Abstract
In contrast with the low frequency of most single epitope reactive T cells in the preimmune repertoire, up to 1 of 1,000 naive CD8+ T cells from A2+ individuals specifically bind fluorescent A2/peptide multimers incorporating the A27L analogue of the immunodominant 26–35 peptide from the melanocyte differentiation and melanoma associated antigen Melan-A. This represents the only naive antigen-specific T cell repertoire accessible to direct analysis in humans up to date. To get insight into the molecular basis for the selection and maintenance of such an abundant repertoire, we analyzed the functional diversity of T cells composing this repertoire ex vivo at the clonal level. Surprisingly, we found a significant proportion of multimer+ clonotypes that failed to recognize both Melan-A analogue and parental peptides in a functional assay but efficiently recognized peptides from proteins of self- or pathogen origin selected for their potential functional cross-reactivity with Melan-A. Consistent with these data, multimers incorporating some of the most frequently recognized peptides specifically stained a proportion of naive CD8+ T cells similar to that observed with Melan-A multimers. Altogether these results indicate that the high frequency of Melan-A multimer+ T cells can be explained by the existence of largely cross-reactive subsets of naive CD8+ T cells displaying multiple specificities.
Keywords: Melan-A-naive-CD8-A2, naive, CD8, A21 peptide multimers, repertoire
Introduction
In recent years numerous tumor-associated antigens recognized by tumor-reactive CD8+ CTLs have been identified. Albeit some of these antigens arise as the consequence of mutations, the majority of them is derived from nonmutated self-proteins either ectopically expressed (1–3) or cell lineage-specific (4–6). Therefore, the analysis of the functional TCR repertoire specific for these self-antigens is highly relevant for the study of immune responses to cancer.
During T cell development in the thymus T cell tolerance is achieved toward antigens expressed by thymic APC. Until recently, the thymus was regarded as the site of tolerance induction to ubiquitously expressed proteins, whereas it was thought that tolerance to tissue specific antigens was achieved through postthymic peripheral mechanisms (7) or that the immune system would simply ignore such antigens (8). Lately, however, this simplistic notion of central versus peripheral tolerance has been challenged by the finding that promiscuous expression of peripheral antigens including lineage specific antigens in the thymus mirrors the peripheral self (9). Thus, repertoire selection toward the majority of peripheral antigens could be achieved similarly to that of the ubiquitously expressed ones.
T cell repertoire selection involves TCR-mediated recognition of self-peptides by immature thymocytes (10) and includes both negative selection of thymocytes that recognize self-peptides with high avidity and positive selection of the ones that recognize self-peptides with low avidity. Survival of positively selected thymocytes after maturation and migration in the periphery will again depend on the delivery of survival signals by self-peptide/MHC ligands (11). The molecular factors determining the size of the preimmune pool of individual epitope specific T cells have not yet been fully elucidated. Whereas it is well accepted that a significant fraction of the repertoire of T cells reactive to self-peptides escape clonal deletion (12), both the number and the nature of the self-peptides that shape in vivo TCR repertoire for individual epitope-specific T cells is largely unknown.
Several lines of evidence, however, indicate that this role is likely to be played by multiple peptides. Indeed, although positive selection can, to some extent, take place in the presence of a single selecting ligand (13), both the number and the sequence of selecting peptides significantly impact on the frequency of single antigen-specific selected precursors. In fetal thymic organ cultures from mice deficient in presenting self-peptides because either β2m or TAP1 deficiency and therefore unable to positively select CD8+ T cells, positive selection was restored by addition of exogenous peptides and β2m. Interestingly, a diverse peptide mixture was more efficient than single peptides at restoring CD8+ T cell development (14, 15). When similar experiments were performed on F1 of TCR transgenic and β2m-deficient mice, it was found that several single amino acid variants of the antigenic peptide were able to mediate a very efficient positive selection of that particular TCR.
All of these variants were, however, either extremely weak or negative in their ability to stimulate mature peripheral T cells (16). Positive selection of single antigen-specific T cells is therefore not only peptide dependent, but also highly specific. In addition, as immature thymocytes are more sensitive to TCR stimulation than mature T cells, the number of peptide ligands able to give weak signals albeit sufficient to promote positive selection is larger than the ones able to activate mature T cells and includes TCR antagonist peptides (16). It follows that the size of individual epitope-specific T cell repertoires in the preimmune T cell pool may primarily be impacted by the proportion of peptides cross-recognized by the TCR of interest and presented by thymic APCs.
The direct analysis of T cells composing antigen-specific preimmune repertoires has thus far been prevented by their low frequency (estimated to be ∼1–5 × 10−5 cells; reference 17). Recently, the development of fluorescent HLA/peptide multimeric arrays (multimers, reference 18) that specifically bind to TCR has tremendously improved our ability to detect antigen specific T cells. However, whereas antigen experienced T cells specific for single antigens are often easily visualized ex vivo by staining with multimers incorporating relevant peptides, naive antigen-specific CD8+ T cells are usually present in the periphery at frequencies that are below multimer detection limits (19).
In humans, one exception is represented by the repertoire of CD8+ T cells reactive to the self-differentiation antigen Melan-A, which is selectively expressed by normal cells of the melanocytic lineage including skin melanocytes and by the large majority of malignant melanomas (5, 20). Phenotypically naive Melan-A multimer+ T cells comprise ∼10−3 cells of circulating CD8+ T cells in A2+ healthy donors and in the majority of A2+ melanoma patients (21). We have recently reported that a proportion of Melan-A multimer+ T cells similar to that detected in the periphery can also be found among CD8+ thymocytes indicating that this large T cell pool is mostly generated by thymic output of a high number of precursors and not due to the peripheral expansion of a few positively selected precursors (22).
The functional diversity of clonotypes composing the repertoire of Melan-A multimer+ T cells ex vivo, however, has not been thus far investigated. In addition, the molecular bases for selection and maintenance of such an aberrantly abundant repertoire of naive antigen specific T cells have remained elusive. To gain insights into these questions, in this study, we have analyzed functionally a large number of circulating Melan-A multimer+ T cell clones isolated ex vivo from an A2+ healthy donor.
The results of this analysis revealed a large functional heterogeneity in terms of functional avidity of antigen recognition and tumor reactivity. An unprecedented finding was that A2/Melan-A multimers specifically stained a significant proportion of clones whose functional avidity for Melan-A peptides was too low to be detectable in a functional CTL assay, but which were able to efficiently recognize peptides from proteins of self- or pathogen origin selected for their potential functional cross-reactivity with Melan-A. Altogether, the reported data indicate that this repertoire is contributed for by largely cross-reactive subsets of T cells displaying multiple specificities and suggest that the degree of cross-reactivity of antigen specific T cells to the pool of peptides accessible to the immune system (degeneracy of antigen recognition) could directly impact on the frequency of reactive precursors.
Materials and Methods
Cells and Antigen Recognition Assay.
