Abstract
Two bacteriophages of an environmental isolate of Vibrio parahaemolyticus were isolated and sequenced. The VP16T and VP16C phages were separated from a mixed lysate based on plaque morphology and exhibit 73 to 88% sequence identity over about 80% of their genomes. Only about 25% of their predicted open reading frames are similar to genes with known functions in the GenBank database. Both phages have cos sites and open reading frames encoding proteins closely related to coliphage lambda's terminase protein (the large subunit). Like in coliphage lambda and other siphophages, a large operon in each phage appears to encode proteins involved in DNA packaging and capsid assembly and presumably in host lysis; we refer to this as the structural operon. In addition, both phages have open reading frames closely related to genes encoding DNA polymerase and helicase proteins. Both phages also encode several putative transcription regulators, an apparent polypeptide deformylase, and a protein related to a virulence-associated protein, VapE, of Dichelobacter nodosus. Despite the similarity of the proteins and genome organization, each of the phages also encodes a few proteins not encoded by the other. We did not identify genes closely related to genes encoding integrase proteins belonging to either the tyrosine or serine recombinase family, and we have no evidence so far that these phages can lysogenize the V. parahaemolyticus strain 16 host. Surprisingly for active lytic viruses, the two phages have a codon usage that is very different than that of the host, suggesting the possibility that they may be relative newcomers to growth in V. parahaemolyticus. The DNA sequences should allow us to characterize the lifestyles of VP16T and VP16C and the interactions between these phages and their host at the molecular level, as well as their relationships to other marine and nonmarine phages.
Surface seawater contains ∼107 phage per ml (6; reviewed in references 19 and 63). Phage lysis affects the marine food web by shunting bacterial biomass into dissolved organic matter. Since dissolved organic matter is essentially assimilated only by other heterotrophic bacteria, phage lysis effectively traps nutrients and energy within the marine microbial food web (19). In contrast, predation by protozoans can transfer bacterial biomass to higher trophic levels in the marine food web. Lysogeny also appears to be common in marine environments, as measured by the fraction of bacterial isolates that are lysed upon treatment with DNA-damaging agents (45). In order to better understand the contribution of phages to the shaping of marine communities, we are seeking a better understanding of the diversity of bacteriophages and their lifestyles.
The extent of phage diversity in any environment is essentially unknown, but it is generally thought that most prokaryotic organisms are parasitized by one or more phages (reviewed in reference 63). Traditional methods of documenting phage diversity require isolating hosts and then screening for infective phages (for marine phages, see references 45 and 58). This method of sampling environmental phages is biased for at least two reasons: first, >99% of microbes have not been cultured by standard techniques (20), and second, some phages (e.g., temperate and chronic or pseudolysogenic phages) may not produce easily identifiable plaques.
Electron microscopy has shown that the three main morphotypes of double-stranded DNA phages, the Myoviridae, Podoviridae, and Siphoviridae, are present in all environments that have been sampled. Pulsed-field gel electrophoresis of the total viral particles isolated from various marine environments and the total viral DNA fraction has been performed (57). In order to determine the actual diversity of phages in various environments and to begin to investigate the consequences of this diversity, it would be desirable to identify different viruses or at least different families of related viruses and to determine the prevalence of each taxon in each environment of interest. While this is significantly more difficult than analysis of organisms which carry ribosomes (in which ribosomal DNA genes can be used as an identifying marker), progress has been made by amplifying specific viral genes or by using degenerate primers to amplify genes expected to appear predominantly in certain viral types (16, 51, 56; M. Breitbart and F. Rohwer, submitted for publication; F. Wolven, F. Rohwer, and A. Segall, unpublished results).
In order to have probes for as many diverse virus types as possible, genome sequences of at least common representative members of the marine bacteriophage community are necessary (46). Currently, only five marine phages, four of which are lytic, have been sequenced; these five phages are PM2 (34), roseophage SIO1 (51), cyanophage P60 (15), Vibrio harveyi phage VHML (43), and Vibrio parahaemolyticus phage VpV262 (23). Only one of these phages may be a classically temperate phage (43).
Here we describe the genomes of two closely related vibriophages that grow on an environmental isolate of V. parahaemolyticus, strain 16. These are the only marine phages whose biogeographic distribution has been documented extensively; Kellogg and Paul found that the vibriophage which they isolated in Tampa Bay in 1994 is distributed very widely (as far as Hawaii), and it was found in the environment at all times of the year (26). This wide distribution of a virus whose host, V. parahaemolyticus, is also very widely distributed in marine environments made the phage a natural target of study. To better understand the lifestyle and influence of this virus on its community, we began by sequencing its genome. We found that the phage lysate which we originally obtained in fact contained two distinct phages that were not separable under the original plating conditions. The genome sequences have given us some hints that these phages may be temperate, although we have not proven this yet. We have also begun to examine the similarities and differences between marine and nonmarine phages by comparing our genomes to phage sequences obtained from uncultured phage DNA sequences obtained from three different environments. This comparison has reconfirmed that sequences of phages having defined lab-adapted hosts are useful in understanding and exploring the diversity of environmental phages.
MATERIALS AND METHODS
Isolation of VP16 DNA.
Vibriophage 16 (VP16) lysate and the phage host, V. parahaemolyticus strain 16, were obtained from John Paul (University of South Florida College of Marine Science). Host cells were grown on Zobell medium plates (5 g of peptone per liter, 1 g of yeast extract per liter, 0.01 g of FePO4per liter, and 1.5 g of agar per liter in 80%, filtered [pore size, 0.45 μm] seawater) at room temperature. A single host colony was used to inoculate 30 ml of Zobell liquid medium and was grown with shaking at room temperature until the density reached ∼2 × 109 cells per ml, as determined by spectrophotometry. This culture was subcultured into 300 ml of fresh Zobell liquid medium and grown for an additional 7 h at room temperature with shaking. Cells were pelleted, resuspended in 20 ml of MSM (32.5 mM NaCl, 12 mM MgSO4, 50 mM Tris, 0.1% gelatin), and then infected with 2 μl of phage lysate and incubated at room temperature for 30 min. Infected cells were transferred to 1 liter of Zobell liquid medium and grown overnight at room temperature with gentle shaking. Lysis was observed within 8 to 12 h of inoculation.
Phages were collected by using a protocol described by Maniatis et al. (33). Host cells potentially carrying mature phage particles were lysed by incubation for 15 min at room temperature with 10 ml of chloroform per liter of medium. Free nucleic acids were degraded with 100 μl of RNase A (10 mg/ml) and 10 μl of DNase I (2,000 Kunitz units (KU)/ml) for 1 h. Sodium chloride was added to a final concentration of 1 M, and the preparation was incubated on ice for 30 min. Cellular debris was pelleted by centrifugation in a Sorvall RC5C centrifuge by using a GSA rotor for 30 min at 9,000 rpm. The supernatant was filtered through one layer of Kim Wipes and mixed with polyethylene glycol 10000 (100 g of polyethylene glycol per liter) to precipitate phage particles. This solution was incubated on ice overnight and then centrifuged at 9,000 rpm for 30 min in a Sorvall GSA rotor. The pellet was resuspended in 5 ml of MSM. Phage particles in MSM were extracted by using 2 volumes of chloroform to remove the polyethylene glycol residue. Several extractions were performed; the aqueous top phase was collected after each extraction and then mixed with 0.75 g of CsCl per ml. The mixture was transferred to ultracentrifuge tubes and spun at 30,000 rpm for 24 h at 4°C by using a Beckman SW41 rotor. A phage band was illuminated with a UV light source, and from this band a total of 2 ml was extracted by using a 20-gauge needle and a syringe. VP16 particles in CsCl were treated with 0.1 volume of 2 M Tris Cl (pH 8.5) and 1 volume of formamide to dissociate the major capsid proteins, which released phage DNA into the solution. VP16 DNA was precipitated once with 100% ethanol and then washed twice with 70% ethanol and pelleted by centrifugation for 15 min at 14,000 rpm in a Sorvall SS34 rotor. The final vibriophage DNA pellet was resuspended in TE (10 mM Tris [pH 7.6], 0.5 mM EDTA). The DNA concentration was determined by spectrophotometry by using the optical density at 260 nm.
