Abstract
The ParA and ParB protein families are well conserved in bacteria. However, their functions are still unclear. In Bacillus subtilis, Soj and Spo0J are members of these two protein families, respectively. A previous report revealed that replication initiated early and asynchronously in spo0J null mutant cells, as determined by flow cytometry. In this study, we examined the cause of this promotion of replication initiation. Deletion of both the soj and spo0J genes restored the frequency of replication initiation to almost the wild-type level, suggesting that production of Soj in the absence of Spo0J leads to early and asynchronous initiation of replication. Consistent with this suggestion, overproduction of Soj in wild-type cells had the same effect on replication initiation as in the spo0J null mutant, and overproduction of both Soj and Spo0J did not. These results indicate that when the ratio of Soj to Spo0J increases, Soj interferes with tight control of replication initiation and causes early and asynchronous initiation. Whereas replication initiation also occurred significantly earlier in the two spo0J mutants, spo0J14 and spo0J17, it occurred only slightly early in the sojK16Q mutant and was delayed in the sojG12V mutant. Although Soj localized to nucleoids in the spo0J mutants, the two Soj mutant proteins were distributed throughout the cell or localized to cell poles. Thus, interestingly, the promotion of replication initiation seems to correlate with localization of Soj to nucleoids. This may suggest that Soj inhibits transcription of some cell cycle genes and leads to early and asynchronous initiation of replication. In wild-type cells Spo0J counteracts this Soj function.
The ParA and ParB protein families are widely conserved in bacteria and plasmids (47). These proteins were first analyzed in low-copy-number plasmids (including prophage) of Escherichia coli, such as F and P1, and were found to be essential for accurate partitioning of the plasmids (9, 17). The P1 ParA protein binds both ADP and ATP (7). The ATP-bound form interacts with the ParB-parS complex (parS:ParB-binding DNA sequence) and plays a direct role in partitioning (4). ParB stimulates a weak ATPase activity of ParA (8) and changes the ATP form to the ADP form that cannot interact with the ParB-parS complex. Thus, ParA-ADP is released from the ParB-parS complex and acts as repressor of the parAB operon by binding to the promoter region (4). The ParB protein was detected as foci within the cell, and its localization depended on ParA and parS (11), as expected from the biochemical data. Using a green fluorescent protein (GFP)-ParB fusion, Li and Austin (28) recently showed that P1 plasmid copies are suddenly separated into two daughter cells from the cell center immediately before cell division.
In the case of the F plasmid, the biochemical activities of the ParA and ParB family members (SopA and SopB) are very similar to those of the P1 proteins (17). The cis-acting sopC region including the SopB-binding sequences was required for localization of SopA and SopB (18). Plasmid molecules were localized at the cell center or the one-quarter and three-quarter positions of cells, and interestingly, plasmid molecules lacking this partitioning system were distributed randomly in cytosolic spaces (38). Finally, the sopABC system stabilized an otherwise unstable oriC plasmid and changed its distribution pattern from random distribution in cytosolic spaces to localization at the mid-cell or the one-quarter and three-quarter positions (39). These cytological data suggest that the parABS system recruits plasmid molecules to a specific position in the cell.
In contrast to the obvious contribution of the ParA and ParB protein families to plasmid partitioning, the functions of bacterial chromosome partitioning counterparts are still ambiguous. In E. coli, neither ParA nor ParB family members are present. In Bacillus subtilis, Soj and Spo0J are members of the ParA and ParB families, respectively, and functions of members of these protein families in bacteria were first analyzed in this organism (20). The spo0J null mutants produced anucleate cells, suggesting that Spo0J is involved in chromosome partitioning, as expected from studies of plasmid-encoded ParA and ParB. However, the frequency of anucleate cells was relatively low, and a later report claimed that the most prominent phenotypic effect in spo0J null mutants was the formation of elongated diffuse nucleoids (1). Spo0J was also involved in correct orientation of the replication origin (oriC) region of the chromosome during an early stage of sporulation (44). However, in a recent report the authors suggested that the random positioning of the oriC region in spo0J null mutant cells may be an indirect effect of asynchronous initiation of replication in the mutant cells (26). In contrast to chromosome partitioning effects, other effects on sporulation are relatively clear. Sporulation defects in spo0J null mutants are due to inhibition of transcription of several early sporulation genes by Soj (6, 43), and the defects are actually suppressed by deletion of soj (20). Thus, Soj acts as a transcriptional repressor, similar to plasmid-coded ParA, and Spo0J counteracts its function in the wild-type cells. Spo0J binds at least eight binding sites (parS) located in the origin-proximal 20% of the chromosome (30) and is localized near the cell quarter positions (13, 29, 45). Based on analogy with the parABS system of plasmids, the Spo0J-parS complex may function to recruit the chromosomal region containing the parS repeats to the cell quarters. However, recently, Lee et al. (26) clearly ruled out this possibility. These authors found that Spo0J-GFP fluorescence was no longer located predominantly in the cell quarters when an array of parS sites was inserted at various chromosomal locations in the absence of six of the eight native parS sites. Thus, they raised another possibility, that the parABS system functions differently on plasmids and chromosomes. Indeed, when the B. subtilis Soj-Spo0J-parS system was inserted into an unstable mini-F plasmid, it stabilized and localized the plasmid to the cell quarters in E. coli cells (47).
In contrast to the situation in B. subtilis, both parA and parB are essential for cell growth in Caulobacter crescentus (33). On the other hand, like B. subtilis, ParB binds to several parS sequences in the oriC region, including a sequence downstream of the parAB operon (9, 33). Both ParA and ParB showed polar localization in predivisional cells, while in B. subtilis Soj moved dynamically between cell poles or nucleoids (32, 42). Overproduction of either ParA or ParB in C. crescentus led to significant defects in chromosome partitioning and cell division (33). In addition, depletion of ParB inhibited cell division (34). Recently, biochemical properties of bacterial members of the ParA and ParB families have been examined in C. crescentus. Interestingly, ParB acted as a nucleotide exchange factor for ParA and thus could switch ParA-ADP to the ATP form (10). An amino-terminal domain of ParB was required for the interaction with ParA and regulation of cell division (12). These results suggest that switching nucleotides is a common and important mechanism for regulation of ParA activities between plasmid and bacterial parABS systems, but its effects on cellular processes appear to be distinct even among bacterial species.