Multimer+ CD8+ T cells (see below) were purified from PBMCs ex vivo by flow cytometry cell sorting and cloned by limiting dilution culture in the presence of PHA, allogenic irradiated PBMC and hrIL-2 as described previously (23). Clones were subsequently expanded by periodical (3–4 wk) restimulation into microtiter plates. Antigen recognition was assessed using chromium release assay (CTL assay). The A2+ human mutant cell line CEMx721.T2 (T2) (24) or the melanoma cell lines Me 275 (A2+ Melan-A+) and NA8-MEL (A2+ Melan-A−) were used as targets. Briefly, after labeling with 51[Cr] during 1 h at 37°C followed by extensive washing, target cells (1,000/well) were incubated with effector cells at the indicated E/T ratio during 4 h at 37°C in V-bottomed microwells in the absence or presence of the indicated synthetic peptide (1 μM). In peptide titration experiments target cells were incubated with effectors at an E/T ratio of 10:1 in the presence of serial dilutions of the indicated peptide. Chromium release was measured in the supernatant of the cultures using a γ-counter. The percentage of specific lysis was calculated as 100 × (experimental-spontaneous release)/(total-spontaneous release).
A2/Peptide Multimers and Flow Cytometry Immunofluorescence Analysis.
PE-conjugated multimeric A2/peptide complexes containing the Melan-A enhanced peptide analogue 26–35 A27L (ELAGIGILTV), the influenza matrix Flu-MA 58–66 (GILGFVFTL), the HIV-1 Pol 476–484 (ILKEPVHGV), and the peptides MSI 44–10 from prostaglandin transporter (LLAGIGTVPI), MSI 44–25 from Pseudorabies virus (VIAGIGILAI) and MSI 44–56 from Chlamydia trachomatis (MLSGIGIFFI) were synthesized as described previously (18, 25). Because of their lower binding capacity to A2, multimers containing Melan-A parental peptides 26–35 (EAAGIGILTV) and 27–35 (AAGIGILTV) were synthesized using A2 molecules incorporating an unpaired cysteine residue at the COOH terminus, that allows site-specific biotinylation at 4°C (26). Samples were stained with multimers at the indicated dose in PBS containing 0.2% BSA during 1 h at room temperature, washed once in the same buffer, stained with mAbs where indicated during 30 min at 4°C, washed again, and analyzed by flow cytometry. Anti-CD8 (SK1) and anti-CD45RA mAbs were purchased from Becton Dickinson. The anti-CCR7 mAb 3D12 was provided by M. Lipp (Max-Dellbrueck Center for Molecular Medicine, Berlin, Germany). Inhibition of A2/Melan-A multimer staining by anti-CD8 Abs was assessed by coincubating clones with A2/Melan-A multimers at the following doses (2/7B12 1 μg/ml, 2/5F1 0.6 μg/ml, 2/7A11 2 μg/ml) resulting in a mean staining intensity of ∼200, together with serial dilutions of the anti-CD8 mAb RPA-T8 (Becton Dickinson) during 1 h at room temperature. Data analysis was performed using CELLQuest™ software.
Results
The Functional Repertoire of Ex Vivo Sorted A2/Melan-A Multimer+ CD8+ T Cell Clones in an A2+ Healthy Donor.
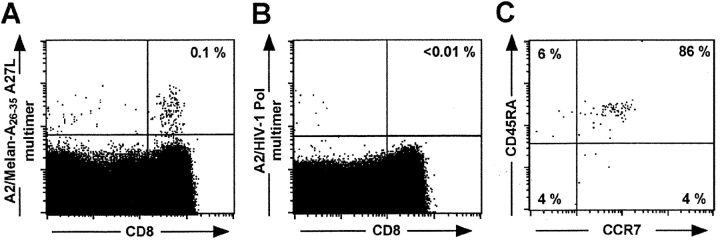
Consistent with what we have previously reported for other A2 expressing individuals (21, 27), ∼10−3 of phenotypically naive circulating CD8+ T cells from the healthy donor HD 421 were specifically stained by A2/Melan-A peptide multimers incorporating the peptide analogue Melan-A 26–35 A27L (A2/Melan-A multimers thereafter, Fig. 1) . To dissect the functional heterogeneity of this repertoire we derived, by limiting dilution cloning of ex vivo flow cytometry sorted A2/Melan-A multimer+ CD8+ T cells, 37 clones that were specifically stained by A2/Melan-A multimers as compared to background staining obtained on CD8+ T cell clones of unrelated specificity (mean fluorescence <10, Fig. 3 A and data not shown). All A2/Melan-A multimer+ CD8+ T cells clones, however, failed to be significantly stained (mean fluorescence <10) with a similar dose of multimers incorporating an unrelated HLA-A2 binder peptide (Flu-MA 58–66) that very brightly stained, at that dose, a Flu-MA–specific clone (mean fluorescence 665, unpublished data).
Figure 1.
Ex vivo detection and phenotyping of circulating A2/Melan-A multimer+ CD8+ T cells in a healthy donor (HD 421). PBMCs from healthy donor HD 421 were stained with anti-CD8 ECD mAb, anti-CD45RA FITC mAbs, anti-CCR7 APC mAb, and PE-labeled A2/peptide multimers (4.5 μg/ml) containing either Melan-A 26–35 analogue A27L (A) or an irrelevant HIV peptide (B). Dot plots are shown for CD8 versus A2/peptide multimer staining (A and B, numbers in the up right quadrant represent the percentage of multimer+ cells among CD8+ T cells) or for CD45RA versus CCR7 staining on gated CD8+ A2/Melan-A multimer+ cells (C, numbers represent the percentage of positive cells among multimer+ CD8+ T cells in the corresponding quadrant).
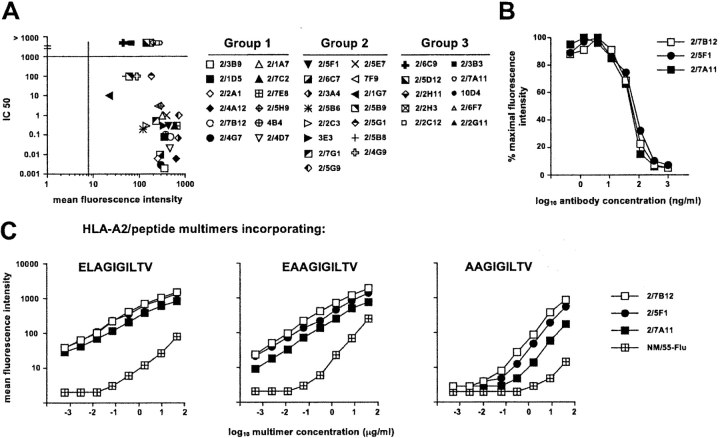
Figure 3.
A2/Melan-A multimer staining of clonal populations. (A) Correlation between avidity of antigen recognition and efficiency of A2/Melan-A multimer staining. Data are shown for all A2/Melan-A multimer+ clonal populations. The mean fluorescence staining obtained by simultaneously staining the 37 clonal populations during 1 h at room temperature with 4.5 μg/ml of A2/Melan-A multimers is shown on the x-axis. The bar on this axis represents the mean fluorescence staining obtained under the same conditions for a clone of unrelated specificity (clone MN/55-Flu specific for Influenza matrix peptide 58–66). The nM concentration of peptide Melan-A 26–35 A27L required to obtain 50% maximal lysis in peptide titration experiments (IC50) is shown in the y-axis. (B) Inhibition of A2/Melan-A multimer staining by anti-CD8 antibodies. Representative clones from each group were stained with A2/Melan-A multimers in the presence of serial dilutions of the anti-CD8 mAb. Results are shown as percentage of maximal staining intensity. (C) Cross-staining of A2/Melan-A multimer+ populations by multimers incorporating Melan-A parental peptides. Staining obtained upon incubation during 1 h at room temperature with serial dilutions of the indicated multimer is shown for one representative clone per group (as in Fig. 2) and for the control clone MN/55-Flu.
