Abstract
An analysis of expression of 137 lipoprotein genes on the course of murine infection revealed a two-step molecular adaptation by Borrelia burgdorferi, the Lyme disease spirochete. For the first step, regardless whether the initial inocula of B. burgdorferi expressed either all (cultured spirochetes) or less than 40 (host-adapted spirochetes) of the 137 lipoprotein genes, the spirochetes were modulated to transcribe 116 of the genes within 10 d after being introduced to the murine host. This step of adaptation was induced by the microenvironment of the host tissue. During the second step, which was forced by host immune selection pressure and occurred between 17 and 30 d after infection, B. burgdorferi down-regulated most of the lipoprotein genes and expressed less than 40 of the 137 genes. This novel adaptation mechanism could be a critical step for B. burgdorferi to proceed to chronic infection, as the pathogen would be cleared at the early stage of infection if the spirochetes failed to undergo this process.
Keywords: Lyme disease, immune evasion, lipoprotein, gene expression, DNA microarray
Introduction
Borrelia burgdorferi, the Lyme disease spirochete, is maintained within a complex enzootic life cycle involving the tick vector and the mammal. The spirochete adapts to these diverse environments, in part, by selective gene expression. Environmental cues such as temperature (1, 2), pH (2, 3), and nutrients or chemicals (4, 5) influence B. burgdorferi gene expression in vitro, and the cultivation of B. burgdorferi in dialysis membrane chambers implanted into rat peritoneal cavities up-regulates several spirochetal genes (6). The most dramatic modifications that occur as B. burgdorferi migrates from ticks to the mammalian host involve lipoprotein gene expression, such as the down-regulation of outer surface protein A (OspA) and the up-regulation of OspC (7, 8). The genome of B. burgdorferi contains more than 150 open-reading frames that encode putative lipoproteins, more than any other known bacterium (9), and many are surface exposed or have direct contact with the environment. These lipoproteins play major roles in pathogenesis and immunity and are likely to be involved in B. burgdorferi adaptation through the spirochete life cycle.
Materials and Methods
Spirochete Isolate.
B. burgdorferi B31 clone 5A11 (a gift from Steven Norris, University of Texas, Houston, TX) was cultivated in Barbour-Stoenner-Kelly H complete medium at 33°C (Sigma-Aldrich). This isolate carries 21 linear and circular plasmids (10).
Mouse Inoculation with Cultured Spirochetes.
Both C3H/HeN (C3H) and C3H/HeN-SCID (SCID) mice (4 to 6-wk-old; National Institutes of Health, Bethesda, MD) were given one single intradermal injection of 105 spirochetes that were grown to stationary phase. Mice were killed at intervals of 1 to 8 wk, starting at 10 d after needle inoculation. Ears and skins were frozen immediately in liquid nitrogen for RNA preparation.
Mouse Inoculation with Host-adapted Spirochetes.
Donor C3H mice were infected by needle inoculation for 4 mo. Ear tissues were cut into small pieces and implanted into the skin at the back of naive C3H mice. Infected mice were killed at 11 to 40 d after infection. Ears and skins were frozen in liquid nitrogen for RNA preparation.
RNA Preparation from Cultured Spirochetes and Mouse Tissues.
Total RNA was purified from spirochetes grown to mid-logarithmic phase or mouse ears and skins using the RNeasy Mini Kit (QIAGEN). To remove DNA contamination, RNA samples were digested in solution with RNase-free DNase I (Life Technologies) at 37°C for 3 h, and then loaded to the RNeasy Mini columns and further treated with RNase-free DNase (QIAGEN) for an additional 30 min at room temperature. Digested samples were repurified and analyzed for potential DNA contamination by PCR.
cDNA Preparation.
RNA preparations were first annealed with reverse oligonucleotide primers of flaB and all the 137 investigated lipoprotein genes in the presence of reverse transcription (RT) buffer (Stratagene). All primer sequences are available by request. dNTPs and reverse transcriptase were added (Stratagene) and transcription was conducted at 42°C for 1 h. RNA–cDNA hybrids were denatured at 95°C for 5 min and RNA chains were hydrolyzed by digestion with 1.0 N NaOH at 65°C for 15 min. After neutralization with 1.0 N HCl, cDNA was purified using the Quik PCR Product Purification Kit (QIAGEN).
PCR.