Additional centrifugation in CsCl equilibrium gradients was performed to remove contaminants of the phage DNA, such as carbohydrates or polysaccharides. In this case, VP16 DNA was mixed with 0.7 g of CsCl per ml and centrifuged at 30,000 rpm for 24 h at 4°C by using a Beckman SW41 rotor. This produced an hourglass-shaped band, and the lower half of the band was extracted with a 20-gauge needle and a syringe. The band material was recentrifuged in CsCl under the conditions described above, which produced two distinct bands. The lower band was extracted, and this DNA was used for the shotgun libraries prepared by physical shearing and random amplification of VP16 DNA.
DNase I, Sau3AI, and nebulizer shotgun libraries.
VP16 DNA was digested with DNase I for different times as described previously (33). Sau3AI reaction mixtures were assembled according to the manufacturer's protocol (New England Biolabs) and incubated for 1 h. Both DNase I digestion and Sau3AI digestion were stopped by adding loading dye containing sodium dodecyl sulfate (SDS) (33). VP16 DNA was also physically sheared by using a nebulizer (Invitrogen Corp.) at 10 lb/in2 for 5, 10, 15, 30, 45, and 60 s.
RASLs.
Shotgun libraries were created by in vitro random amplification of VP16 DNA as described by Rohwer et al. (50). VP16 genomic DNA was randomly amplified by performing a standard PCR with Vent DNA polymerase (New England Biolabs). A standard 50-μl random amplification shotgun library (RASL) PCR mixture consisted of VP16 DNA, Vent polymerase (exopolymerase), deoxynucleoside triphosphates (New England Biolabs), MgSO4 (Sigma), and random 10-mer primers (San Diego State University Microchemical Core Facility). The reaction mixture was subjected to 25 thermal cycles (96°C for 1 min, 35°C for 1 min, 72°C for 3 min) and purified with an UltraClean PCR clean-up kit (MoBio, Solana Beach, Calif.). Randomly amplified DNA was isolated from a 1% agarose gel and was quantitated by spectrophotometry by using the optical density at 260 nm.
Cloning of library fragments and sequencing.
Fragments between 500 and 2,000 bp long generated by either of the methods described above were electrophoresed and cut from a 1% TBE agarose gel stained with ethidium bromide. DNA was extracted with an UltraClean gel spin kit (MoBio), extracted with phenol-chloroform, precipitated with 100% ethanol, washed with 70% ethanol and dried twice, and resuspended in 10 μl of TE. VP16 DNA fragments created by restriction digestion and physical shearing were blunt ended by using T4 DNA polymerase and the Klenow fragment (1 μl each), as well as each deoxynucleoside triphosphate at a concentration of 2.5 mM, 10× T4 DNA polymerase buffer (New England Biolabs), and enough sterile nanopure water to bring the total volume to 25 μl. Then they were ligated into a pCR-BLUNT (Invitrogen) cloning vector according to the manufacturer's protocol. VP16 DNA fragments obtained by random amplification (RASL) (50) were ligated into the pCR-TA Topo cloning vector. Ligated products were heat shocked into Top10 cells and incubated overnight on Luria-Bertani (LB) plates containing 50 μg of kanamycin per ml, 20 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per ml, and 13 μl of a 250 mM stock solution of IPTG (isopropyl-β-d-thiogalactopyranoside). White or light blue colonies were grown in LB liquid medium at 37°C to the log phase. Plasmids were extracted with an UltraClean MINI plasmid prep kit (DocFrugal Scientific Corp.) and bidirectionally sequenced with primers M13F and M13R. Plasmid inserts were sequenced with an ABI 377XL or ABI3100 capillary electrophoresis sequencer (Applied Biosciences, Foster City, Calif.) by using BigDye terminator technology (ABI Prism) at the Microchemical Core Facility, San Diego State University.
Linker amplified shotgun libraries (LASLs).
Purified DNA from clear and turbid VP16 plaques was sent to Lucigen Corporation for additional library construction and sequencing. At Lucigen, VP16 DNA was physically sheared, blunt ended, and ligated into the pSMART cloning vector (Lucigen). Inserts were sequenced from both sides by using Lucigen's Amp primers (AmpL2 [5′ CTGAATGATATCAAGCTTGAA] and AmpR2 [5′ GTATTGAGCGATATCTAGAGAA]).
Primer walking and multiplex PCR.
Many gaps between pairs of contigs were connected by primer walking. Primers designed to extend the ends of contigs were used in standard sequencing reactions performed with VP16 lysate DNA as the template. Gap lengths were estimated by multiplex PCR; pools of primers were used in a single PCR to amplify regions of genomic DNA corresponding to DNA sequences between contigs (51, 61). PCR products purified from a 1% agarose gel were cloned and sequenced.
Contig assembly.
DNA sequence data were assembled into contigs by using the Sequencher 4.1 software package (Gene Codes, Ann Arbor, Mich.). The initial assembly was done under stringent assembly conditions (90% sequence match with a minimum overlap of 50 bases), which yielded contigs consisting of sequences meeting the assembly criteria. Contigs were edited by removing nucleotide gaps and replacing miscalled bases by using a majority rule from the best-quality chromatograms. Several rounds of assembly and editing were performed, and more relaxed parameters were used at each successive assembly procedure (there was a minimum 75% sequence match, and there was a 30-base overlap region) until no more assemblies were found by the software.
GeneMark and SGM.
Open reading frame (ORF) coordinates were obtained by using Heuristic GeneMark (http://opal.biology.gatech.edu/GeneMark/heuristic_hmm2.cgi). A Java program, Small Genome Mapper (SGM), was written and used to parse genome data files (DNA sequence, GeneMark output, and Blast output) and to graphically display an annotated genome map. SGM initially parses a genomic nucleotide sequence into a FastA formatted file consisting of either nucleotide ORF or amino acid sequences (user specified). The blastp program (Net Blast's blastcl3.exe program, downloadable from ftp://ftp.ncbi.nlm.nih.gov/blast/network/netblast/CURRENT/) was executed by using amino acid sequences. ORF nucleotide sequences were compared with the GenBank database by using tblastx and were compared with the blastp output. After completion of Net Blast, SGM parses Blast results, GeneMark output, and the genomic DNA sequence and displays an annotated genome map. ORFs are color coded based on the negative logarithm of the Blast E-value score for the top database hit; the lowest negative logarithmic values corresponding to high levels of similarity are indicated by red, whereas the lowest negative logarithmic values corresponding to low levels of similarity are indicated by dark blue and negative log values in between are indicated by color intensities intermediate between red and blue.
Other bioinformatic analyses.
All ORFs were also analyzed by PSI-BLAST, Pfam, and ProDom. These analyses did not add substantially new information. In addition, the two genomes were compared by using BLAST 2 Sequences, which highlighted closely related segments. About one-half of the segments that were identified by BLAST 2 Sequences as sequences which were not related fell in the middle of ORFs. Since it seemed unlikely that entirely independent insertions had occurred in analogous genes in the two phages, the nucleotide sequences of each pair of ORFs and, independently, the amino acid sequences encoded by the ORFs with such a BLAST gap were analyzed by ClustalX, a global alignment program. Because ClustalX aligns sequences over the entire length, it can accommodate regions in which there is low or no similarity interspersed with regions in which there is high sequence identity or similarity better than BLAST. The quality of the Clustal alignments was scored by expressing the number of identical and similar nucleotide residues as a fraction of the total.
Finder of Identical Sequences (FISeq) program.
A second Java program was written to compare the levels of nucleotide sequence identity between VP16 DNA sequences from particles that were shown to produce clear (VP16C) and turbid (VP16T) plaque morphologies. This program used a method of bisection to find identical sequences (minimum length, 20 nucleotides) in the genomes of VP16C and VP16T and generated a list of their coordinates. The lengths and coordinates of all of the identical sequences were written to a text file and then graphed by using Microsoft Excel.
Electron micrographs of VP16.