In B. subtilis, depletion of Spo0J influenced replication initiation so that it occurred earlier than normal and was asynchronous (26). Mechanisms of regulation of replication initiation are little known in this organism compared to E. coli, although several candidates involved in this regulation have been reported recently (27, 35, 40). In this study, we analyzed this phenomenon to identify the cause of the abnormal replication initiation in the spo0J null mutant in order to help us understand the regulation of replication initiation in B. subtilis. We found that disruption of soj greatly suppresses abnormal initiation of replication in spo0J null mutant cells. Furthermore, overproduction of Soj causes abnormal initiation, indicating that when the ratio of Soj to Spo0J increases, Soj interferes with the tight control of replication initiation so that initiation occurs early and asynchronously.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. subtilis strains used in this study are listed in Table 1. Transformation of B. subtilis cells was carried out as described previously (25). Two E. coli strains, C600 and BL21(DE3)/pLysS (Novagen, Madison, Wis.), were used as cloning hosts, and plasmids were constructed as follows.
TABLE 1.
B. subtilis strains
| Strain | Relevant genotype | Reference, source, or constructiona |
|---|---|---|
| CRK6000 | purA16 metB5 hisA3 guaB | 36 |
| NIS6072 | CRK6000 spo0J-gfp | 19 |
| NIS6074 | CRK6000 spo0J::ermC | 26 |
| NIS6075 | CRK6000 Δsoj | Kobayashib |
| NIS6076 | CRK6000 Δsoj spo0J-gfp | pSM5072 (19) → NIS6075 (Cp) |
| NIS6077 | CRK6000 Δ(soj-spo0J)::tet | pYO3 → CRK6000 (Tc) |
| NIS6078 | CRK6000 amyE::Pspac-soj | pYO4 → CRK6000 (Cp) |
| NIS6079 | CRK6000 amyE::Pspac-soj-spo0J | pYO5 → CRK6000 (Cp) |
| NIS6080 | CRK6000 soj-spo0J::tet amyE::Pspac-sojG12V-spo0J | pYO5 → NIS6076 (Cp, Tc) |
| NIS6081 | CRK6000 soj-spo0J::tet amyE::Pspac-sojG12V-spo0J | pYO31 → NIS6076 (Cp, Tc) |
| NIS6082 | CRK6000 soj-spo0J::tet amyE::Pspac-sojK16Q-spo0J | pYO32 → NIS6076 (Cp, Tc) |
| NIS6083 | CRK6000 soj-spo0J::tet amyE::Pspac-soj-spo0J14 | pYO33 → NIS6076 (Cp, Tc) |
| NIS6084 | CRK6000 soj-spo0J::tet amyE::Pspac-soj-spo0J17 | pYO34 → NIS6076 (Cp, Tc) |
The arrows indicate transformation. The donor plasmid DNA (linear form except pSM5072) and the recipient strain are indicated on the left and right of each arrow, respectively; the antibiotics used for selection are indicated in parentheses (Cp, chloramphenicol; Tc, tetracycline).
K. Kobayashi, unpublished data. An in-frame deletion (from codon 22 to codon 135) was introduced into soj.
pYO3.
The 5′ part of yyaB (nucleotides 4206200 to 4206750 in the Bacillus subtilis Genome Database [BSORF] [http://bacillus.genome.ad.jp/]) and the 3′ part of yyaC (nucleotides 4204075 to 4204604 in BSORF) were amplified by PCR with primers having artificial HindIII, XbaI, and BamHI restriction enzyme sites at the ends. After digestion with the enzymes, the two fragments were connected at the common XbaI sites and then cloned between HindIII and BamHI sites of pBR322. Finally, a tetracycline resistance gene from pBEST307 (21) was inserted into the XbaI site.
pYO4.
The whole soj gene, including its Shine-Dalgarno sequence (nucleotides 4206216 to 4205412 in BSORF), was amplified by PCR with primers containing artificial HindIII and BamHI sites at the ends. After digestion with the enzymes, the resultant fragment was cloned between the HindIII and BamHI sites of pDLT3 (36), which are located downstream of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter (Pspac).
pYO5.
The whole soj-spo0J operon (nucleotides 4206216 to 4204577 in BSORF) was amplified by PCR with forward (YOF5) and reverse (YOR5) primers having artificial BamHI sites at the ends. The operon was cloned into the BamHI site of pDLT3 after digestion with the enzyme.
pYO31 and pYO32.
The sojG12V-spo0J and sojK16Q-spo0J DNA fragments were amplified by PCR with the following long forward primers and the YOR5 reverse primer and cloned into the BamHI site of pDLT3 to construct pYO31 and pYO32, respectively. The long forward primers were 5′-GGAGGATCCGATAGTACATGTTCATGTGAAAGTAGGTGACATCGTGGGAAAAATCATAGCAATTACGAACCAAAAAGtCGGGG-3′ and 5′-GGAGGATCCGATAGTACATGTTCATGTGAAAGTAGGTGACATCGTGGGAAAAATCATAGCAATTACGAACCAAAAAGGCGGGGTCGGCcAAACAACG-3′ (the underlining and lowercase letters indicate artificial BamHI sites and sojG12V and sojK16Q mutations, respectively).
pYO33 and pYO34.
To obtain the soj-spo0J14 DNA fragment, two fragments were first amplified by PCR with two primer sets, YOF5 plus YOR33 and YOF33 plus YOR5. YOF33 (5′-CTTTATTAGAAgcCCTTCAGC-3′) and YOR33 (5′-GCTGAAGGgcTTCTAATAAAG-3′) are complementary to each other (the lowercase letters in the sequences indicate the spo0J14 mutation) (1). These two fragments were combined by performing PCR with the YOF5 and YOR5 primers as described previously (16) and were cloned into the BamHI site of pDLT3 to obtain pYO33. To construct pYO34 containing the spo0J17 mutation (1), primers YOR34 (5′-GCAATTcCCCTCATTAACG-3′) and YOF34 (5′-CGTTAATGAGGGgAATTGC-3′) were used instead of the YOR33 and YOF33 primers described above.
pET-29b-soj and pET-29b-spo0J.
The whole coding region of soj was amplified by performing PCR with primers having artificial NdeI and XhoI sites. After digestion with these enzymes, the DNA fragment was cloned between the NdeI and XhoI sites of pET-29b (Novagen) to produce a Soj-His6 fusion protein in E. coli cells. Similarly, the whole coding region of spo0J was cloned between NdeI and XhoI sites of pET-29b.
Growth conditions.
B. subtilis cells were grown in antibiotic medium 3 (PAB medium; Difco Laboratories, Detroit, Mich.) supplemented with adenine (20 μg/ml) and guanosine (20 μg/ml) (which are required for growth) at 30°C, unless specified otherwise. When necessary, various concentrations of IPTG and drugs were added (chloramphenicol and tetracycline were added at concentrations of 5 and 10 μg/ml, respectively). As addition of drugs decreased the growth rate slightly, cells were grown without drugs for flow cytometry.
Flow cytometry.