For each PCR reaction volume of 50 μl, cDNA that was converted from 25 ng of total spirochetal RNA, 250 ng RNA prepared from SCID mice with acute or chronic infection or from C3H mice with acute infection, or 2,500 ng RNA from C3H mice with chronic infection was applied. The amount of cDNA that was used for each PCR reaction was determined by twofold serial dilution PCR of the flaB gene.
Southern Blotting Confirmation of PCR Products by Microarray Hybridization.
All PCR products were assessed by agarose gel electrophoresis. Those that showed expected sizes were combined and purified using the Quik PCR Product Purification Kit (QIAGEN). Purified DNAs were incorporated with aminoallyl dUTP by nick translation (Life Technologies), fluorescently labeled with the Alexa Fluor 647 kit (Molecular Probes, Inc.), and allowed to hybridize to the lipoprotein DNA microarray (11). The array was scanned with an Axon GenePix 4000A Array Scanner (Axon Instruments, Inc.).
Sequencing of PCR Products.
PCR products of selective genes were gel-purified using the QIAquick Gel Extraction Kit (QIAGEN). Purified DNAs were sequenced.
Passive Immunization of SCID Mice with Anti-B. burgdorferi Antisera.
SCID mice were infected with cultured B. burgdorferi B31 and received three subcutaneous injections of 100-μl pooled mouse antiserum or prebled (control) at 11, 13, and 15 d. Antisera were collected from C3H mice that had been infected with cultured spirochetes for 1 to 6 mo, while prebleed was drawn from naive C3H mice. SCID mice were killed at 2 d after the last passive immunization and ear tissues were frozen in liquid nitrogen for RNA preparation.
Results
Lipoprotein Gene Expression during In Vitro Cultivation.
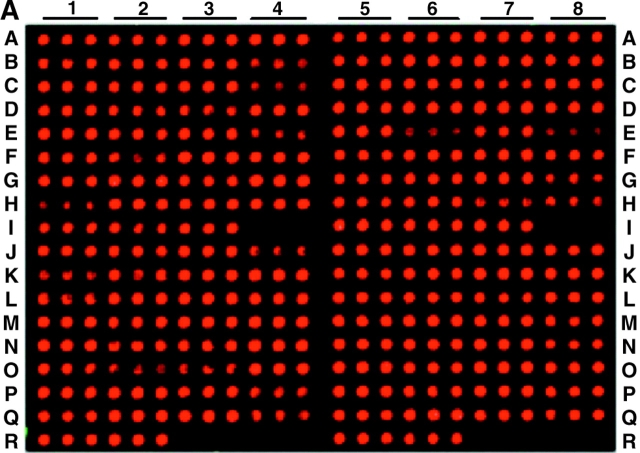
To investigate the molecular adaptation of B. burgdorferi in different environments, we first examined the expression of 137 putative lipoprotein genes during in vitro cultivation. Total RNA was prepared from cultured spirochetes. To determine if any DNA contamination was present, following two DNase digestion treatments, the RNA preparation was amplified by both PCR and RT-PCR using each primer pairs of the first putative lipoprotein genes carried by all of the 20 lipoprotein-carrying genetic elements: one chromosome and 19 plasmids (9). Both RT-PCR and PCR products were assessed by agarose gel electrophoresis. The results obtained from plasmids J (lp38), K (lp36), L (cp32–8), M (cp32–6), N (cp32–9), O (cp32–7), P (cp32–1), Q (lp56), R (cp32–4), and S (cp32–3) were selectively presented in Fig. 1 . The RT-PCR products only showed amplicons with expected sizes, indicating that DNA contamination was not detectable. Then, the 117 remaining lipoprotein genes were individually amplified by RT-PCR, and the flaB gene was used as a control and also amplified. Resultant products were assessed by agarose gel electrophoresis and all showed the expected sizes. To more definitively examine the gene expression, flaB and all 137 lipoprotein PCR products were combined, purified, fluorescently labeled, and allowed to hybridize to a lipoprotein DNA microarray. In vitro cultivated spirochetes expressed virtually all of these 137 (putative) lipoprotein genes (Fig. 2 A). These data were confirmed in a separate experiment.
Figure 1.
Amplification of selected B. burgdorferi lipoprotein genes that are expressed on the plasmids. Total RNA was prepared from cultured B. burgdorferi and analyzed by both PCR (–) and RT-PCR (+). PCR products were separated by ethidium bromide–incorporated agarose gel, and the lack of a product in the (–) column demonstrated that DNA was not present in the RNA preparation.