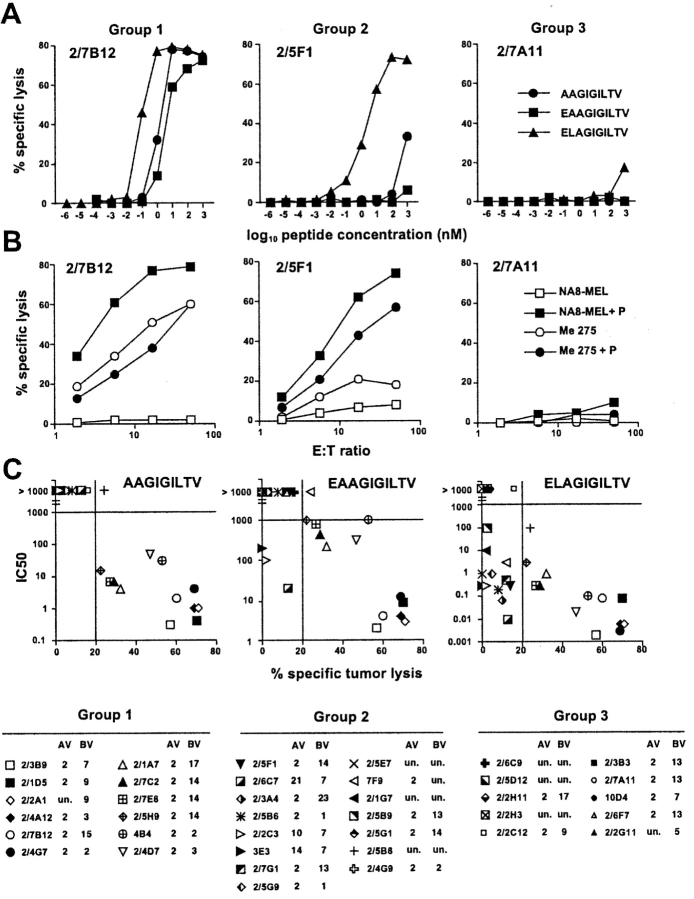
The clones exhibited a diverse BV usage, but a more restricted AV usage (most clones used AV 2, Fig. 2) suggesting some level of TCR structural conservation. All populations analyzed were clonally distinct as clones expressing the same AV and BV segments had different CDR3 regions (unpublished data). Avidity and fine specificity of antigen recognition was assessed by titrating Melan-A parental and analogue peptides in a functional CTL assay on T2 target cells (Fig. 2 A). Tumor recognition was similarly assessed on tumor cell targets expressing or not Melan-A (Fig. 2 B). Three groups of clones were clearly distinguishable according to their functional activity: (1) clones that recognized Melan-A 26–35 A27L analogue and Melan-A parental peptides and specifically lysed Melan-A–expressing tumor cells (e.g., clone 2/7B12) (2) clones that recognized Melan-A 26–35 A27L analogue peptide but only very poorly or not Melan-A parental peptides and failed to specifically lyse Melan-A–expressing tumor cells (e.g., clone 2/5F1) and (3) clones that failed to significantly recognize Melan-A 26–35 A27L analogue peptide and Melan-A parental peptides and failed to lyse Melan-A–expressing tumor cells (e.g., clone 2/7A11).
Figure 2.
Functional avidity of antigen recognition, fine specificity, and tumor-reactivity of ex vivo sorted A2/Melan-A multimer+ CD8+ T cell clones. Data are shown for one representative clone per group. (A) Clonal populations were tested for peptide recognition in chromium release assay using T2 cells as targets at a lymphocyte to target ratio of 10:1 in the presence of serial dilutions of the indicated peptide. (B) Tumor recognition was similarly assessed as the indicated lymphocyte to target cell ratios by using as target cells tumor cell lines Me275 (A2+, Melan-A+) and NA8-MEL (A2+, Melan-A−) in the absence or in the presence of peptide Melan-A 26–35 A27L (1 μM). (C) Correlation between avidity of antigen recognition and tumor reactivity of A2/Melan-A multimer+ clonal populations. Data obtained from the experiments illustrated in A and B are shown for 37 A2/Melan-A multimer+ clonal populations. The nM concentration of the indicated peptide which was required to obtain 50% of maximal lysis in peptide titration experiments (IC50) are shown in y-axis. The percent specific lysis on the Melan-A+ A2+ tumor line Me 275 obtained at an effector to target cell ratio of 50/1 in the absence of exogenously added peptide is shown in x-axis. Values >20% (bar on the X-axis) were considered as significant. Variable α and β (AV and BV) chain region usage of each clone is indicated. Un., unknown.
The proportion of the different groups was of ∼1/3 of the total population for each group. More precisely, 12 clones (32%) fell into group 1, 15 clones (41%) into group 2 and the remaining 10 (27%) into group 3. Avidity of antigen recognition clearly correlated with specific tumor lysis particularly in the case of the Melan-A nonapeptide 27–35 (Fig. 2 C) indicating that, as suggested by mass spectrometric analysis of naturally processed Melan-A peptides (28), this latter could be the main peptide species present on melanoma cells. However, the correlation between functional avidity of antigen recognition and intensity of multimer staining was less stringent (Fig. 3 A). Whereas a general tendency of clones displaying lower avidity (IC 50 > 100 nM) to stain less intensively was observed, exceptions were also found (i.e., clones 2/2C3 or 2/5B6 displayed an avidity at least three orders of magnitude higher than clones 2/5G1 or 2/2C12 but a similar or lower intensity of multimer staining).
In addition, among clones displaying an avidity of antigen recognition <10 nM, clones recognizing the antigen with a difference in avidity of up to three orders of magnitude could display comparable intensity of multimer staining. These results are consistent with our recent data in the Melan-A and in other antigenic systems indicating that the relative efficiency of staining of tumor antigen-specific T lymphocytes with the corresponding fluorescent A2/peptide multimers can considerably vary with staining conditions and does not necessarily correlate with avidity of antigen recognition (29). It is of note that specific staining, for clones belonging to the three groups and including those for which peptide recognition was not detectable (i.e., clone 2/7A11), was obtained with multimers incorporating Melan-A parental peptides albeit, at equivalent dose of multimers used, the staining was less intense, specially in the case of multimers incorporating the nonapeptide (Fig. 3 C). Thus, isolation of clones belonging to the third group was not due to the use of multimers incorporating the A27L analogue. For clones belonging to each of the three groups A2/Melan-A multimer staining was completely inhibited by anti-CD8 Abs (Fig. 3 B).