Chloramphenicol (200 μg/ml) was added to cells growing exponentially to inhibit new rounds of replication. Incubation was continued for an additional 5 h to ensure completion of all rounds started prior to addition of the chloramphenicol. Cells were further treated for flow cytometry, and the number of replication origins in individual cells was determined with a flow cytometer (Bryte HS; Bio-Rad Laboratories, Hercules, Calif.) as described previously (41).
Chemical measurement of DNA and protein.
Nucleic acid and protein fractions were prepared from cells by a slightly modified Schneider method as described in detail previously (22). DNA and protein concentrations in the fractions were determined by colorimetry as described previously (22). The DNA/protein ratio was calculated from the total amounts of DNA and protein in the fractions.
Microscopic observation.
Cells were harvested by centrifugation when the optical density at 600 nm reached 0.4, and they were fixed in 70% ethanol as described previously (15). The fixed cells were stained with a 1-μg/ml solution of 4′,6-diamidino-2-phenylindole (DAPI), and cell morphology and nucleoids were observed by fluorescence microscopy as described previously (15). To determine the average cell length in a log-phase cell population, the cell lengths of 300 cells were measured with the MacSCOPE software (Mitani Corp., Fukui, Japan) by using computerized cell images.
Preparation of anti-Soj antibody and coupling of this antibody to protein A-agarose.
Soj-His6 fusion protein was purified from E. coli cells bearing pET-29b-soj as described in the supplier's manual and was used as an antigen to raise anti-Soj rabbit antiserum by Takara Bio (Ohtsu, Japan). Anti-Spo0J rabbit antiserum was raised similarly.
Coupling of antibody to protein A-agarose beads was carried out as described previously (14). Briefly, 200 μl of rabbit serum was mixed with 100 μl of a protein A-agarose bead suspension (Roche Molecular Biochemicals, Mannheim, Germany), and the preparation was washed well with 0.2 M sodium borate buffer (pH 9.0). The preparation was suspended in 1 ml of the same buffer and reacted with 20 mM dimethyl pimelimidate for 30 min. The reaction was stopped by washing the agarose beads with 0.2 M ethanolamine (pH 8.0). Finally, antibody-linked protein A-agarose was suspended in 1 ml of phosphate-buffered saline (PBS) (pH 7.4) supplemented with 0.05% sodium azide.
In vivo cross-linking and immunoprecipitation.
Cells were grown in PAB medium with or without chloramphenicol at 34°C. When the optical density at 600 nm reached 0.4 to 0.5, cells were collected from 20 ml of culture by centrifugation and washed once with 20 ml of PBS (pH 7.4). The cells were resuspended in the same volume of PBS and treated with a cross-linking reagent, dithiobis(succinimidylpropionate) (DSP) obtained from Pierce (Rockford, Ill.), at a concentration of 50 μM for 30 min at 37°C. After the cells were washed with 20 ml of 20 mM Tris buffer (pH 7.5), they were suspended in 100 μl of a lysis solution containing 10 mM Tris buffer, 10 mM MgCl2, 1 mM Na2-EDTA (pH 7.8), 1 mM phenylmethylsulfonyl fluoride, lysozyme (2 mg/ml), and DNase I (200 μg/ml). After incubation at 37°C for 10 min, 5 μl of 20% sodium dodecyl sulfate (SDS) was added to dissolve the cells.
First, 50 μl of the soluble fraction was precleared by incubating it with 50 μl of a protein A-agarose bead suspension in 1 ml of PBS at 4°C for 4 h with a rotator. The soluble fraction was separated from the agarose beads by centrifugation, and then 100 μl of a Soj antibody-linked protein A-agarose bead suspension was added to the supernatant, which was followed by incubation at 4°C overnight. Proteins bound to the immunoaffinity beads were collected by centrifugation and washed three times with 1 ml of a washing solution containing 50 mM Tris buffer, 500 mM NaCl, 1 mM Na2-EDTA, 1 mM phenylmethylsulfonyl fluoride, and 0.1% Triton X-100. Finally, the immunoprecipitates (IPs) were dissolved in 20 μl of sample loading buffer containing 50 mM dithiothreitol (DTT) to cleave cross-links.
Western blotting.
Cell lysates were prepared as described previously (15). Proteins in the lysates or IPs were separated in an SDS-polyacrylamide constant (10% polyacrylamide) or gradient (10 to 20% polyacrylamide) gel by electrophoresis and were blotted on a nitrocellulose or Hybond-P polyvinylidene difluoride membrane (Amersham Biosciences, Little Chalfont, Buckinghamshire, United Kingdom). Either rabbit polyclonal antibodies or a mouse monoclonal anti-GFP antibody (Roche) was used as the first antibody, and then a goat anti-rabbit or anti-mouse immunoglobulin G-horseradish peroxidase conjugate (Promega, Madison, Wis.) was used as the second antibody. Chemiluminescent signals were detected with ECL Western blotting detection reagents and hyperfilm ECL (Amersham).
RESULTS
Deletion of soj suppresses abnormal replication initiation observed in spo0J null mutant cells.
Initiation of replication is tightly regulated in bacteria so that it occurs at a fixed time in the cell cycle and on all replication origins within the cell simultaneously (synchrony of replication initiation) (27, 37). However, the precise timing and synchrony of initiation were greatly disturbed in a spo0J null mutant (26). Sporulation was defective in this null mutant, and the defect was suppressed by deletion of the soj gene (20). Thus, we thought that the abnormal regulation of replication initiation in the spo0J null mutant might also be suppressed by deletion of soj and tested this possibility.