Figure 2.
Lipoprotein gene expression of B. burgdorferi in in vitro cultivation (A), infected SCID mice (B), and C3H mice (C). flaB and 137 lipoprotein gene fragments were amplified from B. burgdorferi B31 DNA and printed in triplicate on the microarray. The location of each DNA fragment was listed on Fig. 3, and locations I4, I8, R3, R4, R7, and R8 were blank. RNA was prepared from cultured spirochetes (A), the ear tissues of SCID mice that had been infected with cultured B. burgdorferi for 10 d (B), or the ear tissues of C3H mice that had been infected with cultured B. burgdorferi for 24 d (C), and amplified by RT-PCR. Purified PCR products were fluorescently labeled and hybridized to the microarray.
Lipoprotein Gene Expression on the Course of Infection in Immunodeficient Mice.
We next examined lipoprotein gene expression in infected SCID mice. 10 animals were infected with cultured B. burgdorferi and killed at 10, 17, 24, 31, 45, 60, 75, 100, 130, and 180 d. Total RNA was prepared and amplified by RT-PCR. PCR products were analyzed by agarose gel electrophoresis and microarray hybridization. B. burgdorferi quickly down-regulated 21 of the lipoprotein genes within 10 d after introduction into the immunodeficient host (Fig. 2 B). These down-regulated genes included bb0098, bb0424, bb0464, bb0475, bb0664, bb0740, bb0840, bba15, bba62, bbb08, bbe06, bbe08, bbe28, bbg25, bbh32, bbi28, bbi32, bbk04, bbo40, bbq05, and bbr42. The 116 remaining genes were persistently transcribed throughout the entire 6-mo study period, as their mRNA transcripts were detectable by RT-PCR in all of the nine mice that were killed at 17 to 180 d after infection (data not shown). Neither bbo40 (erpM) or bbr42 (erpY) amplicons were revealed by agarose gel electrophoresis but these genes' DNA spots were reactive on the microarray (Fig. 2 B), probably due to the cross-hybridization of other erp gene PCR products with these genes' DNA fragments on the microarray. The sequences of the erp family share high percentages of identity (9). Although the ospA (bba15) mRNA transcript was not detected, surprisingly, ospB (bba16) was continuously expressed throughout infection. The ospB PCR product was sequenced to confirm that this gene was transcribed (data not shown).
Lipoprotein Gene Expression on the Course of Infection in Immunocompetent Mice.
As immune selection pressure could influence the gene expression of surface-exposed, Ab-targeted lipoproteins (12), we investigated B. burgdorferi lipoprotein expression under immune selection pressure. RNA was prepared from 10 immunocompetent C3H mice that were infected with cultured spirochetes for 10 to 180 d, and analyzed by RT-PCR and microarray hybridization. B. burgdorferi expressed a similar set of lipoprotein genes as detected in infected SCID mice (Fig. 2 B) when the mouse was examined at 10 d after infection. At 17 d, however, most of the 116 lipoprotein genes that were expressed at 10 d were down-regulated by immune selection pressure elicited by infection (data not shown). Less than 40 of the lipoprotein genes were continuously expressed when infected C3H mice were examined at 24, 31, 45, 60, 73, 100, 130, and 180 d after infection (Fig. 2 C). These results demonstrated that early and chronic infections constitute two remarkably different environments for B. burgdorferi adaptation and the development of immune selection pressure in the host significantly impacts lipoprotein gene expression.
Molecular Adaptation of B. burgdorferi during Murine Infection.
To further investigate the influence of immune selection pressure on B. burgdorferi adaptation, spirochetes that had been adapted under immune selection pressure were used for inoculation. Donor mice were first infected with cultured B. burgdorferi. 4 mo later, after the spirochetes had adapted and expressed less than 40 lipoprotein genes (Fig. 3) , ear tissues were taken from the donors and transplanted into nine naive immunocompetent mice. B. burgdorferi reexpressed all of the 83 down-regulated lipoprotein genes when five recipient mice were examined at 11 to 22 d after tissue transplantation (Fig. 3). This reexpression persisted for ∼2 wk. B. burgdorferi redown-regulated all of these reexpressed genes within 5 wk after the initial transplantation (Fig. 3), as their mRNA transcripts became undetectable when four recipient mice were examined at 33 d or later. These results further confirmed the influence of immune pressure on B. burgdorferi adaptation.