Images of VP16C and VP16T particles were obtained by transmission electron microscopy (Imaging Facility, San Diego State University) by using a Phillips EM410A electron microscope. VP16 lysates were spread over a Formvar-coated mesh copper grid in the presence of bacitracin (0.75 μM) and stained with 1.5% uranyl acetate. Micrographs were obtained at an accelerating voltage of 60 kV at a magnification of ×85,000 or ×65,000.
Analysis of VP16 phage genomes for the presence of cos sites by restriction enzyme digestion.
Restriction enzyme digests of VP16C and VP16T were used to assess the similarity of the two phages at a gross level. Approximately 1 μg of VP16 DNA was incubated with 1 μl of enzyme (20,000 U/ml) in the appropriate buffer (New England Biolabs) and sterile nanopure H2O in a 20-μl (final volume) mixture for 3 h at 37°C. Two aliquots from each digest were loaded on a 0.8% TBE agarose gel. One sample was loaded directly after digestion. The second aliquot was heated in a 65°C water bath for 10 min and then cooled on ice before it was loaded. A relatively low voltage (40 to 80 V) was used during electrophoresis to minimize heat generation so that the putative noncovalent cos ends held together by base pairing were not denatured.
Obtaining the cos site sequence.
To obtain the cos site sequence of VP16, two VP16 DNA templates were used for PCR amplification with primers that pointed outward from the largest final contig. The encapsidated phage genome template was obtained directly from isolated VP16 lysate DNA which was drop dialyzed against nanopure H2O. The second template, representing the phage DNA as it might be found after infection, consisted of VP16 lysate DNA that was first treated with T4 ligase (New England Biolabs) after incubation at room temperature and then drop dialyzed against nanopure H2O. The ligation reaction was performed to covalently close the phage DNA by sealing the cos site nicks after annealing was allowed. Two oligonucleotides were designed to extend the beginning (CosL [5′ GCTCGCATCCTCGTACAGTC 3′]) and the end (CosR [5′ CGCGCTAAGTGCTTGAAT 3′]) of the putative complete genomic DNA sequence of VP16T and VP16C. cos regions on linear and circular DNA templates were amplified by using primers CosL and CosR and Taq polymerase (Life Technologies, Inc.). PCR products were obtained only after ligation, as expected, and then were purified with an UltraClean 15 kit (MoBio) and sequenced directly. To locate the cos site within the fragment, direct sequencing was performed by using the unligated phage genomes with the CosR and CosL primers individually. The resulting sequence was assembled into a contig by using Sequencher 4.1 and was aligned with the sequence of the PCR fragment obtained from the ligated genome. The sequences overlapped by 15 bases, which comprised the cos site.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the two phage genomes are as follows: AY328852 for VP16T and AY328853 for VP16C.
RESULTS AND DISCUSSION
Isolation and genomic sequencing of VP16T and VP16C.
The original VP16 lysate was obtained from the Paul lab and had been through several plaque purifications. A variety of shotgun libraries were made from this lysate. As discussed below, only the RASL technique worked well with this DNA (50). Sequencing was carried out until approximately 4× coverage of the genome had been obtained. The resulting sequences were assembled by using 20-bp overlaps and a level of identity of 85 to 90%. At this point, we switched to multiplex PCR and primer walking to connect the last contigs. However, these approaches yielded results that were inconsistent with a single genome, including apparently branched contigs. This raised the possibility that the VP16 lysate contained more than one phage.
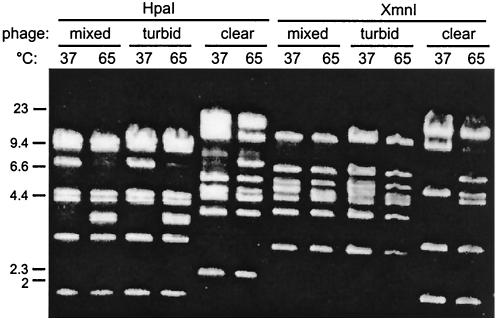
To determine if the VP16 lysate actually contained a mixture of phages, we tested plaque formation on several media. The lysate plated on LB medium supplemented with 2.5% NaCl produced two types of plaques, turbid and clear. The turbid plaques were uniformly cloudy in the middle rather than the traditional turbid plaque made by phage lambda or P22. The plaques maintained their appearance through three rounds of purification. DNA was obtained from lysates prepared by using turbid or clear plaques as inocula, and the restriction digestion patterns of DNA from the original lysate and DNA from the subsequent lysates were compared. As Fig. 1 shows, the DNA isolated from the turbid-plaque lysate was distinct from that isolated from the clear-plaque lysate. The two phages were designated VP16T and VP16C based on plaque appearance. The mixed lysate contained predominantly VP16T.
FIG. 1.
Restriction analysis of phage genomic DNA isolated from the VP16 lysates. Phage DNA was purified from the original lysate (mixed) and from lysates made from clear and turbid plaques from the original lysate after three successive plaque purifications on LB medium containing 2.5% NaCl. The phage DNA was restricted with HpaI or XmnI. After restriction digestion at 37°C, the digests were either kept at 37°C before they were loaded on the agarose gel or incubated for 20 min at 65°C and then loaded on the agarose gel.
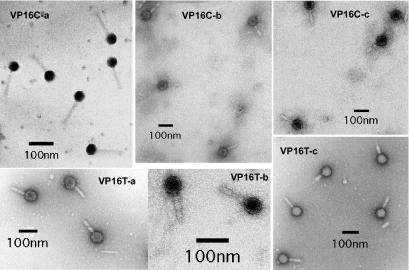
Electron microscopy of the VP16T and VP16C lysates showed that the phage particles were essentially identical to each other; each particle had an icosahedral head about 50 to 60 nm in diameter and an 80- to 100-nm tail (Fig. 2). The tails appeared to consist of a thin internal core sheathed in two thicker collars that were often separated but sometimes were contiguous, as if compression of the tail had brought the two collars together (Fig. 2).
FIG. 2.
Electron micrographs of VP16C and VP16T particles. The micrographs show phage particles from several independent lysates. Although a few of the micrographs showed phage particles like those shown in the VP16C-a panel, the vast majority of micrographs showed phage particles like those shown in the rest of the panels. Note that many of the particles show a disruption in the middle of the tail collar, while the collars are compressed together in a significant fraction of the remaining particles.
New libraries were generated from VP16T and VP16C by using the LASL method. Two hundred clones from each library were sequenced and aligned with the sequences from the original mixed lysate by using a criterion of at least 90% identity over 30 to 50 bp. The turbid and clear phage sequences aligned with different contigs, confirming that the previous library did not contain chimeric fragments with sequences from both phage genomes. The final sequencing to close the two genomes was performed by primer walking by using the VP16T and VP16C genome DNA as templates. The results showed that different phage genomes can be sequenced from mixed lysates if care is taken during the assembly process. In the descriptions below, we refer to putative genes as ORF(number)T and ORF(number)C to indicate their presence in the VP16T and VP16C genomes, respectively. The sequence, map, and predicted ORFs for each phage are available at http://www-rohan.sdsu.edu/∼segurita/vp16/turbid041502/index.html forVP16T and at http://www-rohan.sdsu.edu/∼segurita/vp16/clear061902/index.html for VP16C.
Identification of cos sites.
To determine whether VP16C and VP16T have cohesive ends (cos sites) (reviewed in reference 13), parallel restriction digests were incubated at 37 or 65°C after digestion and then analyzed by agarose gel electrophoresis. If cos ends were present in the genomic DNA packaged in either of the phage particles, incubation at 65°C should have denatured the short complementary single strands and the restriction fragment in which the cos ends resided should have disappeared after denaturation, while two new, smaller fragments should have appeared. This was observed for both the VP16T and VP16C phage genomes (Fig. 1, compare lane 3 with lane 4 for VP16T and lane 11 with lane 12 for VP16C). Because of this feature, the very ends of each genome were sequenced from phage genomic DNA that was first ligated and then amplified by PCR performed with primers designed to point outward from the ends of the single contig comprising the genome of each phage. When the template DNA was circularized by using the cohesive ends, the two primers should have generated a relatively small fragment containing the terminal sequences of each genome. For both phages, these fragments were roughly 500 bases long (data not shown). To determine the sequences of the cohesive ends themselves, the same two primers were used separately as primers in sequencing reactions. The resulting sequences were aligned, and the results revealed an overlap of 15 bases, 14 of which were identical in the two phages (GTTTGGAATCTGACC for VP16T and GTTTGGAATCTGCCC for VP16C; the underlined base is unique in each cos sequence). The overlap of the sequencing primer extension products indicated that the cos sites had 3′ single-stranded extensions.