However, no difference in relative inhibition was seen among the clones irrespectively of the group they belonged to. Thus, CD8 clearly participated to the multimer binding, in agreement with what reported previously (30), but to a similar extent for clones of the three groups. These data clearly demonstrate that clones from group 3 were not isolated because binding A2/Melan-A multimers in a nonantigen-specific manner. The existence of this type of clones was not limited to HD 421 as they were also obtained in the case of other HLA-A2–expressing individuals namely another healthy donor (HD 009) and a melanoma patient (LAU 337, Table II). In the case of HD 009 we obtained results qualitatively similar to those described for HD 421. At variance with the latter, however, a higher proportion of clones was found in group 2 (67%) than in group 1 (11%) or group 3 (22%).
Table II.
Recognition of Natural Melan-A-related Sequences by A2/Melan-A Multimer+ CD8+ T Cell Clones from HD 009 and LAU 337
| HD 009
|
LAU 337 preimmune sample
|
LAU 337 postimmune sample
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Group 1(2 clones) | Group 2(12 clones) | Group 3(4 clones) | Group 1(8 clones) | Group 2(2 clones) | Group 3(3 clones) | Group 1(17 clones) | |||
| Clone | 18 | 10 | 17 | 3F8 | 1E2 | 3D31 | 2A5 | 1B5 | 2A9 |
| MSI 44 peptides | |||||||||
| Human | 1 | 5 | 0 | 2 | 0 | 1 | 8 | 5 | 0 |
| Viral | 2 | 3 | 1 | 5 | 2 | 0 | 19 | 19 | 1 |
| Bacterial | 8 | 2 | 2 | 4 | 2 | 5 | 14 | 9 | 0 |
| Total | 11 | 10 | 3 | 11 | 4 | 6 | 41 | 33 | 1 |
As reported previously (31), Melan-A multimer+ CD8+ T cells from melanoma patient LAU 337 (present among circulating CD8+ T cells at a frequency of 0.1%) exhibited a mixed phenotype composed of both CD45RAbright and CD45RAlow cells at roughly equivalent proportions. This mixed phenotype is found in ∼30% of melanoma patients, most likely as a consequence of a spontaneous response to the tumor, whereas in the remaining 70% Melan-A multimer+ CD8+ T cells exhibit a naive phenotype similarly to healthy donors. Interestingly, upon immunization with peptide Melan-A 26–35 a highly increased frequency of CD45RAlow Melan-A multimer+ T cells (2%) was detectable among circulating CD8+ T cells. Melan-A multimer+ CD8+ T cell clones were isolated by ex vivo sorting of both preimmune and postimmune samples. Out of a total of 13 clones obtained from the preimmune sample, the majority (8) belonged to group 1, 2 clones to group 2, and the remaining 3 to group 3. The increased proportion of clones in group 1 was in good agreement with the antigen experience phenotype of part of the starting population. Also, all Melan-A multimer+ T cell clones obtained from the postimmune samples (a total of 17) fell into group 1.
A2/Melan-A Multimer+ Clones that Fail to Recognize Melan-A Peptides Are Able to Recognize Peptides from Protein of Self- or Pathogen-Origin Selected on the Basis of Their Potential Functional Cross-reactivity with Melan-A.
The molecular basis for the inability of clones from group 3 to recognize Melan-A peptides was investigated by testing the recognition, by 18 of the 37 clones from HD 421 (including 13 clones from the third group), of a panel of 71 natural Melan-A-related peptides selected on the basis of their potential functional cross-reactivity with Melan-A. These sequences were retrieved by using biometric analysis (32) of data previously generated by screening of positional scanning synthetic combinatorial peptide libraries with a Melan-A specific clone and used to identify within proteins contained in a public database sequences with the predicted highest stimulatory values (reference 33, and unpublished data). As summarized in Table I, all but one of the clones tested recognized at least one or multiple peptides, to different extent. Similar results were obtained with clones from HD 009 and LAU 337 (Table II).
Table I.
Recognition of Natural Melan-A–related Sequences by A2/Melan-A Multimer+ CD8+ T Cell Clones from HD 421 (Continued)a
| Group 1 | Group 2 | Group 3 | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide code | Species | Protein | Sequence | 2/4A12 | 2/7B12 | 2/4G7 | 2/5F1 | 2/3A4 | 2/7E8 | 2/3B3 | 2/7A11 | 2/4G9 | 2/6F7 | 2/2G11 | 2/5B9 | 2/6C9 | 10D4 | 2/2C12 | 2/5B8 | 2/5D12 | 2/2H11 | nb positiveb |
| Human | ||||||||||||||||||||||
| MSI 44 -1 |
Homo sapiens | KIAA0935 | RVTDEAGHPV | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | 1 |
| -2 | Homo sapiens | MRP3 | NVADIGLHDV | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | 1 |
| -5 | Homo sapiens | SLC1A1 or EAAT3 or EAAC1 | VLTGLAIHSI | +b | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 2 |
| -9 | Homo sapiens | P47 | RISDIRLFIV | + | − | ++ | − | ++ | − | − | − | − | − | − | + | − | − | ++ | ++++ | − | − | 6 |
| -10 | Homo sapiens | Prostaglandin transporter | LLAGIGTVPI | ++++ | + | +++ | + | ++++ | − | +++ | +++ | − | − | − | − | − | − | +++ | ++++ | +++ | − | 10 |
| -11 | Homo sapiens | ABC transporter MOAT-C | RISDIGLADL | − | − | + | − | − | − | − | − | − | − | − | − | − | − | + | +++ | − | − | 3 |
| -12 | Homo sapiens | KIAA0735 | LISGIGIGGA | − | − | − | − | − | ++++ | − | − | − | − | − | − | − | + | +++ | ++++ | − | + | 5 |
| -13 | Homo sapiens | Hypothetical 20 kD protein |
RISAIILHPN | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | 1 |
| -14 | Homo sapiens | Endothelin-1 receptor |
RVQGIGIPLV | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | 1 |
| -15 | Homo sapiens | G-protein coupled receptor RE2 | RITDLGLSPH | ++ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -17 | Homo sapiens | IGHG1 | RLSELAIFGV | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 1 |
| -18 | Homo sapiens | Monocarboxylate transporter 8 | AVAFIGLHTS | ++ | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | ++ | 3 |
| -21 | Homo sapiens | MRP3 | NVADIGFHDV | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 1 |
| -22 | Homo sapiens | Melan-A/Mart-1 | EAAGIGILTV | ++++ | ++++ | +++ | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | 5 |
| Viral | ||||||||||||||||||||||
| -23 | Herpes simplex virus | Capsid protein P40 | RQAGIAGHTY | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -24 | Little cherry closterovirus | MT and HEL domains |
RVSNIAIATG | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 2 |
| -25 | Pseudorabies virus | Glycoprotein GIII | VIAGIGILAI | ++++ | ++ | +++ | + | ++ | ++ | + | ++ | − | − | − | − | − | − | − | − | − | +++ | 9 |
| -26 | Pseudorabies virus | Glycoprotein C | VIAGIAILAI | + | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | 2 |
