Abstract
Cocultures of Salmonella strains carrying or lacking specific prophages undergo swift composition changes as a result of phage-mediated killing of sensitive bacteria and lysogenic conversion of survivors. Thus, spontaneous prophage induction in a few lysogenic cells enhances the competitive fitness of the lysogen population as a whole, setting a selection regime that forces maintenance and spread of viral DNA. This is likely to account for the profusion of prophage sequences in bacterial genomes and may contribute to the evolutionary success of certain phylogenetic lineages.
Progress in genomic research has revealed the high incidence of phage-related sequences in bacterial genomes. In some isolates, these elements take up more than 10% of the genome and account for a significant portion of sequence diversity between isolates (10-12). A prototypical example, pathogenic Escherichia coli strains from the O157 serogroup contain between 15 and 18 different prophages which account for most of the excess size of the genome of this organism (5.5 Mb) relative to the laboratory K-12 (4.6 Mb) (12). Prophage sequences are also common in Salmonella enterica, a species regrouping a large number of pathogenic serovars with different host ranges and epidemiological properties. The best-studied isolates of S. enterica serovar Typhimurium contain a different complement of about four or five full-size prophages (5, 8, 13). Work in our laboratory has shown most of these elements to be fully functional, e.g., competent to undergo excision and viral development (4, 5). This is the case for lambdoid prophages Gifsy-1 and Gifsy-2, which are found in most, if not all, strains from serovar Typhimurium. Both elements encode a number of factors known or thought to contribute to pathogenicity. In particular, prophage Gifsy-2 encodes, among other proteins, a periplasmic (Cu, Zn) superoxide dismutase (SodC1) and a protein of unknown function (GtgE), both of which are required for systemic infection of mice (4, 7). The selective advantage conferred by these loci is likely to have favored their maintenance in the bacterial chromosome. However, virulence genes occupy less than 15% of Gifsy-1 and Gifsy-2 genomes altogether and cannot easily account for the conservation of the entire elements (approximately 50 kb each) in an active configuration. In the course of our work, it became apparent that the pervasiveness of these two elements might be related to their ability to undergo spontaneous induction.
Phage-driven evolution of Salmonella cocultures.
Salmonella cultures grown to stationary phase contain phage particles in their supernatants (4). Spontaneous prophage induction occurs in a tiny fraction of cells and has no consequences for the majority of the population which is immune to superinfection. Typically, the phage titers range between 104 and 105 PFU/ml in cultures where bacterial counts are on the order of 109 CFU/ml. We considered that the situation might change drastically if the population were not clonal and contained bacteria from a nonlysogenic strain.
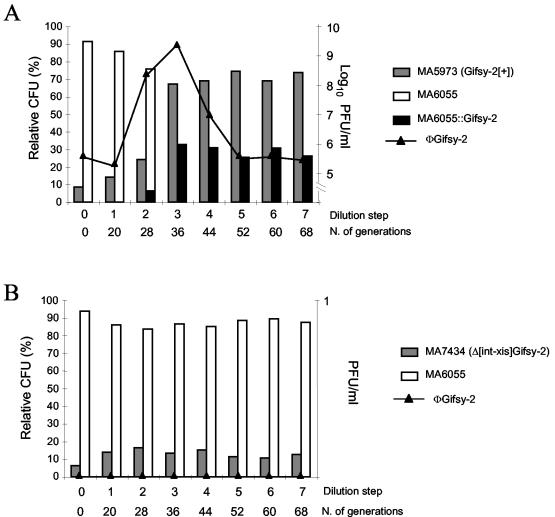
To test this idea, we studied the evolution of cocultures of strains that carried or lacked selected prophages. (A chromosomal araD::MudJ lac fusion in one of the two strains served as a genetic tag for strain identification.) Bacteria were allowed to replicate at 30°C without agitation until stationary phase and then were subcultured under the same conditions. Figure 1A shows the results of one such experiment with strains MA5973 and MA6055, which carry and lack the Gifsy-2 prophage, respectively. One can see that the ratio between the two strains, initially set to the value of 1:10, remains nearly invariant through two subculturing steps and then changes abruptly in favor of the lysogenic strain (Fig. 1A). The change coincides with a sharp increase in the concentration of Gifsy-2 viral particles in the culture. Subsequently, the values level off, as the residual MA6055 population becomes lysogenic for and thus resistant to Gifsy-2 phage (Fig. 1A). Lysogenization was demonstrated by PCR amplification analysis of prophage-chromosome boundaries and/or by the ability of Gifsy-2-resistant MA6055 derivatives to release a phage with Gifsy-2 immunity (data not shown). Clearly, these findings suggest that Gifsy-2 phage determines the fate of the mixed population by selectively killing nonimmune bacteria. A minority of cells, newly lysogenized, escape killing and multiply. To gather support for this model, we isolated a Gifsy-2 int-xis deletion, which prevents prophage excision. The strain carrying such “locked-in” prophage no longer outgrows strain MA6055 in a mixed culture (Fig. 1B), suggesting that the extrachromosomal form of the virus is needed for this effect.