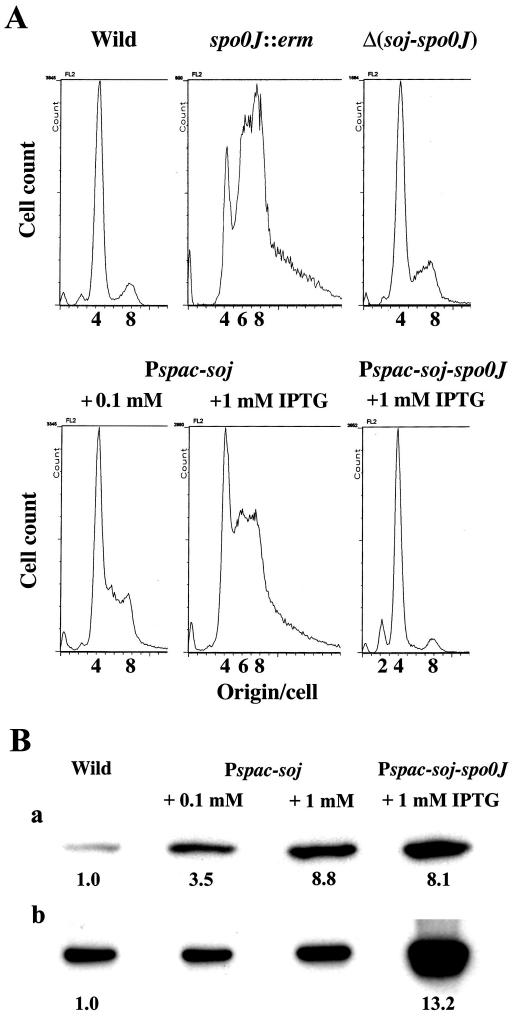
We used flow cytometry to compare the numbers of the replication origins per cell in log-phase cell cultures of the wild-type and spo0J null mutant strains. The results are shown in Fig. 1A. As described previously, in the spo0J null mutant, cells with six replication origins appeared, and the size of the population of cells with eight replication origins increased (26, 46). The increase in the number of cells with eight replication origins was not simply due to cell elongation but rather was due to early initiation (at a smaller cell mass in the cell division cycle) because the amount of mass per origin decreased significantly and the DNA/protein ratio increased considerably in this mutant (Table 2). These results indicate that replication begins early and asynchronously in the spo0J null mutant. On the other hand, when both soj and spo0J were deleted, a DNA histogram obtained by flow cytometry closely resembled the DNA histogram for wild-type cells, although cells with six replication origins were still detected and the number of cells with eight replication origins was slightly more than the number in the wild-type cell culture (Fig. 1A). Consistently, both the amount of mass per replication origin and the DNA/protein ratio in the soj-spo0J deletion mutant were nearly comparable to those in the wild-type strain (Table 2). Furthermore, while spo0J null mutant cells were significantly longer than the wild-type cells, probably because of a delay of cell division, the average cell length of the soj-spo0J deletion mutant was nearly same as that of the wild type (Table 2). Thus, these results indicate that deletion of soj suppresses both the abnormal initiation of replication and the partial inhibition of cell division observed in the spo0J null mutant.
FIG. 1.
Flow cytometric (A) and Western blot (B) analyses of various soj and spo0J mutant B. subtilis cells. (A) DNA histograms obtained by flow cytometry. Usually, more than 20,000 cells were analyzed. The strains analyzed were the wild-type strain (CRK6000 in Table 1) and the spo0J::erm (NIS6074), Δ(soj-spo0J) (NIS6077), Pspac-soj (NIS6078), and Pspac-soj-spo0J (NIS6079) mutants. (B) Both Soj (panel a) and Spo0J (panel b) were detected by Western blotting as described in Materials and Methods. Signals from the same amount of total protein per lane are shown. However, to quantify protein levels, a series of dilutions was prepared for each sample and used for Western blotting. The intensity of each band was quantified by densitometry, and an appropriate dilution that was in a linear range for densitometry was chosen to calculate a protein level relative to that of the wild-type cells. The relative values calculated are shown below the bands.
TABLE 2.
Various parameters for cell division and replication initiation in B. subtilis soj and spo0J mutants
| Strain | Avg cell length (relative)a | Avg no. of origins per cellb | Amt of mass per origin (relative)c | DNA/protein ratiod |
|---|---|---|---|---|
| CRK6000 (wild type) | 1.00 | 4.5 ± 0.1 | 1.00 | 0.041 ± 0.002 |
| NIS7064 (spo0J::ermC) | 1.17 | 8.0 ± 0.1 | 0.67 | 0.060 ± 0.001 |
| NIS6077 [Δ(soj-spo0J)] | 1.03 | 5.1 ± 0.2 | 0.92 | 0.044 ± 0.001 |
| NIS6078 (Pspac-soj) + 1 mM IPTG | 1.16 | 7.0 ± 0.2 | 0.75 | 0.060 ± 0.001 |
| NIS6079 (Pspac-soj-spo0J) + 1 mM IPTG | 1.20 | 4.3 ± 0.1 | 1.27 | 0.034 ± 0.002 |
The average cell length for each strain was obtained by examining 300 cells and is expressed relative to the value for the wild-type cell population.
The average number of replication origins per cell was determined from each DNA histogram of flow cytometry shown in Fig. 1A. The same experiment was repeated three times to obtain the average and standard deviation.
The average cell length was divided by the average number of oriCs per cell to obtain the amount of mass per origin and is expressed relative to the value for the wild type.
Average ± standard deviation obtained from three or four independent experiments.
Overproduction of Soj also leads to early and asynchronous initiation of replication, but overproduction of both Soj and Spo0J does not.
Our results suggested that production of the Soj protein in the absence of Spo0J was the cause of the abnormal replication initiation observed in spo0J null mutant cells and also suggested that a balance between the Soj and Spo0J protein levels might be important for accurate control of replication initiation. To test this possibility, we constructed two strains, one that overproduced only Soj and one that overproduced both Soj and Spo0J. When 0.1 mM IPTG was added to the Soj-overproducing (Pspac-soj) cells, Western blotting showed that the Soj level was about 3.5-fold higher than the wild-type level, and this high level of Soj affected expression of the native spo0J gene very little (Fig. 1B). The tight control of replication initiation was found to be already disturbed by this Soj level, as determined by flow cytometry (Fig. 1A); namely, the proportion of cells with eight replication origins increased significantly compared to the proportion in the wild-type cell population. There also appeared to be cells with six replication origins in the Soj-overproducing strain. When 1 mM IPTG was added, the Soj level increased by about ninefold, as determined by Western blotting (Fig. 1B, panel a), and the interference with the timing of replication initiation became more profound (Fig. 1A). Under these conditions, the amount of mass per replication origin decreased to a level similar to that in the spo0J null mutant, and DNA/protein ratio increased to a value identical to that in the null mutant (Table 2). These results indicate that overproduction of Soj leads to early and asynchronous initiation of replication, as observed in the spo0J null mutant.
When NIS6079 cells (Pspac-soj-spo0J) were grown in the presence of 1 mM IPTG, the Soj and Spo0J levels actually increased by about 8- and 13-fold, respectively (Fig. 1B). Nevertheless, flow cytometry of the overproducing cells revealed a DNA histogram very similar to that of the wild-type cells (Fig. 1A). Since the overproducing cells were elongated (Table 2), this result determined by flow cytometry actually indicated that there was a delay of replication initiation in the cell cycle, in contrast to the enhanced replication initiation in spo0J null mutant and Soj-overproducing cells. Consequently, the amount of mass per replication origin increased in cells overproducing both Soj and Spo0J compared to the amount in wild-type cells, and the DNA/protein ratio decreased slightly in these overproducing cells (Table 2). These results indicate that when the ratio of Soj to Spo0J increases compared to the ratio in the wild-type cells, early and asynchronous initiation of replication occurs, probably by interfering with the tight control mechanism of replication initiation. Spo0J counteracts this effect of Soj because overproduction of both proteins suppresses the enhanced initiation of replication caused by overproduction of Soj alone. This result raises the possibility that Spo0J has an activity opposite that of Soj. Indeed, a delay in replication initiation was observed in cells overproducing both proteins, in which the ratio of Spo0J to Soj was slightly higher than that in the wild-type cells (Fig. 1B). However, no delay in replication initiation was detected in soj deletion mutant cells by flow cytometry and measurement of cell length (data not shown). These results may suggest that a high level of Spo0J, rather than the ratio of Spo0J to Soj, also affects replication initiation.