| -27 | Human rotavirus | Outer capsid protein VP4 |
RLSGIYGLPA | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 1 |
| -28 | Human rotavirus | Outer capsid protein VP4 |
RMSGIYGLPA | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 2 |
| -29 | Canine calicivirus | Capsid protein | NTTDIGIHVV | − | − | ++ | − | − | − | − | − | − | − | − | − | − | − | + | − | − | + | 3 |
| -30 | Variola virus | (XHOI-F, O, H, P, Q) Genes |
MIAGIGISLI | ++++ | ++ | +++ | + | +++ | − | + | ++ | − | − | − | − | − | − | +++ | ++ | + | ++++ | 11 |
| -31 | Bovine herpesvirus type 1 | Capsid protein P40 | REAGIAGHTY | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | + | 2 |
| -32 | Bovine herpesvirus type 2 | DNA-dependent DNA polymerase | RLAGIGLTRA | + | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | − | + | 5 |
| -33 | Vaccinia virus | Protein A49 | RIADIDIKQV | ++ | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | + | 3 |
| -35 | Tobacco necrosis virus | RNA-directed RNA polymerase | RYSGIGGHLL | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 2 |
| -37 | Puma lentivirus 14 | GAG polyprotein | RITGICFHFG | ++ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -38 | Murine cytomegalovirus | Helicase/primase complex protein | RLAGILDHTL | + | − | ++ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 2 |
| -39 | Human cytomegalovirus | Hypothetical protein HVLF2 |
RIAGLLLFQI | ++ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -40 | Mumps virus | Fusion glycoprotein | RFAGIAIGIA | ++ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -41 | Human parainfluenza virus 1 | L protein | RVRGIGIPEV | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -42 | SINP virus | Hypothetical 36.5 kD protein |
CTSIIGIFPV | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | +++ | − | − | 1 |
| -45 | TT virus | Hypothetical 15.9 kD protein |
RAPSIGILPA | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -46 | Human adenovirus type 12 | DNA terminal protein |
RQADIPLPPL | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -47 | Nilaparvata lugens reovirus | 23.6 kD putative nonstructural protein |
NFAGIAILFI | ++ | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | 2 |
| Bacterial | ||||||||||||||||||||||
| -48 | Chlorobium tepidum | Hypothetical 22.8 kD protein |
RLSGHGIHPV | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -49 | Bacillus subtilis | YKOR | RVASIGLHPS | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | 1 |
| -52 | Bacillus subtilis | Hypothetical 48.9 kD protein |
LLAGLAIFPA | − | − | ++ | − | − | − | − | − | − | − | − | − | − | − | +++ | − | − | − | 2 |
| -53 | Rickettsia prowazekii | Cell division protein FTSK homolog | RLSLIGLFPI | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ++ | − | − | 1 |
| -54 | Propionigenium modestum | ATP synthase beta chain |
RIASLGIYPA | ++ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -55 | Aeromonas salmonicida | EPSP synthase | RVTGIGKHSI | + | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 2 |
| -56 | Chlamydia trachomatis | Arginine/ornithine antiporter | MLSGIGIFFI | ++++ | − | +++ | ++ | − | ++++ | ++++ | ++++ | + | + | − | − | − | − | ++++ | ++++ | ++ | − | 11 |
| -57 | Nostoc punctiforme | Carboxyl terminal protease | TRANIAIHPV | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -58 | Streptomyces coelicolor | Putative secreted protein | VLSSIGIFPI | + | ++ | + | − | − | − | ++ | +++ | − | − | +++ | − | − | − | − | ++++ | + | − | 8 |
| -60 | Synechococcus sp. | REPA | RVTGIGLLTG | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -62 | Bacillus pumilus | Anthranilate synthase component I | LVAGIAIGPF | ++ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -63 | Salmonella typhimurium | LTKB homolog | RIADIPIFII | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -64 | Burkholderia cepacia | Hypothetical 23.3 kD protein |
SIADIAIYPW | +++ | − | + | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | 3 |
| -65 | Aquifex geolicus | ATP synthase beta chain |
RLAELGIYPA | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -68 | E. coli | ATP-dependent protease LA | VIADLDIHPV | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| -69 | Azospirillum brasiliense | Glutamate synthase (NADPH) large chain | RISGIGLNGI | + | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | 2 |
| -70 | Rhizobium meliloti | RHSC protein | LIAGHGIHPC | ++ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 2 |
| -71 | Rhizobium sp. | Y4FN probable ABC transporter permease | RSAFIGIDPA | +++ | − | ++ | ++ | − | − | − | − | − | − | − | − | − | − | − | − | − | +++ | 4 |
| -72 | E. coli | K(+)/H(+) antiporter |
LLAGIAIGPW | +++ | − | + | ++ | − | − | − | − | − | − | − | − | − | − | − | − | − | +++ | 4 |
Number of clones recognizing the indicated peptide.
Peptide recognition was assessed in a CTL assay in the presence of 1 mM of the indicated peptide. + > 10% specific lysis, ++ > 20% specific lysis, +++ > 40% specific lysis, ++++ > 60% specific lysis.
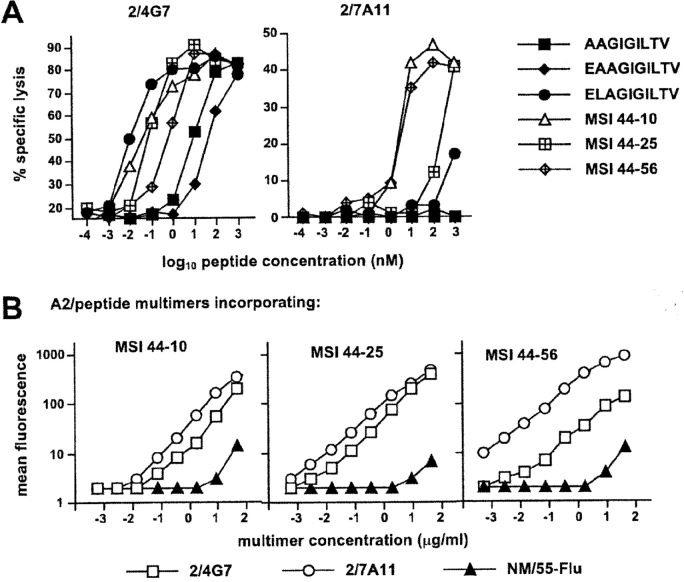
It is noteworthy that clones from each group, including the ones showing little or no peptide recognition, were able to exert efficient lytic activity in redirected lysis experiments (unpublished data). The extent of cross-reactivity between Melan-A and related sequences was sometimes substantial and encompassed tumor-reactive clones. For example, the tumor-reactive clone 2/4G7 was able to recognize some peptides two orders of magnitude more efficiently than Melan-A parental peptides (Fig. 4 A). In addition, Melan-A multimer+ clones that failed to recognize Melan-A parental peptides could recognize some Melan-A–related peptides with relatively good efficiency (i.e., clone 2/7A11 Fig. 4 A).
Figure 4.
Cross-reactivity between Melan-A and natural Melan-A–related peptides. (A) Relative efficiency of recognition of selected natural Melan-A–related peptides by clone 2/4G7 (group 1) and clone 2/7A11 (group 3) was assessed in chromium release assay in the presence of graded dilutions of the indicated peptides. (B) Cross-staining of the above clones and of clone MN/55-Flu used as an internal control by A2/peptide multimers incorporating selected Melan-A–related peptides (peptide MSI 44–10 from prostaglandin transporter, LLAGIGTVPI; MSI 44–25 from Pseudorabies virus, VIAGIGILAI; or MSI 44–56 from Chlamydia trachomatis, MLSGIGIFFI) was assessed by staining cells during 1 h at room temperature with serial dilutions of the indicated multimer.