FIG. 1.
Evolution of the ratio of strains in cocultures of S. enterica serovar Typhimurium strains carrying or lacking the Gifsy-2 prophage. Strains were derived from S. enterica serovar Typhimurium strain ATCC 14028s (3). Cultures (in Luria-Bertani medium) were inoculated with approximately 1,000 CFU from a 1:10 mixture of bacteria lysogenic for Gifsy-2 or lacking the prophage, respectively. Inoculi were verified to be phage-free. After the bacteria reached stationary phase, cultures were diluted 200-fold and incubated further under the same conditions (30°C, no shaking) for 6 to 12 h. The subculturing routine was repeated the indicated number of times (number [N.] of generations). At each dilution step, the culture was analyzed with respect to bacterial composition and phage concentration. (A) The lysogenic strain carries wild-type Gifsy-2. (B) The lysogenic strain contains a Gifsy-2 prophage variant that cannot excise due to an int-xis deletion. Lysogenization by the indicated phage is indicated by a double colon. The full genotypes for competing strains follow: MA5973, lacking Gifsy-1 but carrying Gifsy-2 (Gifsy-2[+]); MA6055, lacking Gifsy-1 and Gifsy-2 but with ara-907 araD901::MudJ; MA7434, lacking Gifsy-1 but with Δ[int-xis]Gifsy-2::kan. (The int-xis/kan swap construct was obtained as described previously [1]. The deleted segment corresponds to the interval between coordinates 1098338 and 1099776 of Salmonella strain LT2 genome sequence [8].) Phage indicator strain MA6684 lacked Gifsy-1 and Gifsy-2 but had galE496 bio-106::Tn10. Panels A and B show the results of one representative experiment of three and two independent experiments, respectively.
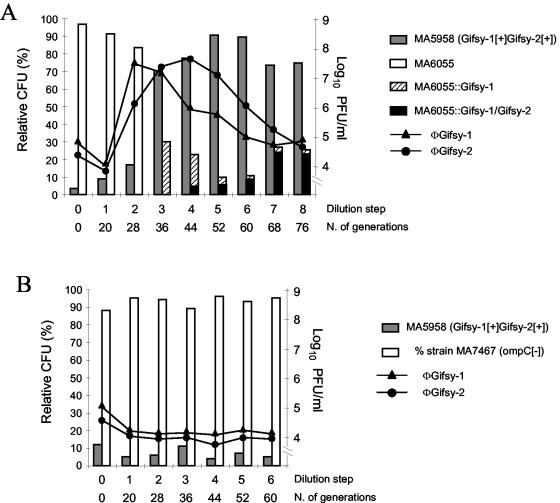
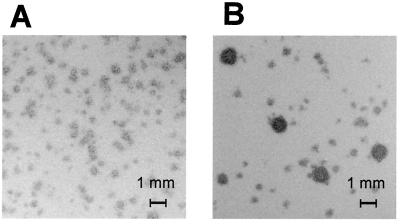
As one might expect, the changes in the ratios of strains are more dramatic when the lysogenic strain releases more than one phage type, since separate integration events are required for the naive strain to survive. For example, the proportion of strain MA5958 (carrying Gifsy-1 and Gifsy-2) in a mixture with strain MA6055 varies from the initially set value of 5% to more than 80% by the fourth dilution step (Fig. 2A). MA6055 bacteria first became lysogenic for Gifsy-1 and then for Gifsy-2, an order that roughly parallels the timing of production of each of the two viruses (Fig. 2A). Lysogenization by the second phage occurred later than it did when Gifsy-2 was the only phage involved (Fig. 1A). This difference could be attributed to the lower spontaneous induction levels of Gifsy-2 in strains that also contain Gifsy-1, combined with a reduced susceptibility of Gifsy-1 lysogens to Gifsy-2 infection (L. Bossi and