Interaction of Soj with Spo0J in vivo.
The fact that Spo0J neutralizes effects of Soj on replication initiation (this study) and sporulation (20) suggests that the two proteins interact with each other. Although genetic evidence supports this possibility (1, 20, 32, 42), a direct physical interaction between the proteins has not been shown. We could not demonstrate a direct interaction between these proteins using the yeast two-hybrid system, and this was probably due to inactivation of proteins by fusion with GAL4 activation or DNA binding domains. Immunoprecipitation was then used to test for a direct interaction in vivo after cells were treated with a cross-linker (DSP). An IP obtained with Soj antibodies was examined by Western blotting to determine whether Spo0J was present in the IP. An spo0J-gfp fusion strain and anti-GFP mouse monoclonal antibody were used to detect Spo0J protein in this study, so that the second antibody (anti-mouse horseradish peroxidase conjugate) used in Western blotting did not react with antibody molecules released from the anti-Soj rabbit antibody-linked protein A-agarose used for immunoprecipitation.
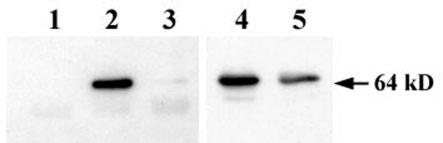
As shown in Fig. 2, lane 4, the anti-GFP antibody did not react with cellular proteins other than Spo0J-GFP. In this lane, whole-cell lysate from spo0J-gfp cells cross-linked in vivo was loaded after the cross-links were cleaved by treatment with DTT. A Soj complex was then immunoprecipitated from the cross-linked cell lysate by Soj antibody, and proteins in the complex were separated in an SDS-polyacrylamide gel after cleavage of the cross-links with DTT (lane 2). As expected, Spo0J-GFP was detected in the Soj complex. The coprecipitation was not simply due to nonspecific binding of the GFP to the protein A-agarose or immunoglobulin molecules because Spo0J-GFP was barely detectable in an IP from Δsoj spo0J-gfp cells (lane 3). The Spo0J-GFP level in the cells was slightly reduced by deletion of soj (lanes 4 and 5), but it was not low enough to explain why Spo0J-GFP was not detected in the IP (lane 3). Furthermore, Spo0J was also reproducibly detected by its antibody in the IP from wild-type cells, but seven other proteins, Bex, DnaA, DnaB, DnaD, DnaI, YabA, and YqeH, were not detected by their antibodies in the same IP (data not shown). These results suggest that only proteins which interact specifically with Soj are probably cross-linked under these conditions. Thus, these results indicate that Soj and Spo0J do interact with each other in vivo.
FIG. 2.
Spo0J-GFP levels in IPs and whole-cell lysates from several B. subtilis strains. In all samples, cross-links made by the DSP cross-linker were cleaved with DTT before loading into an SDS-polyacrylamide gel. Immunoprecipitation was carried out with protein A-agarose beads coupled with anti-Soj rabbit antibody. Spo0J-GFP was detected by Western blotting with anti-GFP monoclonal antibody and horseradish peroxidase-conjugated second antibody. Lanes 1 to 3, IPs from wild-type (CRK6000), spo0J-gfp (NIS6072), and Δsoj spo0J-gfp (NIS6076) cells, respectively; lanes 4 and 5, whole-cell lysates from spo0J-gfp and Δsoj spo0J-gfp cells, respectively.
Effects of deletion or overproduction of Soj and Spo0J on chromosome segregation, chromosome compaction, and cell division.
The sporulation defect and abnormal replication initiation in spo0J null mutants are both suppressed by deletion of soj (20; this study). However, other phenotypes of the spo0J mutants, such as defects in chromosome partitioning or chromosome compaction, were not suppressed by a soj deletion (1, 20). To examine whether these cellular processes were disturbed by overproduction of the Soj and Spo0J proteins, we observed cell and nucleoid morphologies after cells were stained with DAPI.
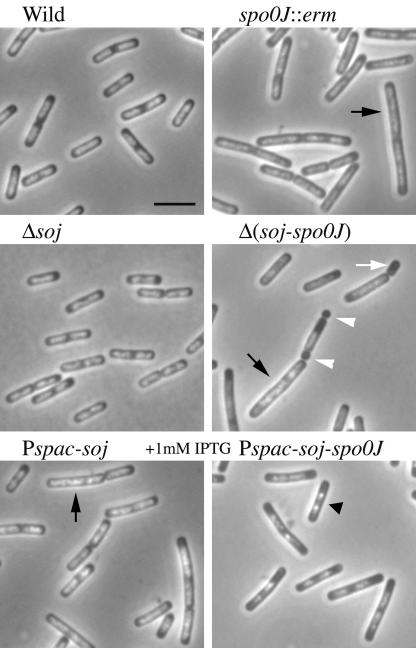
As reported previously (1), elongated cells with less compact nucleoids were often observed in an spo0J null mutant (spo0J::erm [Fig. 3]). Abnormal replication initiation observed in the spo0J null mutant was improved by deletion of soj to a great extent (Fig. 1A). However, elongated cells with less compact nucleoids were still found in the soj-spo0J double deletion mutant (Fig. 3), which is consistent with previous reports (1, 20). Furthermore, anucleate cells were also detected in this double deletion mutant (Fig. 3). These results indicate that the defect in chromosome segregation observed in spo0J null mutants could be separated from the defect in regulation of replication initiation. Surprisingly, minicells were also detected in this double deletion mutant (Fig. 3) at a very low frequency (about 1.3%: 8 minicells in 603 cells). This was not reported previously. Such cells were not detected in 630 spo0J null mutant cells or in 671 soj null mutant cells. These results suggest that cell division is partially affected by deletion of both genes, perhaps partly because of loss of the putative interaction between Soj and MinD suggested previously (2).
FIG. 3.
DAPI-stained images of various soj and spo0J mutant cells. The strains used were the wild-type strain (CRK6000 in Table 1) and the spo0J::erm (NIS6074), Δsoj (NIS6075), Δ(soj-spo0J) (NIS6077), Pspac-soj (NIS6078), and Pspac-soj-spo0J (NIS6079) mutants. The black arrows, white arrowheads, and black triangle indicate elongated cells with diffuse nucleoids, minicells, and a cell with highly condensed nucleoids, respectively. The white arrow indicates an anucleate cell. Scale bar = 3 μm.