G+C content and codon preferences.
VP16T and VP16C were 49,575 and 47,537 bp long, respectively. The G+C content of each genome was 59%. In comparison, the G+C of V. parahaemolyticus is 46%, and thus there is a 13% difference between the phage and host DNA. Although large differences in G+C contents are not unusual in defective (cryptic) prophages (in fact, this feature was originally used to identify some of these elements, which generally have lower G+C contents than the rest of the host genome), the G+C contents in other phage-host systems are much more similar (Table 1). We compared the codon usage of all of the ORFs of the two V. parahaemolyticus chromosomes with the codon usage of all of the ORFs of the two phages using the General Codon Usage Analysis program available at http://bioinf.may.ie/gcua/download.html (36). This program calculates a relative synonymous codon usage value for each codon in an ORF. The results showed that in most cases, codon usage was different in the phage genomes and the V. parahaemolyticus genome. In 44 of 59 codons (the codons for methionine and tryptophan and the three stop codons were excluded from this analysis), the third position was biased, as expected for the G+C content of the genome; i.e., the VP16T and VP16C codons were more likely to have G or C at the third position, while the V. parahaemolyticus codons were more likely to have A or U at this position (data not shown). Twelve codons occurred equally frequently in the phage genomes and in the V. parahaemolyticus genome. The occurrence of the remaining three codons, UUG (Leu), CUA (Leu), and AUA (Ile), was biased strongly in the direction opposite from that expected based on the G+C content of the VP16T and VP16C genomes; UUG was present less frequently than expected, while CUA and AUA were present more frequently than expected. The UUG and AUA codons appeared in 71 to 73.5% of the ORFs, while CUA appeared in 89% of the ORFs in both phages. A comparison of the UUG and CUA codons was useful, since these codons code for the same amino acid and have the same G+C content and neither codon is rare. In V. parahaemolyticus, the frequency of UUG is 23.3/1,000 codons, and the frequency of CUA is 15.8/1,000 codons (this frequency is based on 242 coding sequences [codon usage database maintained by Y. Nakamura at http://www.kazusa.or.jp]). In comparison, in the VP16T phage the frequency of UUG was 8.6/1,000 codons, and the frequency of CUA was 17.3/1,000 codons. The isoleucine codon (AUA) is a rare codon in V. parahaemolyticus (5.6/1,000 codons, about 20% as frequent as either of the remaining two Ile codons). In the VP16T phage, however, the frequency of this codon was 10.2/1,000 codons.
TABLE 1.
Difference between phage and host G+C contents
| Host
|
Phage
|
Difference (%) | ||
|---|---|---|---|---|
| Organism | G+C content (%) | Phage | G+C content (%)a | |
| V. parahaemolyticus | 46 | VP16T and VP16C | 59 | 13 |
| V. parahaemolyticus | 46 | VpV262 | 46 | 0 |
| Haemophilus influenzae | 39 | HP1 | 40 | 1 |
| E. coli | 51 | 47-52 | 1-4 | |
| S. thermophilus | 40 | 38-39 | 1-2 | |
| Lactobacillus sp. | 50 | 50-51 | 1 | |
| S. enterica | 47 | P22 | 50 | 3 |
| B. subtilis | 41 | 40-43 | 1-2 | |
| M. tuberculosis | 62 | 62-64 | 0-2 | |
Where a range is given, the G+C values for several phage genomes were included in the analysis.
Overall, the bias for G or C in the third position occurs in almost 3:1 codons of all phage ORFs, while in V. parahaemolyticus G or C occurs in the third position in only 44.5% of the codons (http://www.kazusa.or.jp). The difference in codon usage between the VP16 phages and V. parahaemolyticus presumably reflects the life history and the host range of the two phages (27) and suggests that these phages may have acquired the ability to infect V. parahaemolyticus only recently.
SGM.
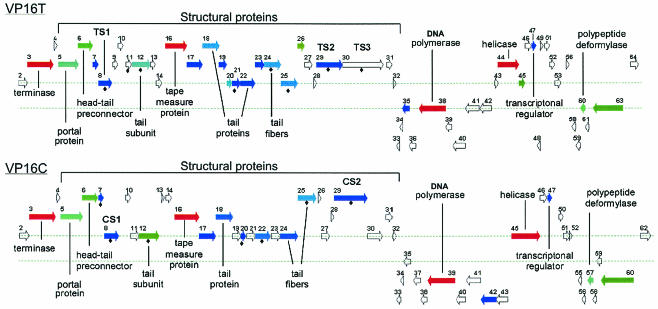
The coordinates of the putative ORFs were identified by using GeneMark (7) and were used as input into the Java computer program SGM (see Materials and Methods). As part of the SGM algorithm, the amino acid sequences encoded by the phage ORFs were input into a batch BLAST program (National Center for Biotechnology Information) (4, 5) to generate a table of the top 11 BLAST hits for each ORF. In addition, the SGM algorithm created a map of the genome in which each ORF was identified by a proportional arrow positioned along the DNA genome both according to the coding strand and according to the translation frame (Fig. 3). Each arrow was color coded; red arrows indicated ORFs that were most similar to other entries in the GenBank database (i.e., had very low expect values), and dark blue arrows indicated ORFs that were not very similar to other GenBank entries (i.e., had high expect values), while arrows with intermediate color intensities indicated intermediate similarity scores. ORFs that exhibited no similarity to any GenBank entry appeared as arrows outlined with gray. For clarity, only ORFs whose products were longer than 70 amino acids were shown on the map. Based on output from the GeneMark algorithm, the phages carried 62 (VP16C) or 64 (VP16T) ORFs, and only 10 to 14 of these ORFs (a little under 25% of the coding capacity) were similar to GenBank entries whose functions may be inferred from similarity to other genes. Many more putative genes exhibited similarity to hypothetical ORF entries in the GenBank database. A complete list of ORFs carried by both phages is shown in Table 2, along with the corresponding most similar GenBank entries. If the closest GenBank hit was an ORF encoding a hypothetical protein but a less similar entry had a known or predicted function or was similar to a phage gene, that entry is also shown in Table 2.
FIG. 3.
Genomic maps of VP16T and VP16C. ORFs are color coded based on the BLAST E-values and are labeled largely based on the BLAST hits shown in Table 2. BLAST hits returning hypothetical proteins or weak E-values (≤1e−2) are shown as unlabeled color-coded ORFs. Gray ORFs do not exhibit homology to GenBank entries. An ORF labeled with a solid diamond is a VP16 ORF that was identified by the neural network as an ORF that encodes a possible capsid protein. The amino acid sequences of VP16T proteins were found in VP16T ORF sequences labeled TS1 (ANELATGWVQ), TS2 (LIKLT), and TS3 (ADRHIL). Similarly, amino acid sequences of VP16C proteins were encoded in the VP16C ORF sequences labeled CS1 (ANELATGWVQ) and CS2 (LVKLS). ORFs encoding sequences shorter than 70 amino acids are not included in the map for ease of display, but all ORFs are listed in Table 2.
TABLE 2.