N. Figueroa-Bossi, unpublished results). One factor that could affect the experimental results shown in Fig. 2A is the appearance of recombinant viruses, namely, phages with Gifsy-1 plaque morphology and Gifsy-2 immunity and vice versa (Fig. 3 and data not shown). Accumulation of these new species in the late stages of the incubation might account for the broader shape of the phage curves in Fig. 2A. Finally, inactivation of OmpC, the receptor of the two Gifsy phages (6), in the nonlysogenic strain, completely prevents all culture composition changes (Fig. 2B), further confirming that viral infection is required for these changes in the composition of the culture.
FIG. 2.
Evolution of the ratio of strains in cocultures of S. enterica serovar Typhimurium strains carrying or lacking the Gifsy-1 and Gifsy-2 prophages. The experiment was performed as described in the legend to Fig. 1. (A) The naive strain, MA6055, is susceptible to infection by both Gifsy-1 and Gifsy-2 phages. (B) The naive strain, MA7467, carries an ompC gene disruption that renders the strain resistant to Gifsy-1 and Gifsy-2 infection. Lysogenization by the indicated phage(s) is indicated by a double colon. The full genotypes of competing strains follow: MA5958, carrying Gifsy-1 and Gifsy-2 (Gifsy-1[+]Gifsy-2[+]); MA6055, see the legend to Fig. 1; MA7467, lacking Gifsy-1 and Gifsy-2 but with ompC159::Tn10 ara907 araD901::MudJ. The full genotypes of phage indicator strains follow: MA6685, lacking Gifsy-1 but carrying Gifsy-2 galE496 bio-106::Tn10; MA7416, carrying Gifsy-1 and lacking Gifsy-2 but carrying galE496 bio-106::Tn10. Panel A shows the result of one representative experiment of two independent experiments.
FIG. 3.
Plaques from phage with Gifsy-2 immunity in early and late subcultures. Supernatants from cocultures of strains MA5958 and MA6055 were diluted suitably and plated on a lawn of strain MA7416 (Fig. 2). (A) Phage from dilution step 3 in Fig. 2A. (B) Phage from dilution step 6 in Fig. 2A. The larger plaques have the size and morphology typical of Gifsy-1 phage.
Since both Gifsy-1 and Gifsy-2 phages have small burst sizes, it was of interest to examine the effects of a prophage with a more productive burst, such as Salmonella phage P22. A strain lysogenic for phage P22 was isolated and cocultured with a strain lacking the prophage. P22-mediated changes in the ratios of strains were not more dramatic than those previously observed with Gifsy lysogens (data not shown); however, a relevant difference was that changes occurred significantly earlier (in the first subculture [data not shown]). No putative recombinant phages were observed.
Phage as weapons in competing strains.