Overproduction of Soj resulted in elongated cells with less compact nucleoids, similar to the spo0J null mutant cells (Fig. 3). In contrast, in cells overproducing both Soj and Spo0J, the nucleoids appeared to be more compact than those in the wild-type cells (Fig. 3). In these overproducing cells, the ratio of Spo0J to Soj was slightly higher than the ratio in the wild-type cells (Fig. 1B). However, the compactness of nucleoids was almost the same in both wild-type cells and soj null mutant cells producing only Spo0J (Fig. 3). Thus, the more compact nucleoid structure observed when there was overproduction of Soj and Spo0J may have been due to a high Spo0J level rather than an increased ratio of Spo0J to Soj (Fig. 1B). Alternatively, the apparent increased compactness may have reflected less DNA within cells as the DNA/protein ratio in cells overproducing both Soj and Spo0J was slightly less than the ratio in the wild-type cells (Table 2).
Replication initiation in several soj and spo0J point mutants.
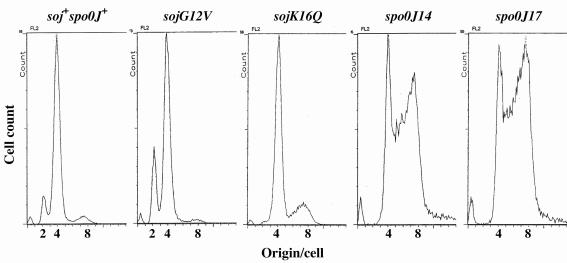
To examine which biochemical function of Soj was related to interference with tight regulation of replication initiation, we analyzed several soj and spo0J point mutants with different phenotypes by flow cytometry. In the mutant cells, expression of the mutant soj-spo0J operon inserted at the amyE locus was controlled by the IPTG-inducible spac promoter, and the native operon was deleted (Table 1). When 50 μM IPTG was added, the protein levels in the mutants were comparable to those in the wild-type cells, as determined by Western blotting (data not shown). Thus, cells were grown in the presence of 50 μM IPTG for flow cytometry. NIS6080 (soj-spo0J::tet amyE::Pspac-soj+-spo0J+) control cells produced a DNA histogram similar to that produced by the wild-type (CRK6000) cells (Fig. 1A and 4).
FIG. 4.
Analysis of replication initiation in several soj and spo0J mutants by flow cytometry. DNA histograms were obtained by flow cytometry for the following strains: soj+ spo0J+ strain NIS6080, sojG12V strain NIS6081, sojK16Q strain NIS6082, spo0J14 strain NIS6083, and spo0J17 strain NIS6084. The cells were grown in the presence of 50 μM IPTG because expression of both the soj and spo0J genes is controlled by the IPTG-inducible spac promoter in these mutant strains and at this IPTG concentration, the levels of both proteins were nearly comparable to those in the wild-type strain.
The ATPase activity of Soj is greatly reduced in the sojG12V mutant, and thus, the mutant protein, SojG12V, probably exists as an ATP-bound form within the cell (42). This mutant protein localized to the cell poles but lost the ability to jump between poles (42). In contrast, another mutant protein, SojK16Q, was perhaps defective in nucleotide binding and was distributed throughout the cell (42). These two soj mutations appeared to have opposite effects on replication initiation, although the effects were only slight. Replication initiation was slightly delayed in the sojG12V mutant but occurred slightly early in the sojK16Q mutant (Fig. 4 and Table 3).
TABLE 3.
Various parameters for cell division and replication initiation in several soj and spo0J mutants of B. subtilis with point mutationsa
| Strain | Avg cell length (relative) | Avg no. of origins per cell | Amt of mass per origin (relative) |
|---|---|---|---|
| NIS6080 (Pspac-soj+-spo0J+) | 1.00 | 4.4 ± 0.4 | 1.00 |
| NIS6081 (Pspac-sojG12V-spo0J+) | 1.07 | 3.8 ± 0.1 | 1.24 |
| NIS6082 (Pspac-sojK16Q-spo0J+) | 1.01 | 5.0 ± 0.3 | 0.89 |
| NIS6083 (Pspac-soj+-spo0J14) | 0.99 | 6.2 ± 0.3 | 0.70 |
| NIS6084 (Pspac-soj+-spo0J17) | 0.95 | 6.7 ± 0.3 | 0.62 |
Two spo0J mutants with point mutations (spo0J14 and spo0J17) were used in this study, and in the mutant cells Soj was associated with all the nucleoids in the cells without jumping (1), as it was in the spo0J null mutant cells (32, 42). Replication initiation also occurred significantly early in these two spo0J mutants (Fig. 4 and Table 3), as in spo0J null mutants. This enhancement of replication initiation was not due to an increased ratio of Soj to Spo0J caused by degradation of the mutant Spo0J proteins because the levels of both of the proteins in the spo0J17 mutant were comparable to those in the control strain, NIS6080, as determined by Western blotting (data not shown). An interaction between Spo0J and Soj was also observed in the spo0J17 mutant (data not shown). Thus, these results raise the possibility that the stable association of Soj with nucleoids correlates with the abnormal replication initiation observed in the spo0J mutants.
DISCUSSION
B. subtilis Soj and Spo0J are members of the ParA and ParB protein families, respectively, which are well conserved in both plasmids and bacteria (20, 47). Based on this conservation, both B. subtilis proteins were first believed to have functions analogous to those of plasmid proteins. However, in a recent paper the authors described significant apparent differences in function between the plasmid and chromosomal ParA-ParB systems, although the biochemical activities could be quite similar (26). In fact, in addition to chromosome segregation, the ParA (Soj)-ParB (Spo0J) systems of B. subtilis and C. crescentus are also required for accurate initiation of replication and cytokinesis, respectively (26, 34). Thus, the precise functions of bacterial ParA-ParB systems are still unclear. To determine the primary roles of Soj and Spo0J in B. subtilis, it is necessary to elucidate why there is not tight regulation of replication initiation in spo0J null mutant cells. The present study showed that the increased ratio of Soj to Spo0J that occurs in a spo0J null mutant is responsible for the abnormal replication initiation and that association of Soj with nucleoids under these conditions seems to correlate with the abnormal replication initiation.