ORFs in the genomes of VP16T and VP16C
| VP16T
|
VP16C
|
||||
|---|---|---|---|---|---|
| ORF/ stranda | Encoding nucleotides | Function of similar hit and expect value (% identity)b | ORF/ strand | Encoding nucleotides | Function and expect value (% identity)b |
| 1/− | <3-116 | 1/− | <2-85 | ||
| 2/+ | 115-828 | Likely small subunit of terminasec | 2/+ | 124-837 | (70% identical to ORF2C) |
| 3/+ | 812-2839 | Large subunit of phage lambda terminase gpA, 3e-17 (22); closest phage hit bacteriophage WO, 3e-50 (27); closest bacterial hit, R. solanacearum (28) | 3/+ | 821-2842 | phi WO, 1e-51 (27) |
| 4/+ | 2880-3158 | 4/+ | 2883-3161 | ||
| 5/+ | 3224-4861 | Portal protein gp4 of phi 21, 6e-13 (22); minor capsid (portal) protein gpB of phage lambda, 6e-10 (19); closest bacterial hit, R. solanacearum, 5e-12 (21) | 5/+ | 3227-4864 | gpB of phage lambda, 2e-14 (21) |
| 6/+ | 4845-6032 | Similar to head-tail preconnector protein of Vibrio shiloii, 2e-19 (34); closest phage hit gpC of phage lambda, 4e-9 (27) | 6/+ | 4848-6035 | Same (31) |
| 7/+ | 6047-6451 | 7/+ | 6051-6458 | ||
| 8/+ | 6466-7560 | Hypothetical protein gp348 of Sfi11 (S. thermophilus), 5e-4 (25) | 8/+ | 6472-7566 | gp348 of Sfi11, 0.005 (24) |
| 9/+ | 7610-7999 | 9/+ | 7954-8046 | ||
| 10/+ | 8007-8393 | 10/+ | 8046-8432 | ||
| 11/+ | 8603-9064 | 11/+ | 8425-9114 | ||
| 12/+ | 9077-10579 | Tail sheath protein L of phage Mu, 3e-12 (24) | 12/+ | 9127-10629 | gpL of phage Mu, 1e-21 (27) |
| 13/+ | 10592-10975 | 13/+ | 10815-11024 | ||
| 14/+ | 11053-11454 | 14/+ | 11109-11510 | ||
| 15/+ | 11550-11654 | 15/+ | 11606-11710 | ||
| 16/+ | 11769-13514 | Hypothetical protein (Haemophilus somnus), 6e-52 (31); ORF43, V. harvey phage VHML, 4e-49 (32), putative tail length tape measure protein | 16/+ | 11759-13570 | Same, 7e-41 (29) |
| 17/+ | 13514-14767 | 17/+ | 13570-14823 | ||
| 18/+ | 14760-16013 | Putative tail protein, E. coli O157:H7, 3e-10 (25); gpP tail protein of phage Mu, 2e-009 (23) | 18/+ | 14816-16069 | 43-kDa tail protein, H. influenzae, 6e-9 (24) |
| 19/+ | 15992-16636 | GTG start codon | 19/+ | 16048-16692 | |
| 20/+ | 16648-17055 | Hypothetical protein NP518999.1 of R. solanacearum, 4e-11 (30) | 20/+ | 16705-17112 | Hypothetical protein of R. solanacearum, 3e-5 (26) |
| 21/+ | 17059-17724 | 21/+ | 17116-17781 | ||
| 22/+ | 17734-18921 | Tail protein of phi V (Shigella flexneri), 4e-008 (28); hypothetical protein ymfP (b1152) in prophage e14 region of E. coli K-12, 7e-8 (29) | 22/+ | 17791-18960 | Hypothetical protein, E. coli O157:H7, 2e-9 (22) |
| 23/+ | 18914-19636 | 23/+ | 18973-19695 | ||
| 24/+ | 19649-20983 | Hypothetical protein NP519813.1 of R. solanacearum, 3e-10 (71); putative tail fiber-related protein NP520042.1 of R. solanacearum (78) | 24/+ | 19708-21042 | Same, 8e-9 (79% identical to ORF24T); putative tail fiber-related protein NP520042.1 of R. solanacearum, 1e-7 (70) |
| 25/+ | 20986-22320 | Hypothetical protein NP519813.1 of R. solanacearum, 5e-10 (43) | 25/+ | 21045-22394 | Same, 4e-010 (81% identical to ORF25T); putative tail fiber-related protein NP520042.1 of R. solanacearum, 1e-007 (33) |
| 26/+ | 22341-22865 | Hypothetical protein NMB1012 (imported) of Neisseria meningitidis, 2e-28 (39); secretion activator protein NP539912.1, Bacillus melitensis, 3e-15 (30) | |||
| 26/+ | 22557-22808 | ||||
| 27/+ | 22862-23458 | 27/+ | 22810-23406 | ||
| 28/+ | 23566-23802 | 28/+ | 23525-23758 | ||
| 29/+ | 23804-25897 | 29/+ | 23763-26282 | Probable HA-related protein NP519008 of R. solanacearum, 0.011 (28) | |
| 30/+ | 25901-29173 | 30/+ | 26287-27408 | ||
| 31/+ | 29411-29884 | 31/+ | 27638-28117 | ||
| 32/+ | 29899-30168 | 32/− | 28126-28392 | ||
| 33/− | 30182-30433 | 33/− | 28411-28722 | ||
| 34/− | 30408-30647 | 34/− | 28712-28936 | ||
| 35/− | 30619-31185 | eac protein of phage P22, 0.027 (19) | 35/− | 28908-29468 | No similarity (70% identical to ORF35T) |
| 36/− | 31178-31705 | 36/− | 29461-29640 | ||
| 37/− | 29633-30187 | ||||
| 38/− | 30193-30678 | ||||
| 37; 38/− | 31763-31951; 31948-34092 | DNA, polymerase of phi SPO2 of B. subtitis, 4e-70 (35 and 31) | 39/− | 30737-32797 | Same, 3e-57 (31) |
| 39/− | 34089-34602 | 40/− | 32857-33582 | First 520 of 726 nucleotides are similar to ORF39T | |
| 40/− | 34667-35680 | 41/− | 33653-34714 | ||
| 41/− | 35692-36744 | 42/− | 34732-35859 | Phage-related protein of X. fastidiosa 9a5c, 8e-004 (26) | |
| 42/− | 36823-37686 | 43/+ | 35863-36714 | ||
| 42.1/− | 37785-37858 | Same as ORF44C, not called by GeneMark | 44/+ | 36801-37004 | 90% likelihood of HTH domain |
| 43+ | 37969-38226 | ||||
| 44/+ | 38219-39871 | Hypothetical protein of phi A2 (Lactobacillus casei), 6e-50 (32); helicase, phi PSA (Listeria monocytogenes), 7e-47 (32); putative DEAH family helicase, Lactobacillus phage phi adh, 3e-46 (29) | 45/+ | 36997-39153 | Same, 9e-49 (34) |
| 45/+ | 39868-40374 | Hypothetical protein NP416862 of E. coli K-12, 1e-19 (45); closest phage hit, unknown protein, phi V (S. flexneri), 3e-19 (44) | |||
| 46/+ | 40374-40886 | 46/+ | 39153-39662 | ||
| 47/+ | 40932-41267 | Putative transcriptional regulator NP 335121, M. tuberculosis CDC1551, 2e-5 (33) | 47/+ | 39708-40043 | Same, 4e-5 (34) |
| 48/− | 41324-41575 | ||||
| 49/+ | 41628-41840 | ||||
| 48/+ | 40031-40216 | ||||
| 50/+ | 41837-41995 | 49/+ | 40402-40560 | ||
| 51/+ | 41982-42344 | 50/+ | 40580-40912 | ||
| 52/+ | 42341-42781 | ||||
| 53/+ | 42778-43266 | 51/+ | 40909-41361 | ||
| 54/+ | 43280-43480 | 52/+ | 41365-41574 | ||
| 55/+ | 43483-43674 | 53/+ | 41578-41766 | ||
| 56/+ | 43676-43909 | 54/+ | 41823-42002 | ||
| 57/+ | 43969-44112 | ||||
| 58/− | 44109-44387 | 55/− | 41999-42265 | ||
| 59/− | 44384-44731 | 56/− | 42262-42570 | ||
| 60/− | 44728-45141 | Putative polypeptide deformylase, S. coelicolor, 3e-015 (37) | 57/− | 42704-43117 | Polypeptide deformylase, Aquifex aeolicus, 2e-13 (38) |
| 61/− | 45138-45422 | 58/− | 43114-43416 | ||
| 62/− | 45371-45778 | 59/− | 43413-43784 | ||
| 63/− | 45805-48180 | Hypothetical protein NP489430 of Nostoc sp. PCC7120, 2e-19 (27); virulence-associated protein E of D. nodosus, 3e-15 (28) | 60/− | 43781-46156 | Same, 3e-20 (27) |
| 61/+ | 46198-46371 | ||||
| 64/+ | 48776-49468 | 62/+ | 46690-47442 | (86% identical to ORF64T) | |
ORFs of the two phages whose nucleotide sequences are similar or which encode similar amino acid sequences are on the same line. ORFs that are not similar to each other are on different lines.