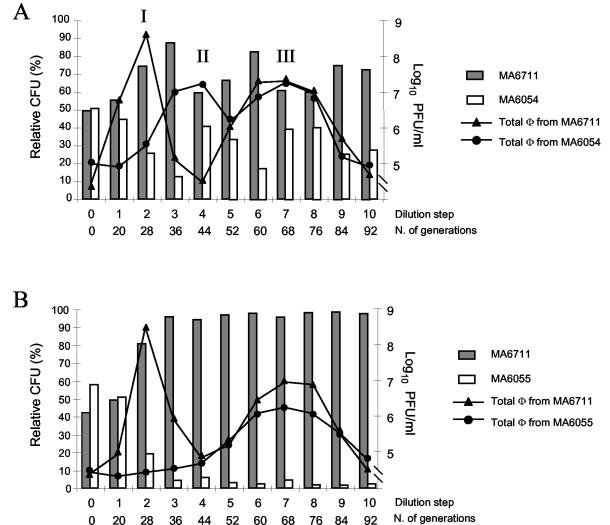
All of the above experiments were performed with strains that are closely related (derived from strain ATCC 14028s), differing only in the presence or absence of selected prophages. To gain further insight into the possible role of this phenomenon in Salmonella evolution, cocultures were made with isolates with different origins. Epidemic strain MA6711 (lysotype DT104) was mixed with one of the two ATCC 14028-derived strains, MA6054 (carrying Gifsy-1 and Gifsy-2) or MA6055 (lacking Gifsy-1 and Gifsy-2). Although the full prophage repertoires of the two lines are not known, we expected each of the competing strains to be sensitive to one or more phages released by the other. This assumption proved correct, as the coculture conditions supported a series of viral bursts, which in turn correlated with alternating changes in the ratios of the strains.
As shown in Fig. 4, strain MA6711 clearly outgrows the ATCC 14028-derived strains in the initial period (ratio changes from 1:1 to approximately 9:1 by the third subculturing step). This change coincides with the proliferation of a phage from MA6711, which we found to be related to P22 and which probably corresponds to the previously described PDT104 (15) (peak I in Fig. 4A and B). The MA6711/MA6054 ratio changed after the third subculturing step (dominance of MA6711 drops to less than 2:1), concomitant with the accumulation of a phage released by MA6054 and identified as Gifsy-1 (peak II in Fig. 4A). No such trend inversion is observed in the MA6711-MA6054 coculture, consistent with the absence of Gifsy-1 in strain MA6054 (Fig. 4B). Thus, Gifsy-1 effectively counteracts the outgrowth of the DT104 strain (although DT104 carries a Gifsy-1-related prophage, it is not immune to Gifsy-1 from strain ATCC 14028 [unpublished data]). New phage bursts occur after the fifth subculturing step (peak III in Fig. 4A and B). These latecomers originate from both competing strains and represent mixtures of different viral species, including a Gifsy-2-related phage from strain MA6711 and phage Gifsy-3 from strain ATCC 14028 (5; data not shown). Overall, this activity appears to favor the DT104 strain slightly (Fig. 4A). A limited survey of individual colonies at the end of the experiment in Fig. 4A revealed the presence of MA6711 derivatives lysogenic for Gifsy-1 from strain ATCC 14028 and MA6064 derivatives lysogenic for the presumptive PDT104 phage (data not shown).
FIG. 4.
Evolution of the ratio of strains in cocultures of S. enterica serovar Typhimurium strains from unrelated lineages. Strain MA6711 (a gift from S. Uzzau, University of Sassari, Sassari, Italy) is a phage-type DT104 strain from an epidemic outbreak in Zimbabwe. The experiment was performed as described in the legend to Fig. 1, except that strains were inoculated in a 1:1 ratio. Phage titers were measured by plating aliquots from culture supernatants onto bacterial lawns from strains MA6711 (total phage from MA6054 or MA6055) and MA5958 (total phage from MA6711). (A) Strains harbor the wild-type prophage complements of the respective lineages. (B) The ATCC 14028s-derived strain (MA6055) lacks the Gifsy-1 and Gifsy-2 prophages.
Conclusions.
The data presented here suggest that prophages can improve the competitive fitness of the host strain by virtue of their capacity to grow lytically and destroy bacteria from rival strains. The effect is exerted at the population level, since the individual bacteria that suffer induction are presumably killed in the process, an altruistic suicide reminiscent of features of multicellular systems (16). In this particular case, the biological cost appears minimal, as the fraction of phage-producing cells can be estimated to be less than 10−5 of the population. Thus, spontaneous phage release acts as a selection regimen that forces maintenance and spread of the lysogenic condition in bacterial genomes. This model can also account for maintenance of prophage segments that confer immunity or exclusion, since these elements will protect the carrier strain from superinfecting phages. For example, the resistance of S. enterica serovar Typhi to Gifsy-2 phage results from a defective prophage that expresses Gifsy-2 immunity determinants (5). Like the latter, this element is located in the chromosomal interval between pncB and pepN genes (2). Replacing this segment by the corresponding region from the Gifsy-2-cured S. enterica serovar Typhimurium strain renders S. enterica serovar Typhi susceptible to infection (5; data not shown).