The sporulation defect of spo0J null mutants was due to repression of transcription of several early sporulation genes by Soj (6, 20, 43). Thus, the abnormal replication initiation in the spo0J null mutant and Soj-overproducing cells could be also caused by an Soj transcriptional repressor. The following observations may actually support this possibility. In C. crescentus, ParB acts as a nucleotide exchange factor, and thus, ADP bound to ParA cannot be replaced by ATP without ParB, leading to a high ParA-ADP level within the cell (10). Indeed, high ParA-ADP levels were detected in ParA-overproducing and parB deletion mutant cells (10, 12). ParA-ADP is an active form that acts as a transcriptional repressor in the P1 plasmid (4). In B. subtilis, Soj-ADP may also directly bind to a small unwound region within the replication origin (oriC), as well as to promoter sequences as a transcriptional repressor, because this form exhibits nonspecific single-stranded DNA binding activity in C. crescentus (10). This binding may assist extending the unwound region within oriC further, resulting in early initiation of replication. Alternatively, Soj-ADP may interact with possible modulators of replication initiation, including Bex, YqeH, and YabA, which have recently been identified (35, 40), and it may interfere with their activities. Actually, it has been speculated that in C. crescentus ParA might directly inhibit the assembly of the cell division machinery (34). However, preliminary experiments with B. subtilis have not supported this possibility so far (Moriya, unpublished data).
Loss of proper higher-order structure of the oriC region of the chromosome may be considered another possibility to explain why tight regulation of replication initiation is destroyed by a deficiency of Spo0J or overproduction of Soj. Participation of the Soj-Spo0J system in nucleoid organization has been postulated and demonstrated in previous studies (1, 5, 26, 30). Actually, less compact nucleoids were frequently observed in spo0J null mutant and Soj-overproducing cells (1; this study). Intriguingly, in this study, nucleoids that looked more compact were observed in some cells overproducing both Soj and Spo0J. Since overproduction of Soj caused diffusion of nucleoids, the apparently more compact structure might be formed by overproduction of Spo0J. Thus, Soj and Spo0J may affect chromosome compaction in opposite ways. Replication initiation occurred earlier in spo0J null mutant and Soj-overproducing cells than in the wild-type cells, as determined by flow cytometry and, in contrast, was delayed in cells overproducing both Soj and Spo0J. These results may imply that there is a correlation between higher-order structure of the chromosome and control of replication initiation.
Replication initiation is tightly regulated, so that it takes place only once per cell cycle. In E. coli, at least two main negative regulatory mechanisms are known. These involve sequestration of newly replicated origins in the cell membrane by SeqA (31) and inactivation of DnaA by Hda and DnaN (23, 24). These mechanisms act after replication initiation to suppress extra and consecutive initiation events. In B. subtilis, these types of negative regulatory mechanisms have not been reported so far. However, in a recent report the authors suggested that a new gene product, YabA, regulates replication initiation negatively like E. coli Hda, although YabA exhibited no significant homology with DnaA, unlike Hda (40). Preliminary results indicated that when YabA levels were lowered, early and asynchronous initiation of replication occurred, as observed in spo0J null mutant cells (Y. Ogura and M. Hayashi, unpublished data). Furthermore, the same phenotype in replication initiation was observed with decreased levels of two new GTP-binding proteins, Bex and YqeH (35). These results may indicate that the potential for the next round of replication initiation is already present at an early stage of the replication cycle in B. subtilis but is not liberated until the proper time comes, as proposed previously (37, 41). Three modulators of replication initiation, Bex, YqeH, and YabA, may join the regulatory mechanism suppressing release of the initiation potential, and thus, replication initiation advances when the levels of these proteins are decreased. Thus, the regulation of replication initiation in B. subtilis might be significantly different than that in E. coli. The mechanism of replication initiation regulation in B. subtilis appears to resemble the mechanism of replication initiation regulation in eukaryotes, in which the prereplicative complex is assembled on the replication origins at the M-G1 transition of the cell cycle and is maintained during the G1 phase until it is activated at the onset of the S phase (3). Further investigation is needed to prove this interesting model proposed for regulation of replication initiation in B. subtilis.
Acknowledgments
We thank T. Morimoto for help with flow cytometry.
This work was supported by grants-in-aid for scientific research (B) from the Japan Society for the Promotion of Science to S.M. and N.O. and by a large grant from the Australian Research Council to E.J.H. (grant A10009161). E.J.H. was the recipient of an ARC QEII research fellowship.
REFERENCES
- 1.Autret, S., R. Nair, and J. Errington. 2001. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol. 41:743-755. [DOI] [PubMed] [Google Scholar]
- 2.Autret, S., and J. Errington. 2003. A role for division-site-selection protein MinD in regulation of internucleoid jumping of Soj (ParA) protein in Bacillus subtilis. Mol. Microbiol. 47:159-169. [DOI] [PubMed] [Google Scholar]
- 3.Bell, S. P. 2002. The origin recognition complex: from simple origins to complex functions. Genes Dev. 16:659-672. [DOI] [PubMed] [Google Scholar]
- 4.Bouet, J.-Y., and B. E. Funnell. 1999. P1 ParA interacts with the P1 partition complex at parS and an ATP-ADP switch controls ParA activities. EMBO J. 18:1415-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britton, R. A., and A. D. Grossman. 1999. Synthetic lethal phenotypes caused by mutations affecting chromosome partitioning in Bacillus subtilis. J. Bacteriol. 181:5860-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cervin, M. A., G. B. Spiegelman, B. Raether, K. Ohlsen, M. Perego, and J. A. Hoch. 1998. A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis. Mol. Microbiol. 29:85-95. [DOI] [PubMed] [Google Scholar]
- 7.Davey, M. J., and B. E. Funnell. 1997. Modulation of the P1 plasmid partition protein ParA by ATP, ADP, and P1 ParB. J. Biol. Chem. 272:15286-15292. [DOI] [PubMed] [Google Scholar]
- 8.Davis, M. A., K. A. Martin, and S. J. Austin. 1992. Biochemical activities of the ParA partition protein of the P1 plasmid. Mol. Microbiol. 6:1141-1147. [DOI] [PubMed] [Google Scholar]
- 9.Draper, G. C., and J. W. Gober. 2002. Bacterial chromosome segregation. Annu. Rev. Microbiol. 56:567-597. [DOI] [PubMed] [Google Scholar]
- 10.Easter, J., Jr., and J. W. Gober. 2002. ParB-stimulated nucleotide exchange regulates a switch in functionally distinct ParA activities. Mol. Cell 10:427-434. [DOI] [PubMed] [Google Scholar]