Interestingly, some peptides (i.e., peptides MSI 44–10 from prostaglandin transporter, MSI 44–25 from Pseudorabies virus or MSI 44–56 from Chlamydia trachomatis) were recognized by the majority of the clones. Cross-reactivity was also clear in terms of multimer staining: indeed, A2/peptide multimers incorporating some among the most frequently recognized Melan-A–related peptides were able to specifically stain both clone 2/4G7 and clone 2/7A11 (Fig. 4 B). Because of the degenerate recognition of some of the natural Melan-A–related peptides by the majority of the Melan-A multimer+ clones and because of the ability of A2/peptide multimers incorporating those peptides to cross-stain Melan-A multimer+ clones belonging to different groups, we speculated that a significant proportion of circulating Melan-A multimer+ CD8+ T cells might be cross-stained by multimers incorporating natural Melan-A-related peptides. In agreement with this hypothesis, A2/MSI 44–56 multimers specifically stained a frequency of circulating naive CD8+ T cells from HD 421 similar to that stained by Melan-A multimers (Fig. 5) . Similar results were obtained with multimers incorporating other frequently recognized peptides (i.e., MSI 44–10, MSI 44–25) albeit the proportion of the multimer+ population varied depending on both the peptide and the dose of multimer used (unpublished data).
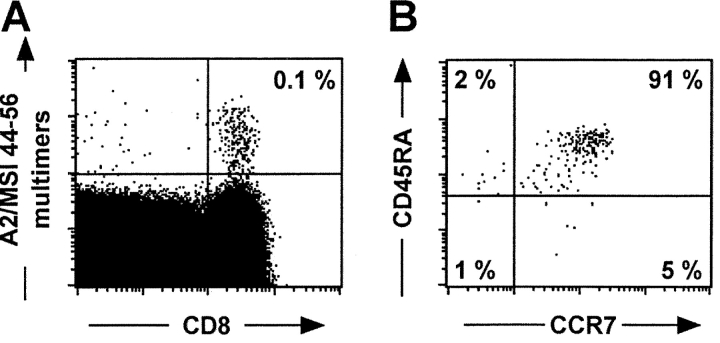
Figure 5.
Ex vivo detection and phenotyping of circulating A2/MSI 44–56 multimer+ CD8+ T cells in HD 421. PBMCs from healthy donor HD 421 were stained with A2/peptide multimers containing MSI 44–56 together with other mAbs as in Fig. 1. Dot plots are shown for CD8 versus multimer staining (A) or for CD45RA versus CCR7 staining on gated CD8+ A2/Melan-A multimer+ cells (B).
Discussion
This study represents the first functional dissection of a normal self-antigen specific naive CD8+ T cell repertoire. Because of the nature of the antigen under study the results of this analysis are relevant for the understanding of immune responses to tumors in humans. Ex vivo isolated Melan-A multimer+ T cell clones displayed a large heterogeneity in terms of avidity of antigen recognition and tumor reactivity. Tumor-reactive clones (that represented a minority of the total population) were among the ones displaying higher avidity of recognition of Melan-A parental peptides. The latter, however, was generally low (in the group of highly tumor reactive clones, average IC50 at ∼1 nM for Melan-A 27–35) as compared with CTL specific for virally derived antigens (IC50 often in the pM range). Clones that recognized the parental peptides with lower avidity generally failed to significantly recognize antigen-expressing tumor cells. These data are consistent with the idea of the T cell repertoire specific for this self-antigen being a residual repertoire, the large majority of high avidity self-reactive T cells being most likely deleted in the thymus.
A surprising and unprecedented finding was that a significant proportion of ex vivo sorted Melan-A multimer+ T cell clones failed to significantly recognize the antigenic peptides. However, because of the ability of multimers to bind T cells bearing specific TCR irrespectively of their function, dissociation between multimer staining and T cell function is predictable, and has already been documented by us and others (29, 34, 35). Whereas in some cases the observed dissociation was to be attributed to an inherent inability of multimer+ cells to exert effector functions (anergy; reference 34), it appears that, more in general, the efficiency of multimer staining may be primarily affected by the kinetics of TCR–ligand interaction (29, 34, 35).
Among different possibilities, we favored the hypothesis that clones from group 3 could display some degree of specificity for Melan-A peptides (sufficient to bind A2/Melan-A multimers) but a functional avidity too low to be detectable in a functional assay.
This type of T cells could have been undetected in previous studies as, even when using multimers, specific T cells are generally isolated either ex vivo or in vitro from populations that have already undergone antigen-driven selection. Indeed, if such clones could well be a normal component of any antigen specific naive repertoire, it is likely that, because of their unresponsiveness to antigen, they would not normally become part of the memory pool. They could however, at least in principle, be stimulated by cross-reactive sequences of pathogen or self-origin such as some of the ones identified here. Thus, staining by multimers could be, in some cases, highly specific but physiologically irrelevant. These considerations further underline the need of routinely associating a complete analysis of T cell functions to multimer staining.
In agreement with our hypothesis most Melan-A multimer+ clones, including those unable to recognize Melan-A peptides in functional assay, efficiently recognized several Melan-A-related peptides selected on the basis of their potential functional cross-reactivity with Melan-A. Reciprocally, A2/peptide multimers incorporating some of the most frequently recognized peptides specifically stained a proportion of naive CD8+ T cells comparable to A2/Melan-A peptide multimers. These data suggest that TCR from the majority of clones able to recognize Melan-A-related natural peptides could be structurally related with those able to recognize Melan-A parental peptides. Further support for this hypothesis issues from the finding that the majority of these clones use several TCR BV regions but a conserved AV region (unpublished data). Altogether, these data indicate that, due to the degeneracy of Melan-A antigen recognition, largely cross-reactive subsets of T cells displaying multiple and overlapping specificities but sharing common structural features contribute to form the abundant repertoire of Melan-A multimer+ CD8+ T cells.
Degeneracy of antigen recognition by Melan-A–specific T cells was reported previously (36) and proposed to be at the origin of the high frequency of Melan-A–specific T cells in the periphery. The proposed molecular mechanism was molecular mimicry, i.e., CTL responses to Melan-A might be augmented by T cell encounters with Melan-A–like peptides derived from sources other than Melan-A and most likely of pathogen origin. The same authors later reported that several Melan-A analogs of self-origin showed antagonistic behavior toward Melan-A (37). Based on this observation they proposed that encounters with self-peptide analogs of Melan-A may contribute to the peripheral maintenance of these CTL, while ultimately impairing the efficacy of antitumor T cell responses.
Both our data and their interpretation, however, are clearly at variance with those previous reports in that (a) degeneracy of antigen recognition of Melan-A like sequences is larger than anticipated and encompasses clones that are specifically stained by multimers incorporating Melan-A peptides but fail to recognize the latter in functional assays; and (b) the large majority of both Melan-A multimer+ T cells as well as T cells specifically stained by A2/peptide multimers incorporating the most frequently recognized peptides from pathogen origin are naive. Together with the observation that the proportion of Melan-A multimer+ T cells is similar in the thymus and in the periphery these data indicate that, although it cannot formally be excluded that some pathogen derived peptide could stimulate in vivo specific responses cross-reactive with Melan-A, this would not be a frequent event, and molecular mimicry would not account for the high frequency of Melan-A–specific T cells. Also, no peripheral tolerance is observed, and Melan-A–specific precursors can be efficiently activated both in vitro and in vivo by stimulation with antigen.