Concomitant with the effect on population dynamics, phage multiplication results in the generation of a new strain by lysogenic conversion. This is relevant to the ways in which Salmonella bacteria evolve and diversify. The standing coculture conditions set up in this study must not be uncommon in nature, particularly in the case of enteric pathogens with animal reservoirs. Interactions between strains could occur in the gut of an infected host or, perhaps more often, within fecal matter disseminated in areas of high animal density. One could expect that strains circulating in such promiscuous environments would be subjected to the strongest selective pressures for increasing and diversifying their prophage repertoires. Phage recombination in multiply infected bacterial cells may contribute to the exchange, making it a powerful mechanism for reassorting sequences and generating new strains and phages. This scenario raises the intriguing possibility that the emergence of epidemic strains reflects phage selection as opposed to acquisition of novel pathogenic properties. This seems conceivable, given that in Salmonella, emergence of a new strain is usually indicated by the appearance of a new phage type (also called lysotype), a taxonomic definition (representing the strain sensitivity to a reference collection of bacteriophages) that is likely to reflect the prophage composition of the isolate (9, 14). One could speculate that the dominance of a clone could result from a change in its prophage complement that improves the capacity of the strain to elicit phage-mediated killing of competitors. If this were true, the phage type would not be simply a taxonomic record but would constitute one of the bases of the evolutionary success of a strain.
Acknowledgments
We thank Antony Poteete for the gift of P22 phage and Sergio Uzzau for strain MA6711.
This work was supported in part by the Centre National de la Recherche Scientifique, France. During their stay in France, J. A. Fuentes and G. Mora were supported by the ECOS-Sud program (University “René Descartes,” France/CONICYT Chile).
REFERENCES
- 1.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng, W., S. R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figueroa-Bossi, N., and L. Bossi. 1999. Inducible prophages contribute to Salmonella virulence in mice. Mol. Microbiol. 33:167-176. [DOI] [PubMed] [Google Scholar]
- 5.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-272. [DOI] [PubMed] [Google Scholar]
- 6.Ho, T. D., and J. M. Slauch. 2001. OmpC is the receptor for Gifsy-1 and Gifsy-2 bacteriophages of Salmonella. J. Bacteriol. 183:1495-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho, T. D., N. Figueroa-Bossi, M. Wang, S. Uzzau, L. Bossi, and J. M. Slauch. 2002. Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:5234-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 9.Mmolawa, P. T., R. Willmore, C. J. Thomas, and M. W. Heuzenroeder. 2002. Temperate phages in Salmonella enterica serovar Typhimurium: implications for epidemiology. Int. J. Med. Microbiol. 291:633-644. [DOI] [PubMed] [Google Scholar]
- 10.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 11.Ohnishi, M., J. Terajima, K. Kurokawa, K. Nakayama, T. Murata, K. Tamura, Y. Ogura, H. Watanabe, and T. Hayashi. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. USA 99:17043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 13.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabsch, W., S. Mirold, W. D. Hardt, and H. Tschape. 2002. The dual role of wild phages for horizontal gene transfer among Salmonella strains. Berl. Munch. Tierarztl. Wochenschr. 115:355-359. [PubMed] [Google Scholar]
- 15.Schmieger, H., and P. Schicklmaier. 1999. Transduction of multiple drug resistance of Salmonella enterica serovar typhimurium DT104. FEMS Microbiol. Lett. 170:251-256. [DOI] [PubMed] [Google Scholar]
- 16.Strassmann, J. E., Y. Zhu, and D. C. Queller. 2000. Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature 408:965-967. [DOI] [PubMed] [Google Scholar]