The first function is the most closely related GenBank entry. The other function(s) is the most closely related phage hit, if it is not the top hit. The percentages of identity are the percentages of identical residues between the ORFs and each GenBank gene.
See text.
The genomic organizations of the two phages are very similar, although not identical. The genomes are shown as linear molecules with the cos site at the left end of the map in Fig. 3. Since these two phages are expected to generate circular genomes which serve as transcription templates shortly after infection, the gene organization along the genomes suggests that each phage has two regulatory regions, each with two divergent promoters, which drive a total of four major transcription units. Each phage has a gene, ORF47, which is weakly similar to the genes encoding a transcriptional activator of Mycobacterium tuberculosis and the tetracycline repressor, a protein with a helix-turn-helix (HTH) DNA binding motif (25). A phylogenetic tree of the proteins encoded by these ORFs and the top PSI-BLAST hits related to them showed that the proteins clustered with the phage P22 analog of the cII protein of bacteriophage lambda (data not shown), which is consistent with the possibility that the ORF47-encoded proteins could be part of the transcriptional regulatory loop if these phages are indeed temperate phages. When the proteins encoded by all ORFs were analyzed by using an algorithm predicting the likely presence of HTH motifs (17), the protein encoded by ORF44C was given a 90% probability of having such a domain (in comparison, the proteins encoded by ORF47 sequences from both phages had only a 50% chance of having an HTH motif). Although GeneMark did not identify the homolog of ORF44C in VP16T, there is a similar ORF between ORF42T and ORF43T, which we designated ORF42.1T. Phage VP16T has an additional putative transcription regulator; the product of ORF26T is similar to a secretion activator protein from Zymomonas mobilis that is related to the PhoB/OmpR family of transcriptional regulators, having winged HTH DNA binding domains (35). This ORF has no analog in the VP16C genome. We are cloning and expressing these proteins to test their functions as transcription modulators, and we are beginning to investigate the differences in the transcriptional patterns and the similarities and differences in the regulation of the two phages' life cycles.
Replication functions of the VP16 phages.
Two divergent transcription units to the right of the structural operon (including genes presumed to be involved in morphogenesis and assembly of phage particles) contain genes predicted to be involved in the DNA replication of the genome. In VP16T, ORF37T and ORF38T are homologous to ORF39C of VP16C and most similar to the gene that encodes the DNA polymerase of phage SPO2 of Bacillus subtilis. In VP16C, the putative 686-amino-acid polymerase is encoded by one ORF; in VP16T, it is encoded by two immediately adjacent ORFs, ORF38T, which encodes 714 amino acids, and ORF37T, which encodes 62 amino acids that are nearly identical (only four differences) to the C-terminal 62 amino acids encoded by ORF39C. Since we have many overlapping good-quality sequencing reads for this region, it is unlikely that the splitting of the polymerase ORF is due to sequencing errors. One possibility is that the 62-amino-acid tail is dispensable for polymerase function and that a relatively recent mutational event in VP16C that separated this domain from the main body of the polymerase was not detrimental. A related possibility is that this small domain folds independently and can function in trans with the main polymerase protein. Interestingly, ORF39C starts with a GTG codon, just like its nearest relative, the phage SPO2 polymerase gene; in contrast, ORF38T starts with an ATG codon.
ORF44T and ORF45C, which are on the strand opposite the strand containing the DNA polymerase ORFs, are most similar to each other and to the genes encoding DEAH helicases (Fig. 3 and Table 2). In the same putative transcription unit as the genes encoding the polymerases are ORF35T, which exhibits a low level of similarity to the eac gene of Salmonella phage P22, and ORF35C, which is 70% identical to this gene. The eac gene is expressed early in the infection cycle of P22, but its function is unknown (M. Susskind, personal communication).
Structural and packaging genes.
On the basis of similarities to sequences in the GenBank database, the largest putative operon in each phage consists mostly of structural and packaging genes (ORF3, ORF5, ORF6, ORF12, ORF16, ORF18, ORF22, ORF24, and ORF25); we refer to this operon as the structural operon. ORF3 of phage VP16T and ORF3 of phage VP16C encode proteins with very significant levels of similarity (e-18) to the large subunit (gpA) of terminase, a protein complex encoded by phage lambda and its relatives that recognizes the cos site and separates unit-length phage genomes during the packaging stage of the replication cycle (for a review see reference 13). The proteins encoded by these ORFs are 88.5% identical to each other at the amino acid level. In the well-characterized lambdoid phages, terminase also contains a small subunit, Nu1, which is encoded next to the large subunit (13). Interestingly, the proteins encoded by ORF2T and ORF2C are about 70% identical to each other at the amino acid level, and based on their position downstream of cos and relative to the gene encoding the putative large terminase subunit, we suspect that these two ORFs encode the small terminase subunit. A comparison of the secondary structure predictions for the phage lambda Nu1 translated region with those for ORF2T and ORF2C performed with the SOPMA program (21) showed that these regions share a large number of structural features and that the dispositions of these features along the primary sequence are the same, suggesting that our prediction may be correct. A global alignment was obtained by using ClustalX and the protein sequences encoded by the genes encoding the terminase small subunits of phages λ and 21, the λ-related prophage Qin, and the Gifsy-1 phage of Salmonella enterica and ORF2 of both VP16T and VP16C. The alignment highlighted 12 identical residues, 35 residues belonging to very similar amino acid groups, and 18 residues belonging to less similar amino acid groups (data not shown). Based on these analyses, we concluded that ORF2T and ORF2C encode the small subunits of a terminase protein. The cos site of each of these phages starts at the 5′ end of the phage, upstream of both terminase-encoding subunits. Several other ORFs in the large putative operon have significant similarities to genes encoding structural proteins of other phages, including gpB and gpC of phage lambda (ORF5 and ORF6, respectively) and the tail sheath protein gpL and tail protein gpP of phage Mu (ORF12 and ORF18, respectively). The sequence encoded by ORF12C also exhibits amino acid similarity with a hypothetical protein encoded on chromosome I of V. parahaemolyticus (32); the region of homology is represented six times in the bacterial ORF as nearly perfect 192-residue tandem repeats (84% identity in all repeats). This arrangement is an extreme form of the two 39-residue tandemly repeated segments found in the tail fiber genes of bacteriophages P2 (H protein), phage 186 (K protein), and Mu (22, 37, 53, 65). These repeats are sufficiently divergent to make recombination between them less likely; this, coupled with possible functional selection, presumably accounts for the continued presence of the repeats.
In the same region, the phages also have two ORFs that are similar to each other, ORF24 and ORF25. ORF24T and ORF24C are 79% identical at the amino acid level, and ORF25T and ORF25C are 81% identical. In addition, ORF24T is 35% identical to ORF25T, while ORF24C is 38% identical to ORF25C; these ORFs are similar enough to be clearly homologous genes that probably arose by duplication followed by divergence. Based on the relative identities of the four genes, this duplication must have preceded the divergence of the two phages from each other. We do not know the function of these ORFs, but they are similar to genes that encode phage tail fiber proteins.
We found only one other case of directly repeated DNA in phage genomes in the literature, a 305-nucleotide region that includes the origin of replication of phage Sfi19 (31). The pair of sequences is more than 95% identical, but the two repeats are separated from each other by six ORFs. The importance of two replication origins is unknown. The ORF24 and ORF25 sequences appear to be the longest tandemly repeated sequences identified so far (1,335 nucleotides/gene). A computer analysis of all the completed phage genomes in the GenBank database revealed that repeated sequences, tandem, inverse, or separated from each other by intervening segments, occur rarely (R. Edwards, unpublished data). This suggests that selection to streamline phage genomes is as strong as has been suspected or is counteracted by low levels of recombination and/or strong selection for maintenance of the repeats.
