1997. Molecular characterization of a gene in the rat homologous to human CD94. Eur. J. Immunol. 27:2080–2086. [DOI] [PubMed] [Google Scholar]
- 25.Berg, S.F., E. Dissen, I.H. Westgaard, and S. Fossum. 1998. Molecular characterization of rat NKR-P2, a lectin-like receptor expressed by NK cells and resting T cells. Int. Immunol. 10:379–385. [DOI] [PubMed] [Google Scholar]
- 26.Nicolls, M.R., G.G. Aversa, N.W. Pearce, A. Spinelli, M.F. Berger, K.E. Gurley, and B.M. Hall. 1993. Induction of long-term specific tolerance to allografts in rats by therapy with an anti-CD3-like monoclonal antibody. Transplantation. 55:459–468. [DOI] [PubMed] [Google Scholar]
- 27.Hünig, T., H.J. Wallny, J.K. Hartley, A. Lawetzky, and G. Tiefenthaler. 1989. A monoclonal antibody to a constant determinant of the rat T cell antigen receptor that induces T cell activation. Differential reactivity with subsets of immature and mature T lymphocytes. J. Exp. Med. 169:73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dallman, M.J., M.L. Thomas, and J.R. Green. 1984. MRC OX-19: a monoclonal antibody that labels rat T lymphocytes and augments in vitro proliferative responses. Eur. J. Immunol. 14:260–267. [DOI] [PubMed] [Google Scholar]
- 29.McMaster, W.R., and A.F. Williams. 1979. Identification of Ia glycoproteins in rat thymus and purification from rat spleen. Eur. J. Immunol. 9:426–433. [DOI] [PubMed] [Google Scholar]
- 30.McKnight, A.J., and B.J. Classon. 1992. Biochemical and immunological properties of rat recombinant interleukin-2 and interleukin-4. Immunology. 75:286–292. [PMC free article] [PubMed] [Google Scholar]
- 31.Chambers, W.H., N.L. Vujanovic, A.B. DeLeo, M.W. Olszowy, R.B. Herberman, and J.C. Hiserodt. 1989. Monoclonal antibody to a triggering structure expressed on rat natural killer cells and adherent lymphokine-activated killer cells. J. Exp. Med. 169:1373–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams, A.F., G. Galfrè, and C. Milstein. 1977. Analysis of cell surfaces by xenogeneic myeloma-hybrid antibodies: differentiation antigens of rat lymphocytes. Cell. 12:663–673. [DOI] [PubMed] [Google Scholar]
- 33.Brideau, R.J., P.B. Carter, W.R. McMaster, D.W. Mason, and A.F. Williams. 1980. Two subsets of rat T lymphocytes defined with monoclonal antibodies. Eur. J. Immunol. 10:609–615. [DOI] [PubMed] [Google Scholar]
- 34.Damoiseaux, J.G., E.A. Dopp, W. Calame, D. Chao, G.G. MacPherson, and C.D. Dijkstra. 1994. Rat macrophage lysosomal membrane antigen recognized by monoclonal antibody ED1. Immunology. 83:140–147. [PMC free article] [PubMed] [Google Scholar]
- 35.Barsumian, E.L., C. Isersky, M.G. Petrino, and R.P. Siraganian. 1981. IgE-induced histamine release from rat basophilic leukemia cell lines: isolation of releasing and nonreleasing clones. Eur. J. Immunol. 11:317–323. [DOI] [PubMed] [Google Scholar]
- 36.Reynolds, C.W., E.W. Bere, Jr., and J.M. Ward. 1984. Natural killer activity in the rat. III. Characterization of transplantable large granular lymphocyte (LGL) leukemias in the F344 rat. J. Immunol. 132:534–540. [PubMed] [Google Scholar]
- 37.Spruyt, L.L., M.J. Glennie, A.D. Beyers, and A.F. Williams. 1991. Signal transduction by the CD2 antigen in T cells and natural killer cells: requirement for expression of a functional T cell receptor or binding of antibody Fc to the Fc receptor, Fc gamma RIIIA (CD16). J. Exp. Med. 174:1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pende, D., S. Parolini, A. Pessino, S. Sivori, R. Augugliaro, L. Morelli, E. Marcenaro, L. Accame, A. Malaspina, R. Biassoni, et al. 1999. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J. Exp. Med. 190:1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunt, S.V., and M.H. Fowler. 1981. A repopulation assay for B and T lymphocyte stem cells employing radiation chimaeras. Cell Tissue Kinet. 14:445–464. [DOI] [PubMed] [Google Scholar]
- 40.Arase, H., T. Saito, J.H. Phillips, and L.L. Lanier. 2001. Cutting edge: the mouse NK cell-associated antigen recognized by DX5 monoclonal antibody is CD49b (alpha 2 integrin, very late antigen-2). J. Immunol. 167:1141–1144. [DOI] [PubMed] [Google Scholar]
- 41.Vaage, J.T., C. Naper, G. Lovik, D. Lambracht, A. Rehm, H.J. Hedrich, K. Wonigeit, and B. Rolstad. 1994. Control of rat natural killer cell-mediated allorecognition by a major histocompatibility complex region encoding nonclassical class I antigens. J. Exp. Med. 180:641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berg, S.F., E. Dissen, I.H. Westgaard, and S. Fossum. 1998. Two genes in the rat homologous to human NKG2. Eur. J. Immunol. 28:444–450. [DOI] [PubMed] [Google Scholar]
- 43.Naper, C., S. Hayashi, G. Løvik, L. Kveberg, E.C. Niémi, B. Rolstad, E. Dissen, J.C. Ryan, and J.T. Vaage. 2002. Characterization of a novel killer cell lectin-like receptor (KLRH1) expressed by alloreactive rat natural killer cells. J. Immunol. 168:5147–5154. [DOI] [PubMed] [Google Scholar]
- 44.Boyington, J.C., A.N. Riaz, A. Patamawenu, J.E. Coligan, A.G. Brooks, and P.D. Sun. 1999. Structure of CD94 reveals a novel C-type lectin fold: implications for the NK cell-associated CD94/NKG2 receptors. Immunity. 10:75–82. [DOI] [PubMed] [Google Scholar]
- 45.Tormo, J., K. Natarajan, D.H. Margulies, and R.A. Mariuzza. 1999. Crystal structure of a lectin-like natural killer cell receptor bound to its MHC class I ligand. Nature. 402:623–631. [DOI] [PubMed] [Google Scholar]
- 46.Li, P., D.L. Morris, B.E. Willcox, A. Steinle, T. Spies, and R.K. Strong. 2001. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat. Immunol. 2:443–451. [DOI] [PubMed] [Google Scholar]
- 47.Radaev, S., B. Rostro, A.G. Brooks, M. Colonna, and P.D. Sun. 2001. Conformational plasticity revealed by the cocrystal structure of NKG2D and its class I MHC-like ligand ULBP3. Immunity. 15:1039–1049. [DOI] [PubMed] [Google Scholar]