- 11.Erdmann, N., T. Petroff, and B. E. Funnell. 1999. Intracellular localization of P1 ParB protein depends on ParA and parS. Proc. Natl. Acad. Sci. USA 96:14905-14910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figge, R. M., J. Easter, Jr., and J. W. Gober. 2003. Productive interaction between the chromosome partitioning proteins, ParA and ParB, is required for the progression of the cell cycle in Caulobacter crescentus. Mol. Microbiol. 47:1225-1237. [DOI] [PubMed] [Google Scholar]
- 13.Glaser, P., M. E. Sharpe, B. Raether, M. Perego, K. Ohlsen, and J. Errington. 1997. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 11:1160-1168. [DOI] [PubMed] [Google Scholar]
- 14.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Hassan, A. K. M., S. Moriya, M. Ogura, T. Tanaka, F. Kawamura, and N. Ogasawara. 1997. Suppression of initiation defects of chromosome replication in Bacillus subtilis dnaA- and oriC-deleted mutants by integration of a plasmid replicon into the chromosomes. J. Bacteriol. 179:2494-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiraga, S. 2000. Dynamic localization of bacterial and plasmid chromosomes. Annu. Rev. Genet. 34:21-59. [DOI] [PubMed] [Google Scholar]
- 18.Hirano, M., H. Mori, T. Onogi, M. Yamazoe, H. Niki, T. Ogura, and S. Hiraga. 1998. Autoregulation of the partition genes of mini-F plasmid and the intracellular localization of their products in Escherichia coli. Mol. Gen. Genet. 257:392-403. [DOI] [PubMed] [Google Scholar]
- 19.Imai, Y., N. Ogasawara, D. Ishigo-oka, R. Kadoya, T. Daito, and S. Moriya. 2000. Subcellular localization of Dna-initiation proteins of Bacillus subtilis: evidence that chromosome replication begins at either edge of nucleoids. Mol. Microbiol. 36:1037-1048. [DOI] [PubMed] [Google Scholar]
- 20.Ireton, K., N. W. Gunther IV, and A. D. Grossman. 1994. spoOJ is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176:5320-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itaya, M. 1992. Construction of a novel tetracycline resistance gene cassette useful as a marker on the Bacillus subtilis chromosome. Biosci. Biotechnol. Biochem. 56:685-686. [DOI] [PubMed] [Google Scholar]
- 22.Kadoya, R., A. K. M. Hassan, Y. Kasahara, N. Ogasawara, and S. Moriya. 2002. Two separate DNA sequences within oriC participate in accurate chromosome segregation in Bacillus subtilis. Mol. Microbiol. 45:73-87. [DOI] [PubMed] [Google Scholar]
- 23.Katayama, T., T. Kubota, K. Kurokawa, E. Crooke, and K. Sekimizu. 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94:61-71. [DOI] [PubMed] [Google Scholar]
- 24.Kato, J., and T. Katayama. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunst, F., T. Msadek, and G. Rapoport. 1994. Signal transduction network controlling degradative enzyme synthesis and competence in Bacillus subtilis, p. 1-20. In P. J. Piggot, C. P. Moran, Jr., and P. Youngman (ed.), Regulation of bacterial differentiation. American Society for Microbiology, Washington, D.C.
- 26.Lee, P. S., D. C.-H. Lin, S. Moriya, and A. D. Grossman. 2003. Effects of the chromosome partitioning protein Spo0J (ParB) on oriC positioning and replication initiation in Bacillus subtilis. J. Bacteriol. 185:1326-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemon, K. P., S. Moriya, N. Ogasawara, and A. D. Grossman. 2002. Chromosome replication and segregation, p. 73-86. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 28.Li, Y., and S. Austin. 2002. The P1 plasmid is segregated to daughter cells by a ‘capture and ejection’ mechanism coordinated with Escherichia coli cell division. Mol. Microbiol. 46:63-74. [DOI] [PubMed] [Google Scholar]
- 29.Lin, D. C.-H., P. A. Levin, and A. D. Grossman. 1997. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 94:4721-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, D. C.-H., and A. D. Grossman. 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92:675-685. [DOI] [PubMed] [Google Scholar]
- 31.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413-426. [DOI] [PubMed] [Google Scholar]
- 32.Marston, A. L., and J. Errington. 1999. Dynamic movement of the ParA-like Soj protein of B. subtilis and its dual role in nucleoid organization and developmental regulation. Mol. Cell 4:673-682. [DOI] [PubMed] [Google Scholar]
- 33.Mohl, D. A., and J. W. Gober. 1997. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell 88:675-684. [DOI] [PubMed] [Google Scholar]
- 34.Mohl, D. A., J. Easter, Jr., and J. W. Gober. 2001. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol. Microbiol. 42:741-755. [DOI] [PubMed] [Google Scholar]
- 35.Morimoto, T., P. C. Loh, T. Hirai, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2002. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 148:3539-3552. [DOI] [PubMed] [Google Scholar]
- 36.Moriya, S., K. Kato, H. Yoshikawa, and N. Ogasawara. 1990. Isolation of a dnaA mutant of Bacillus subtilis defective in initiation of replication: amount of DnaA protein determines cells' initiation potential. EMBO J. 9:2905-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moriya, S., Y. Imai, A. K. M. Hassan, and N. Ogasawara. 1999. Regulation of initiation of Bacillus subtilis chromosome replication. Plasmid 41:17-29. [DOI] [PubMed] [Google Scholar]
- 38.Niki, H., and S. Hiraga. 1997. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell 90:951-957. [DOI] [PubMed] [Google Scholar]
- 39.Niki, H., and S. Hiraga. 1999. Subcellular localization of plasmids containing the oriC region of the Escherichia coli chromosome, with or without the sopABC partitioning system. Mol. Microbiol. 34:498-503. [DOI] [PubMed] [Google Scholar]
- 40.Noirot-Gros, M.-F., E. Dervyn, L. J. Wu, P. Mervelet, J. Errington, S. D. Ehrlich, and P. Noirot. 2002. An expanded view of bacterial DNA replication. Proc. Natl. Acad. Sci. USA 99:8342-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogura, Y., Y. Imai, N. Ogasawara, and S. Moriya. 2001. Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J. Bacteriol. 183:3833-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quisel, J. D., D. C.-H. Lin, and A. D. Grossman. 1999. Control of development by altered localization of a transcription factor in B. subtilis. Mol. Cell 4:665-672. [DOI] [PubMed] [Google Scholar]
- 43.Quisel, J. D., and A. D. Grossman. 2000. Control of sporulation gene expression in Bacillus subtilis by the chromosome partitioning protein Soj (ParA) and Spo0J (ParB). J. Bacteriol. 182:3446-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharpe, M. E., and J. Errington. 1996. The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol. Microbiol. 21:501-509. [DOI] [PubMed] [Google Scholar]
- 45.Sharpe, M. E., and J. Errington. 1998. A fixed distance for separation of newly replicated copies of oriC in Bacillus subtilis: implications for coordination of chromosome segregation and cell division. Mol. Microbiol. 28:981-990. [DOI] [PubMed] [Google Scholar]
- 46.Webb, C. D., A. Teleman, S. Gordon, A. Straight, A. Belmont, D. C.-H. Lin, A. D. Grossman, A. Wright, and R. Losick. 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88:667-674. [DOI] [PubMed] [Google Scholar]
- 47.Yamaichi, Y., and H. Niki. 2000. Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:14656-14661. [DOI] [PMC free article] [PubMed] [Google Scholar]