Figure 3.
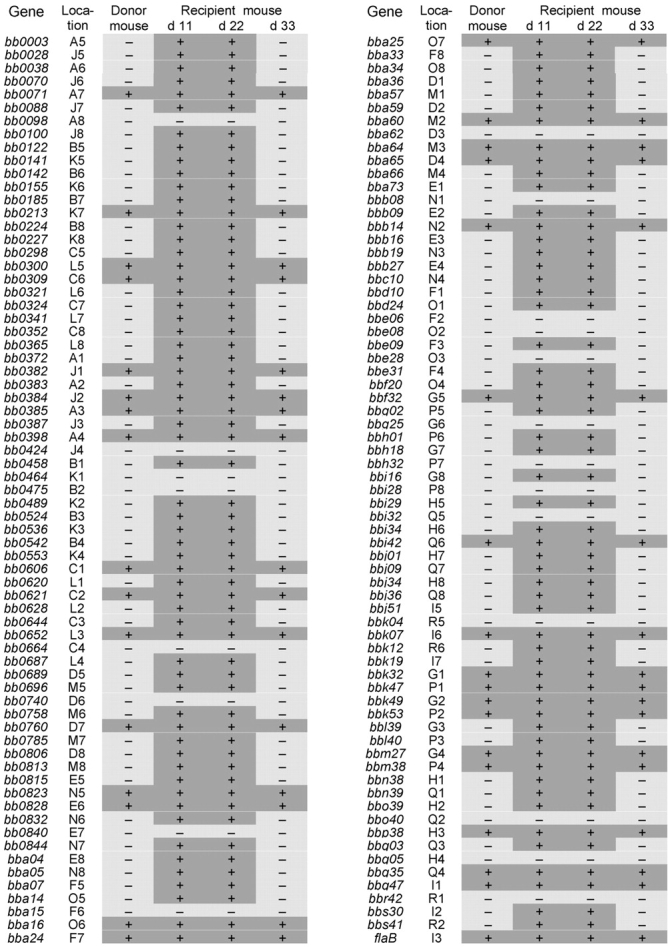
Molecular adaptation of B. burgdorferi in the murine host. C3H mice that had been infected with cultured B. burgdorferi for 4 mo were used as donors. Naive C3H mice received ear tissue implants and were killed 11, 22, and 33 d later. RNA was prepared from ears and analyzed for the expression of the 137 putative lipoprotein genes and flaB by RT-PCR and microarray hybridization. “Location,” location of each gene's DNA fragment on the microarray; “+,” expressed; “–,” not expressed.
It took 5 wk for host-adapted B. burgdorferi to finish the adaptation to the immune selection pressure but only 3.5 wk for the cultured spirochetes to complete the same process (Figs. 2 C and 3). The temporal difference may be due to the fact that cultured B. burgdorferi expressed all of the lipoproteins while the host-adapted spirochetes had down-regulated all of the adaptable genes and thus must reexpresses to trigger the immune response. In addition, the spirochete inoculation dose may also be a factor as the tissue implantation introduced fewer organisms than the needle inoculation.
Dominant Role of the Humoral Response in B. burgdorferi Adaptation.
B. burgdorferi is primarily an extracellular pathogen and the humoral response has been shown to play a critical role in immunity and disease regression. To carefully examine the role of Abs in B. burgdorferi molecular adaptation, four infected SCID mice were passively immunized with antisera from infected immunocompetent mice and two received normal mouse sera as controls. Almost all the lipoprotein genes that were down-regulated by immune selection pressure in immunocompetent mice were no longer expressed in the four passively immunized SCID mice but expressed in the two controls (data not shown). Several of the genes, however, were not down-regulated in the passive transfer studies including bb0122, bb0155, bba34, bbe31, bb01, bbh18, and bbi29. The fact that passive immunization could not provide as high a level of Ab as those elicited by infection might be the reason why some of the 83 down-regulated genes in infected immunocompetent mice were still expressed at a detectable level in the passively immunized SCID mice. Alternatively, the host cellular immune response may also play a role in B. burgdorferi adaptation.