It is likely that Melan-A–related peptides of self-origin described here only represent a small subgroup of the existing ones. Why, in the presence of this large array of cross-reactive self-peptides the Melan-A multimer+ T cells remain mostly inactive? The natural self-peptides recognized in vitro by Melan-A multimer+ T cells with an increased or comparable efficiency than Melan-A parental peptides are most likely not efficiently generated by intracellular processing as their presence in the thymus would result in T cell deletion. In contrast, cross-reactive peptides recognized in vitro with very low to undetectable avidity could be among the ones that positively select Melan-A multimer+ T cells in the thymus. A similar set of peptides could be present in the periphery and provide to Melan-A multimer+ T cells activation signals that are sufficient for their survival but not for their activation.
In conclusion, the repertoire of Melan-A multimer+ T cells would differ from any other antigen specific repertoire only in terms of size possibly due to the high frequency of Melan-A–related sequences in proteins (36) and therefore to the high proportion of TCR able to interact with those sequences. The existence of such a degenerate recognition of Melan-A–like sequences by a significant proportion of different but structurally related TCRs also indicates that a large proportion of self-peptides could contribute to the repertoire selection of Melan-A multimer+ cells in the thymus as well as to their maintenance in the periphery.
Both the nature and the number of Melan-A cross-reactive sequences present on thymic APCs as well as their role in negative and positive selection of Melan-A multimer+ cells remain to be determined. An interesting implication of the results of this study is that the frequency of T cells specific for a self-antigen in the naive repertoire could be determined by the degree of structural proximity between the antigenic peptide and the pool of self-peptides accessible to the immune system. Because of the clear implications in the study of autoimmune diseases as well as of tumor-specific responses, this hypothesis deserves further investigation.
Acknowledgments
We would like to thank Dr. Donata Rimoldi for providing the melanoma lines and for helpful discussions, N. Montandon for excellent technical assistance, and M. van Overloop for assistance with manuscript preparation.
V. Dutoit was supported by a grant from Mixture Science. V. Rubio-Godoy was supported by the Swiss Cancer League grant SLK782-2-1999.
References
- 1.Boel, P., C. Wildmann, M.L. Sensi, R. Brasseur, J.C. Renauld, P. Coulie, T. Boon, and P. van der Bruggen. 1995. BAGE: a new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity. 2:167–175. [DOI] [PubMed] [Google Scholar]
- 2.Van den Eynde, B., O. Peeters, O. De Backer, B. Gaugler, S. Lucas, and T. Boon. 1995. A new family of genes coding for an antigen recognized by autologous cytolytic T lymphocytes on a human melanoma. J. Exp. Med. 182:689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Bruggen, P., C. Traversari, P. Chomez, C. Lurquin, E. De Plaen, B. Van den Eynde, A. Knuth, and T. Boon. 1991. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 254:1643–1647. [DOI] [PubMed] [Google Scholar]
- 4.Brichard, V., A. Van Pel, T. Wolfel, C. Wolfel, E. De Plaen, B. Lethe, P. Coulie, and T. Boon. 1993. The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J. Exp. Med. 178:489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coulie, P.G., V. Brichard, A. Van Pel, T. Wolfel, J. Schneider, C. Traversari, S. Mattei, E. De Plaen, C. Lurquin, J.P. Szikora, et al. 1994. A new gene coding for a differentiation antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J. Exp. Med. 180:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox, A.L., J. Skipper, Y. Chen, R.A. Henderson, T.L. Darrow, J. Shabanowitz, V.H. Engelhard, D.F. Hunt, and C.L. Slingluff, Jr. 1994. Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science. 264:716–719. [DOI] [PubMed] [Google Scholar]
- 7.Stockinger, B. 1999. T lymphocyte tolerance: from thymic deletion to peripheral control mechanisms. Adv. Immunol. 71:229–265. [DOI] [PubMed] [Google Scholar]
- 8.Miller, J.F.A.P., and W.R. Heath. 1993. Self-ignorance in the peripheral T-cell pool. Immunol. Rev. 133:131–150. [DOI] [PubMed] [Google Scholar]
- 9.Derbinski, J., A. Schulte, B. Kyewski, and L. Klein. 2001. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat. Immunol. 2:1032–1039. [DOI] [PubMed] [Google Scholar]
- 10.Ashton-Rickardt, P.G., L. Van Kaer, T.N.M. Schumacher, H.L. Ploegh, and S. Tonegawa. 1993. Peptide contributes to the specificity of positive selection of CD8+ T cells in the thymus. Cell. 73:1041–1049. [DOI] [PubMed] [Google Scholar]
- 11.Ernst, B., D.S. Lee, J.M. Chang, J. Sprent, and C.D. Surh. 1999. The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity. 11:173–181. [DOI] [PubMed] [Google Scholar]
- 12.Bouneaud, C., P. Kourilsky, and P. Bousso. 2000. Impact of negative selection on the T cell repertoire reactive to a self-peptide: a large fraction of T cell clones escapes clonal deletion. Immunity. 13:829–840. [DOI] [PubMed] [Google Scholar]
- 13.Surh, C.D., D.S. Lee, W.P. Fung-Leung, L. Karlsson, and J. Sprent. 1997. Thymic selection by a single MHC/peptide ligand produces a semidiverse repertoire of CD4+ T cells. Immunity. 7:209–219. [DOI] [PubMed] [Google Scholar]
- 14.Ashton-Rickardt, P.G., A. Bandeira, J.R. Delaney, L. Van Kaer, H.P. Pircher, R.M. Zinkernagel, and S. Tonegawa. 1994. Evidence for a differential avidity model of T cell selection in the thymus. Cell. 76:651–663. [DOI] [PubMed] [Google Scholar]
- 15.Hogquist, K.A., M.A. Gavin, and M.J. Bevan. 1993. Positive selection of CD8+ T cells induced by major histocompatibility complex binding peptides in fetal thymic organ culture. J. Exp. Med. 177:1469–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogquist, K.A., S.C. Jameson, W.R. Heath, J.L. Howard, M.J. Bevan, and F.R. Carbone. 1994. T cell receptor antagonist peptides induce positive selection. Cell. 76:17–27. [DOI] [PubMed] [Google Scholar]
- 17.Mason, D. 1998. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today. 19:395–404. [DOI] [PubMed] [Google Scholar]