Structural genes may be harder to identify on the basis of similarity to other genes because of lower sequence conservation due to their architectural functions; they often do not encode an enzymatic function with a concomitant demand for active site conservation. For example, it was not possible to identify any of the structural proteins of roseophage SIO1 from its genome sequence (51) until the sequences of other phages, which are related both to roseophage and to coliphages T3 and T7, became available (15, 25). Artificial neural networks (ANN) have been used successfully in the past to predict common amino acid contents of proteins exhibiting levels of sequence similarity too low to detect by BLAST or similar algorithms (2, 3, 41). Neural networks detect common information content in a set of sequences with known functions and are then used to predict whether an unknown ORF is part of the same set. For example, Abremski and Hoess (1) used an ANN to identify a new active site residue found in the phage lambda integrase family in a very poorly conserved region of the proteins. We designed an ANN in an attempt to identify capsid proteins. This network will be described in detail elsewhere (V. Seguritan, J. Donald, P. Salamon, F. Rohwer, and A. Segall, unpublished data). Our neural network was trained with sequences downloaded from GenBank and annotated as major capsid proteins or capsid proteins; sequences with the word probable or putative in the annotation were rejected from the training set. The trained neural network did identify several ORFs as ORFs that encode putative capsid proteins, including VP16T ORF8, ORF11, ORF12, ORF21, ORF24, ORF25, ORF29, and ORF30 and VP16C ORF6, ORF7, ORF11, ORF19, ORF23, ORF24, and ORF28. The protein encoded by one of these ORFs, ORF12, is similar to gpL, the tail sheath protein of phage Mu. The proteins encoded by the other ORFs are either similar to hypothetical proteins with no known function or not similar to any GenBank entries. The identified ORFs are interspersed among other ORFs that encode proteins with similarity to structural proteins of other phages (Fig. 3 and Table 2), increasing our confidence that the entire left half of the genome encodes phage assembly and packaging functions.
To test the predictions of the neural network and to investigate the similarity between VP16T and VP16C, we also compared some of the structural proteins of the two phages using polyacrylamide gel electrophoresis followed by protein sequencing. The patterns of the proteins obtained from the two lysates were closely related but not identical (Fig. 4). The most abundant species (same size in both phages), as well as the other marked species in Fig. 4, were excised from a polyvinylidene difluoride membrane to which the capsid proteins were transferred. The sequences of 5 or 10 N-terminal amino acids from these proteins were obtained by Edman degradation. The sequences of the most abundant structural proteins of the two phages, TS1 and CS1, were the same (ANELATGWVQ) and were present in the protein encoded by ORF8, an ORF also identified by the neural network as encoding a major capsid protein (Fig. 3). The protein encoded by ORF8 is also similar to a hypothetical protein of Streptococcus thermophilus phage Sfi11 (Table 2). The amino acid sequences LIKLT and LVKLS of TS2 and CS2, respectively, are encoded by ORF29 in both phages (Fig. 3) and were also identified by the neural network, but they did not exhibit a high level of similarity to any phage proteins (Table 2). Finally, the TS3 protein band (amino acid sequence, ADRHIL) was present in the sequence encoded by ORF30T, which was found only in the VP16T genome.
FIG. 4.
SDS-polyacrylamide gel electrophoresis analysis of VP16C and VP16T phage capsid proteins. TS and CS are structural proteins from the turbid- and clear-plaque phages, respectively. Twenty or 30 μl of phage lysate was mixed with Laemmli buffer containing SDS, boiled for 10 min, and loaded on a 4 to 20% polyacrylamide gradient gel that was electrophoresed with Tris-glycine runing buffer. Color-coded molecular weight markers were electrophoresed in parallel.
Other phage-encoded functions.
ORF63T and ORF60C are most similar to vapE, a gene encoding a virulence-associated protein in a prophage of Dichelobacter nodosus (9, 14); while this protein has been annotated as a helicase, it does not exhibit any similarity to other helicases, as determined by blastp or PSI-BLAST analysis. Using the same algorithms, we investigated whether ORF63T and ORF60C may encode an integrase, but we found no similarity with either tyrosine recombinases or serine recombinases when we performed BLASTP or ClustalX analyses. However, these two ORFs do encode appropriately positioned residues that make up the active site of the tyrosine recombinases (lambda integrase family) (see Materials and Methods). While we are very intrigued by this finding, we also point out that the VP16 ORFs are much longer than the average tyrosine recombinase gene and that the proteins are predicted to be much more acidic, with pIs of 5.59 for the ORF63T-encoded protein and 5.68 for the ORF60C-encoded protein, than most integrase family proteins, whose pI values are >9. Since some temperate phages integrate into tRNA genes (12, 29, 54), we also used the algorithm tRNAscan-SE (30) to determine homology to tRNA genes. Such phages have attP sites that exhibit identity to parts of the tRNA gene that contains the attB sequence, thereby avoiding disruption of a possibly essential tRNA. However, we found no homology to any tRNA in either phage genome.
Many phages have acquired a variety of bacterium-derived genes that are not widespread in phage genomes. For example, the Roseobacter SIO1 genome encodes a PhoH homolog (51), and the Salmonella Gifsy-2 phage genome encodes a superoxide dismutase necessary for the complete virulence phenotype of S. enterica serovar Typhimurium (18). Both VP16T and VP16C have genes, ORF60T and ORF57C, respectively, that may encode polypeptide deformylases, which are enzymes that process the formyl methionine amino acid at the beginning of each polypeptide chain. This function could increase the translation efficiency of phage proteins. VP16T and VP16C are the first examples of phages that carry this enzyme.
Two other ORFs, ORF40T and ORF41C, stood out in both phage genomes. The second half of each of the putative proteins is extremely glutamine rich; there are over 100 glutamines in the last 170 amino acids. The glutamines are interspersed with WG, NNGG, YE, and YG. At the DNA level, the last half of each of the genes consists of repeated triplets and tandem repeats of about 50 bases. We have not found any genes similar to these ORFs, although the proteins encoded by androgen receptor genes are also very glutamine rich. We do not know whether this similarity is accidental due to the low complexity of these protein regions or is related to their functions.
What type of phage are the VP16 phages?
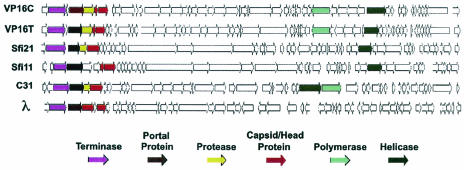
Based on genome organization, the presence of cos sites, and several predicted structural proteins, the two VP16 phages most resemble the siphoviruses, particularly the Sfi21 and Sfi11 dairy viruses that infect S. thermophilus (11). All viruses belonging to the phage λ supergroup possess a large segment that encodes phage packaging, assembly, and structural proteins, a region that encodes replication functions, and a region that encodes transcription regulatory functions. A comparison of the maps of several siphophages is shown in Fig. 5. Many, but not all, of the λ supergroup phages encode an integrase and an excisionase; in this respect, the vibriophages more closely resemble Sfi11, which does not encode an integrase (as far as we can detect currently), than Sfi21.
FIG. 5.
Comparison of genome maps of several siphophages. Sfi21 and Sfi11 are S. thermophilus phages. L5, M. tuberculosis phage L5; C31, Streptococcus coelicolor φC31; λ, coliphage lambda.