Discussion
B. burgdorferi expressed all of the 137 lipoprotein genes we investigated during in vitro cultivation (Fig. 2 A). Once the spirochete was introduced into the mammalian host, however, 21 of the lipoprotein genes were quickly down-regulated (Fig. 2 B). As expected, one of the down-regulated genes was ospA. B. burgdorferi does not prominently express ospA during mammalian infection but overexpresses this lipoprotein within the tick vector (7, 8) to facilitate the adherence of B. burgdorferi to the tick midgut (13). When OspA is expressed in the reservoir host, it would elicit strong immune responses including the development of OspA Ab. As a consequence, acquisition of the spirochetes from mammals by larval ticks would be impaired or even blocked (14). Therefore, B. burgdorferi does not generally express ospA during infection of its natural mammalian host in order to maintain the enzootic cycle. OspA Ab may sometimes be found in patients with Lyme disease (15); however, humans are an aberrant host and not important for spirochete persistence. Most of the 20 remaining down-regulated lipoprotein genes are hypothetical. It remains to be addressed if they are, like ospA, also tick-specifically expressed.
A group of ∼80 lipoprotein genes were persistently expressed during murine infection when no immune selection pressure was present. ospC (bbb19) is the best known member of this group. OspC has been identified as an Ab-targeted, surface-exposed Ag during murine infection (12). To avoid immune clearance by OspC Ab during mammalian infection, B. burgdorferi shuts down ospC (12). Although the other members of this group also appeared to be down-regulated by immune selection pressure elicited during infection, it remains to be addressed whether they are, like OspC, surface-exposed, Ab-targeted Ag. Some of them might be neither surface-exposed nor Ab-targeted but could be codown-regulated with other Ab-targeted Ag(s).
The third group of lipoproteins were expressed throughout mammalian infection. Some of them were significantly down-regulated by immune selection pressure but their expression was detectable. They may be critical for the survival of B. burgdorferi in the mammalian host. As expected, VlsE (BBF32) was continuously expressed since it is able to undergo antigenic variation during murine infection (16). Three of the four-member Bmp family (17), BmpB, BmpC, and BmpD (BB0382, BB0384 and BB0385), but not BmpA (BB0383), were also persistently found. Although the functions of these lipoproteins are unknown, they are homologous to TmpC, a putative outer or cytoplasmic membrane lipoprotein of Treponema pallidum, the spirochetal agent of syphilis (18), indicating the importance of these lipoproteins in the spirochetes' pathogenesis. The best known members of this group are decorin-binding proteins A and B (DbpA and DbpB; BBA24 and BBA25; reference 19) and fibronectin-binding protein (BBK32; reference 20). These three lipoproteins have been shown in vitro to be surface exposed and be able to bind extracellular matrices (19, 20). Recent evidence suggests that Dbps interact with their ligand and enhance the infectivity of B. burgdorferi during murine infection (21). It remains to be addressed if host decorin or fibronectin can coat B. burgdorferi via the Dbps and fibronectin-binding protein, and thus protect the pathogen from attack by specific Ab. ErpK (BBM38), ErpA (BBP38), and ErpX (BBQ47) were expressed in chronically infected immunocompetent mice, albeit some of them were transcribed at a very low level or even undetectable in some cases. Interestingly, like BBA24, BBA25, and BBK32, the Erp lipoproteins also are able to bind a host component, complement inhibitor factor H (22).
Some of our findings strikingly differ from those that have been obtained with cultured B. burgdorferi. For instance, ospA and ospB are transcribed as a single mRNA unit in cultured B. burgdorferi (23). Our study indicated that B. burgdorferi expressed ospB but not ospA throughout murine infection. Hübner and colleagues have reported an RpoN-RpoS regulatory pathway that controls the expression of both dbpA and ospC (24). Our study, however, clearly showed that whereas B. burgdorferi expressed both genes in SCID mice, immune selection pressure shut down ospC but not dbpA. The differential expression of these two lipoproteins has already been reported in the tick vector (25). B. burgdorferi has coevolved with the mammalian (immunocompetent) host and tick vector for a long period of time. The in vitro artificial conditions probably constitute an aberrant environment where B. burgdorferi changes protein expression profiles and can more easily lose plasmids, infectivity, and pathogenicity (26).