- 18.Altman, J.D., P.A.H. Moss, P.J.R. Goulder, D.H. Barouch, M.G. McHeyzer-Williams, J.I. Bell, A.J. McMichael, and M.M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science. 274:94–96. [DOI] [PubMed] [Google Scholar]
- 19.Valmori, D., M.J. Pittet, C. Vonarbourg, D. Rimoldi, D. Lienard, D. Speiser, R. Dunbar, V. Cerundolo, J.C. Cerottini, and P. Romero. 1999. Analysis of the cytolytic T lymphocyte response of melanoma patients to the naturally HLA-A*0201-associated tyrosinase peptide 368-376. Cancer Res. 59:4050–4055. [PubMed] [Google Scholar]
- 20.Kawakami, Y., S. Eliyahu, C.H. Delgado, P.F. Robbins, K. Sakaguchi, E. Appella, J.R. Yannelli, G.J. Adema, T. Miki, and S.A. Rosenberg. 1994. Identification of a human melanoma antigen recognized by tumor-infiltrating lymphocytes associated with in vivo tumor rejection. Proc. Natl. Acad. Sci. USA. 91:6458–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pittet, M.J., D. Valmori, P.R. Dunbar, D.E. Speiser, D. Lienard, F. Lejeune, K. Fleischhauer, V. Cerundolo, J.C. Cerottini, and P. Romero. 1999. High frequencies of naive Melan-A/MART-1-specific CD8+ T cells in a large proportion of human histocompatibility leukocyte antigen (HLA)-A2 individuals. J. Exp. Med. 190:705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zippelius, A., M.J. Pittet, P. Batard, N. Rufer, M. de Smedt, P. Guillaume, K. Ellefsen, D. Valmori, D. Liénard, J. Plum, et al. 2002. Thymic selection generates a large T cell pool recognizing a self-peptide in humans. J. Exp. Med. 195:485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valmori, D., M.J. Pittet, D. Rimoldi, D. Lienard, R. Dunbar, V. Cerundolo, F. Lejeune, J.C. Cerottini, and P. Romero. 1999. An antigen-targeted approach to adoptive transfer therapy of cancer. Cancer Res. 59:2167–2173. [PubMed] [Google Scholar]
- 24.Salter, R.D., J. Alexander, F. Levine, D. Pious, and P. Cresswell. 1985. Evidence for two trans-acting genes regulating HLA class II antigen expression. J. Immunol. 135:4235–4238. [PubMed] [Google Scholar]
- 25.Romero, P., P.R. Dunbar, D. Valmori, M. Pittet, G.S. Ogg, D. Rimoldi, J.-L. Chen, D. Liénard, J.C. Cerottini, and V. Cerundolo. 1998. Ex vivo staining of metastatic lymph nodes by Class I major histocompatibility complex tetramers reveals high numbers of antigen-experienced tumor-specific cytolytic T lymphocytes. J. Exp. Med. 188:1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalergis, A.M., E.C. Goyarts, E. Palmieri, S. Honda, W. Zhang, and S.G. Nathenson. 2000. A simplified procedure for the preparation of MHC/peptide tetramers: chemical biotinylation of an unpaired cysteine engineered at the C-terminus of MHC-I. J. Immunol. Methods. 234:61–70. [DOI] [PubMed] [Google Scholar]
- 27.Pittet, M.J., D. Speiser, D. Liénard, D. Valmori, P. Guillaume, V. Dutoit, D. Rimoldi, F. Lejeune, J.-C. Cerottini, and P. Romero. 2001. Expansion and functional maturation of human tumor antigen-specific CD8+ T cells following vaccination with antigenic peptide. Clin. Cancer Res. 7:7965–8035. [PubMed] [Google Scholar]
- 28.Skipper, J.C., P.H. Gulden, R.C. Hendrickson, N. Harthun, J.A. Caldwell, J. Shabanowitz, V.H. Engelhard, D.F. Hunt, and C.L. Slingluff, Jr. 1999. Mass-spectrometric evaluation of HLA-A*0201-associated peptides identifies dominant naturally processed forms of CTL epitopes from MART-1 and gp100. Int. J. Cancer. 82:669–677. [DOI] [PubMed] [Google Scholar]
- 29.Dutoit, V., V. Rubio-Godoy, M.A. Doucey, P. Batard, D. Lienard, D. Rimoldi, D. Speiser, P. Guillaume, J.C. Cerottini, P. Romero, and D. Valmori. 2002. Functional avidity of tumor antigen-specific CTL recognition directly correlates with the stability of MHC/peptide multimer binding to TCR. J. Immunol. 168:1167–1171. [DOI] [PubMed] [Google Scholar]
- 30.Daniels, M.A., and S.C. Jameson. 2000. Critical role for CD8 in T cell receptor binding and activation by peptide/major histocompatibility complex multimers. J. Exp. Med. 191:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valmori, D., V. Dutoit, V. Schnuriger, A.L. Quiquerez, M.J. Pittet, P. Guillaume, V. Rubio-Godoy, P.R. Walker, D. Rimoldi, D. Lienard, et al. 2002. Vaccination with a Melan-A peptide selects an oligoclonal T cell population with increased functional avidity and tumor reactivity. J. Immunol. 168:4231–4240. [DOI] [PubMed] [Google Scholar]
- 32.Zhao, Y., B. Gran, C. Pinilla, S. Markovic-Plese, B. Hemmer, A. Tzou, L.W. Whitney, W.E. Biddison, R. Martin, and R. Simon. 2001. Combinatorial peptide libraries and biometric score matrices permit the quantitative analysis of specific and degenerate interactions between clonotypic TCR and MHC peptide ligands. J. Immunol. 167:2130–2141. [DOI] [PubMed] [Google Scholar]
- 33.Pinilla, C., V. Rubio-Godoy, V. Dutoit, P. Guillaume, R. Simon, Y. Zhao, R.A. Houghten, J.C. Cerottini, P. Romero, and D. Valmori. 2001. Combinatorial peptide libraries as an alternative approach to the identification of ligands for tumor-reactive cytolytic T lymphocytes. Cancer Res. 61:5153–5160. [PubMed] [Google Scholar]
- 34.Lee, P.P., C. Yee, P.A. Savage, L. Fong, D. Brockstedt, J.S. Weber, D. Johnson, S. Swetter, J. Thompson, P.D. Greenberg, et al. 1999. Characterization of circulating T cells specific for tumor-associated antigens in melanoma patients. Nat. Med. 5:677–685. [DOI] [PubMed] [Google Scholar]
- 35.Rubio-Godoy, V., V. Dutoit, D. Rimoldi, D. Lienard, F. Lejeune, D. Speiser, P. Guillaume, J.C. Cerottini, P. Romero, and D. Valmori. 2001. Discrepancy between ELISPOT IFN-γ secretion and binding of A2/peptide multimers to TCR reveals interclonal dissociation of CTL effector function from TCR-peptide/MHC complexes half-life. Proc. Natl. Acad. Sci. USA. 98:10302–10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loftus, D.J., C. Castelli, T.M. Clay, P. Squarcina, F.M. Marincola, M.I. Nishimura, G. Parmiani, E. Appella, and L. Rivoltini. 1996. Identification of epitope mimics recognized by CTL reactive to the melanoma/melanocyte-derived peptide MART-1(27–35). J. Exp. Med. 184:647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loftus, D.J., P. Squarcina, M.B. Nielsen, C. Geisler, C. Castelli, N. Odum, E. Appella, G. Parmiani, and L. Rivoltini. 1998. Peptides derived from self-proteins as partial agonists and antagonists of human CD8+ T-cell clones reactive to melanoma/melanocyte epitope MART1(27–35). Cancer Res. 58:2433–2439. [PubMed] [Google Scholar]