There are no existing mechanisms that can be used to explain how immune selection pressure down-regulates specific genes. A previous study indicates that B. burgdorferi is able to generate multiple phenotypes during murine infection (12). Different phenotypes may express unique sets of lipoproteins and these phenotypes can interconvert. B. burgdorferi infection elicits the immune responses including those to surface-exposed lipoproteins once they are expressed. The specific Abs selectively eliminate the phenotypes that express these Ab-targeted Ags. The Ab-targeted phenotypes are continuously generated and then eliminated during infection. Some of the lipoprotein genes might exhibit this “down-regulation phenomenon.” Coexistence of multiple phenotypes of B. burgdorferi has already been observed in the tick vector (27) as well as in vitro cultivation (28). This selection hypothesis can explain some but not all of the observations that have been obtained to date. For instance, if Ab-targeted phenotypes were continuously generated, specific lipoprotein mRNAs could be positive by sensitive RT-PCR even though these newly “borne” spirochetes were immediately eliminated. Moreover, if Ab-targeted phenotypes were continuously generated, newly synthesized lipoproteins would constantly prime the immune system. As a consequence, the specific Ab response to Ab-targeted lipoproteins should be maintained. However, the analysis of the Ab response to selected lipoproteins, such as OspC, in infected white-footed mice and human patients with Lyme disease does not support this view. The OspC Ab titer substantially wanes while infection continues in white-footed mice (29). In human patients, the OspC Ab titer may fall below a detectable level as the infection persists, and then reappear months later (30). If OspC-expressing spirochetes were continuously generated, the anti-OspC response should persist throughout the infection. Therefore, at least, some of the lipoprotein genes could be turned off by specific Ab by mechanisms that have not been known yet.
Our study has clearly demonstrated a two-step process of molecular adaptation by B. burgdorferi during early murine infection. The first-step adaptation is induced by the microenvironment of the host tissues. Regardless whether cultured B. burgdorferi, which expressed all of the 137 lipoprotein genes, or host-adapted organisms, which expressed less than 40 of them, were used for inoculation, the spirochetes were modulated to transcribe the same set of 116 lipoprotein genes within 10 d after infection. During the second step of adaptation, immune selection pressure forced spirochetes to down-regulate more than 80 of these 116 genes. This adaptation is a fundamental for B. burgdorferi to establish chronic infection, as the pathogen would be cleared by the host immune system at the very early stage of infection if the spirochete failed to undergo the process.
Acknowledgments
We thank Steven Norris (University of Texas, Houston, TX) for providing clonal isolate B31 5A11. The excellent technical assistance of Debbie Beck is gratefully acknowledged.
This study was supported by grants from the National Institutes of Health and American Heart Association. E. Fikrig is the recipient of a Burroughs Wellcome Clinical Scientist Award in Translational Research.
References
- 1.Stevenson, B., T.G. Schwan, and P.A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:4535–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramamoorthy, R., and D. Scholl-Meeker. 2001. Borrelia burgdorferi proteins whose expression is similarly affected by culture temperature and pH. Infect. Immun. 69:2739–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll, J.A., C.F. Garon, and T.G. Schwan. 1999. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 67:3181–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babb, K., N. El-Hage, J.C. Miller, J.A. Carroll, and B. Stevenson. 2001. Distinct regulatory pathways control expression of Borrelia burgdorferi infection-associated OspC and Erp surface proteins. Infect. Immun. 69:4146–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang, X., T.G. Popova, M.S. Goldberg, and M.V. Norgard. 2001. Influence of cultivation media on genetic regulatory patterns in Borrelia burgdorferi. Infect. Immun. 69:4159–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akins, D.R., K.W. Bourell, M.J. Caimano, M.V. Norgard, and J.D. Radolf. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Invest. 101:2240–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwan, T.G., J. Piesman, W.T. Golde, M.C. Dolan, and P.A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA. 92:2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montgomery, R.R., S.E. Malawista, K.J.M. Feen, and L.K. Bockenstedt. 1996. Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo: exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J. Exp. Med. 183:261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Institute for Genomic Research website. http://www.tigr.org.
- 10.Purser, J.E., and S.J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA. 97:13865–13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang, F.T., F.K. Nelson, and E. Fikrig. 2002. DNA microarray assessment of putative Borrelia burgdorferi lipoprotein genes. Infect. Immun. 70:3300–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang, F.T., M.B. Jacobs, L.C. Bowers, and M.T. Philipp. 2002. An immune evasion mechanism of spirochetal persistence in Lyme borreliosis. J. Exp. Med. 195:415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pal, U., A.M. de Silva, R.R. Montgomery, D. Fish, J. Anguita, J.F. Anderson, Y. Lobet, and E. Fikrig. 2000. Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J. Clin. Invest. 106:561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Silva, A.M., D. Fish, T.R. Burkot, Y. Zhang, and E. Fikrig. 1997. OspA antibodies inhibit the acquisition of Borrelia burgdorferi by Ixodes ticks. Infect. Immun. 65:3146–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akin, E., G.L. McHugh, R.A. Flavell, E. Fikrig, and A.C. Steere. 1999. The immunoglobulin (IgG) antibody response to OspA and OspB correlates with severe and prolonged Lyme arthritis and the IgG response to P35 correlates with mild and brief arthritis. Infect. Immun. 67:173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang, J.R., J.M. Hardham, A.G. Barbour, and S.J. Norris. 1997. Antigenic variation in Lyme disease Borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 89:275–285. [DOI] [PubMed] [Google Scholar]
- 17.Ramamoorthy, R., L. Povinelli, and M.T. Philipp. 1996. Molecular characterization, genomic arrangement, and expression of bmpD, a new member of the bmp class of genes encoding membrane proteins of Borrelia burgdorferi. Infect. Immun. 64:1259–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schouls, L.M., H.G. van der Heide, and J.D. van Embden. 1991. Characterization of the 35-kilodalton Treponema pallidum subsp. pallidum recombinant lipoprotein TmpC and antibody response to lipidated and nonlipidated T. pallidum antigens. Infect. Immun. 59:3536–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, B.P., E.L. Brown, D.W. Dorward, L.C. Rosenberg, and M. Höök. 1998. Decorin-binding adhesins from Borrelia burgdorferi. Mol. Microbiol. 30:711–723. [DOI] [PubMed] [Google Scholar]
- 20.Probert, W.S., and B.J.B. Johnson. 1998. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi B31. Mol. Microbiol. 30:1003–1015. [DOI] [PubMed] [Google Scholar]
- 21.Brown, E.L., R.M. Wooten, B.J.B. Johnson, R.V. Iozzo, A. Smith, M.C. Dolan, B.P. Gao, J.J. Weis, and M. Höök. 2001. Resistance to Lyme disease in decorin-deficient mice. J. Clin. Invest. 107:845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevenson, B., N. El-Hage, M.A. Hines, J.C. Miller, and K. Babb. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70:491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergstrom, S., V.G. Bundoc, and A.G. Barbour. 1989. Molecular analysis of linear plasmid-encoded major surface proteins, OspA and OspB, of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 3:479–486. [DOI] [PubMed] [Google Scholar]
- 24.Hübner, A., X. Yang, D.M. Nolen, T.G. Popova, F.C. Cabello, and M.V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA. 98:12724–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagman, K.E., X. Yang, S.K. Wikel, G.B. Schoeler, M.J. Caimano, J.D. Radolf, and M.V. Norgard. 2000. Decorin-binding protein A (DbpA) of Borrelia burgdorferi is not protective when immunized mice are challenged via tick infestation and correlates with the lack of DbpA expression by B. burgdorferi in ticks. Infect. Immun. 68:4759–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norris, S.J., J.K. Howell, S.A. Garza, M.S. Ferdows, and A.G. Barbour. 1995. High- and low-infectivity phenotypes of clonal populations of in vitro cultured Borrelia burgdorferi. Infect. Immun. 63:2206–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohnishi, J., J. Piesman, and A.M. de Silva. 2001. Antigenic and genetic heterogeneity of Borrelia burgdorferi populations transmitted by ticks. Proc. Natl. Acad. Sci. USA. 98:670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanson, M.S., N.K. Patel, D.R. Cassatt, and N.D. Ulbrandt. 2000. Evidence for vaccine synergy between Borrelia burgdorferi decorin binding protein A and outer surface protein A in the mouse model of Lyme borreliosis. Infect. Immun. 68:6457–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwan, T.G., K.K. Kime, M.E. Schrumpf, J.E. Coe, and W.J. Simpson. 1989. Antibody response in white-footed mice (Peromyscus leucopus) experimentally infected with the Lyme disease spirochete (Borrelia burgdorferi). Infect. Immun. 57:3445–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fung, B.P., G.L. McHugh, J.M. Leong, and A.C. Steere. 1994. Humoral immune response to outer surface protein C of Borrelia burgdorferi in Lyme disease: role of the immunoglobulin M response in the serodiagnosis of early infection. Infect. Immun. 62:3213–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]