Recently, Rohwer and Edwards (49) compared all of the translated ORFs of 105 phages, whose genomes were then completely sequenced, and generated a proteomic tree based on this comparison. Their comparison included the VP16T genome, which was labeled V. parahaemolyticus TB16 (49). This phage grouped most closely with two siphophages, Lactococcus lactis phage Tuc2009 and B. subtilis phage 105, as well as several unclassified phages. A more recent analysis, which included 167 sequenced phage genomes, showed that the VP16T and VP16C phages group most closely with the Escherichia coli myophages P2 and 186 and are slightly more distantly related to several siphophage groups (Edwards and Rohwer, unpublished data). Based on this analysis, the genome organization, and the similarity of several predicted proteins to proteins encoded by well-studied lambdoid phages, we predict that our phages most likely have the lifestyle of the siphophages. The hallmark of many, but not all, of the siphophages is that they are temperate (that is, they establish lysogenic relationships with their hosts).
Lysogeny should be a very attractive lifestyle in the marine environment, where phage particles may be fully infectious for only about 24 h (24, 42, 59, 64). Based on the turbid plaque phenotype and several features of the VP16T and VP16C genomes similar to features of many temperate phages, we tried to isolate VP16 lysogens of V. parahaemolyticus, but we have not yet found conditions in which these two phages establish lysogeny (17a; Y. Xu and A. Segall, unpublished results). Our assays involved isolating phage-resistant V. parahaemolyticus clones and then testing for the presence of phage sequences either by Southern analysis of putative lysogen colonies or by PCR of cultures with several sets of phage-specific primers. These assays should have detected either integrated or nonintegrated prophages. Three possibilities for these results are (i) that we did not find the appropriate physiological conditions for the establishment of lysogeny; (ii) that the VP16 phages may establish lysogeny with another strain of Vibrio or even with another bacterial host but not with strain 16 of V. parahaemolyticus; and (iii) that these phages are not temperate phages. We have tested several other Vibrio species, including V. harveyi, V. fisheri, and V. anguillarum, but we did not observe VP16 plaques on any of them (Xu and Segall, unpublished).
Comparison of the VP16 genomes with the V. parahaemolyticus chromosomes.
The VP16 phage coding sequences were compared directly with the two translated V. parahaemolyticus chromosomes by tblastn analysis. Only the following three phage genes showed similarities with the host genome: ORF12C (but not ORF12T), the helicase genes ORF44T and ORF45C, and the polypeptide deformylase genes ORF60T and ORF57C. The similarities of the genes encoding the putative phage deformylases were with the two cellular deformylase genes, one on each of the two chromosomes. The helicases encoded by the ORFs are similar to many cellular helicases, with expect values ranging from 3e-26 to more than 0.1. Most of the similar bacterial genes are very unlikely to be or categorically are not prophage encoded, based on the absence of other phage-related neighboring genes. However, the phage genes encoding helicases did exhibit sequence similarity with a gene encoded by a putative prophage on chromosome I of the host. The gene encoding the cellular protein (NP797451.1; 22% identity to ORF45C and 21% identity to ORF44T) was annotated as a putative helicase gene, and it maps next to a putative immunity repressor gene (NP797452.1) and near an integrase family gene (NP797456.1). The host gene similar to ORF12C (NP797766.1; 29% identity) is annotated as a gene encoding a hypothetical protein but has features similar to tail fiber genes, as discussed above. However, no other phage-related genes are in its vicinity.
VP16 phage evolution: potential sites of recombination and nucleotide identity in VP16C and VP16T.
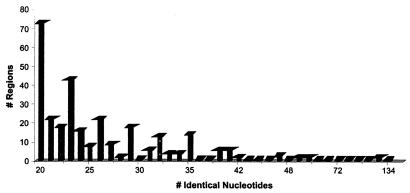
Many of the VP16T and VP16C ORFs are similar to each other (Table 2). A more detailed comparison of the genomes was performed by using the National Center for Biotechnology Information program BLAST 2 Sequences (60). This analysis revealed that these phages exhibit 73 to 88% identity at the DNA level over roughly 80% of the two genomes. Within these regions, the longest uninterrupted stretch of identical DNA is 134 bp long (as determined by the FISeq program [see Materials and Methods]), while the average region of identity is just under 28 bp long (Fig. 6). These identical regions are expected to serve as substrates for recombination events catalyzed by the host's RecBC and RecF pathways. The minimum substrate length (minimal efficient processing segment) necessary for detectable RecBC-mediated recombination is about 23 to 27 bp, and the minimum substrate length for RecF-mediated recombination in wild-type E. coli is 44 to 90 bp (55). Alternatively, recombinants could result from strand invasion of replicative intermediates, like that which occurs during late replication of phage T4 (for reviews see references 38 and 39). The remaining 20% of each genome consists of sequences whose levels of homology are significantly less than 70% (closer to 50%) and of several phage-specific inserted segments (∼3.7 kb in one phage and 5 kb in the other). We do not know how frequently the two phages recombine, but the fact that our early sequencing from a mixed lysate did not yield chimeric sequences suggests that recombination is infrequent.
FIG. 6.
Plot of frequency of occurrence versus length of identical sequences in the two phages. Only sequences consisting of at least 20 contiguous identical nucleotides were considered.
E. coli and S. enterica exhibit ∼85% nucleotide sequence identity and have been estimated to have diverged as long as 100 million years ago (44). While it is unclear that molecular clocks can be applied to phage genomes, this suggests that the two VP16 phages may have diverged from each other at about the same time as E. coli and S. enterica, based upon the 73 to 88% level of nucleotide identity. The differences in their genomes presumably prevent efficient homologous recombination and the creation of hybrids between the two genomes if they coinfect the same host cell, thereby maintaining the two phages as distinct entities. Only 1 of 15 bases is different in the cohesive end sequences of the two phages, but this single base change is sufficient to restrict packaging of each genome into the corresponding head. The extent of the interactions between the two phages during replication, assembly, and packaging or at the regulatory level remains to be tested experimentally.
Finally, we compared the two phage genomes to a set of 3,003 DNA sequences isolated from uncultured phages growing in marine environments (10; M. Breitbart, B. Felts, S. Kelley, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer, submitted for publication). Roughly 75% of these sequences are not similar to any sequences in the nonredundant GenBank database. Our comparison revealed several similarities between our phage sequences and those from the uncultured libraries (Table 3). Hits were observed to sequences from all three libraries, which were obtained from sites in San Diego, including the Mission Bay water column, Mission Bay sediment, and the Pacific Ocean off Scripps Pier. Some of the hits are more closely related to VP16T than to VP16C. An example of this is ORF21T and ORF22T. ORF21T is nearly identical, both at the amino acid level and at the nucleotide level, to one of the clones in the Mission Bay sediment library. ORF22T is similar to the same clone. Moreover, the relative position and orientation of ORF21 and ORF22 in the VP16 phages are the same as the position and orientation of the Mission Bay sediment clone. ORF21C and ORF22C are related to the same clone (Table 3). In other cases, a phage ORF is similar to multiple clones but the levels of identity are different, suggesting that the environmental samples contain more than one phage with genes related to the genes in VP16. In several cases, the similarities of the VP16T and VP16C ORFs give hints about the possible functions of the sequences from the environmental libraries, which are themselves distantly related to genes with known functions.
TABLE 3.
Comparison of VP16T and VP16C ORFs with clones from three uncultured marine phage libraries (Scripps Pier, Mission Bay water, and Mission Bay sediment)
| VP16T
|
VP16C
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Library or clonea | Similarityb | ORF | Score (bits)/ E value | Identityc | ORF | Score (bits)/ E value | Identityc | VP16 blastd |
| MBSP9F5 | NSS | 3 | 137/1e-033 | 78/207 (37) | 3 | 129/5e-031 | 66/163 (44) | Terminase |
| MB61p11B3 | Phage, prophage | 3 | 106/4e-024 | 82/281 (29) | 3 | 120/2e-028 | 86/280 (30) | Terminase |
| MBSP14E11 | Phage, prophage | 3 | 54/2e-008 | 64/258 (24) | Terminase | |||
| MB61p1E10 | Phage, prophage | 3 | 44/3e-005 | 20/43 (46) | Terminase | |||
| MBSP1C7 | Phage, prophage | 6 | 44.5/8e-006 | 45/164 (27) | 6 | 47/2e-006 | 44/173 (25) | |
| SIO51p7G4R | NSS | 8 | 118/5e-028 | 58/142 (40) | 8 | 112/2e-026 | 58/142 (40) | Capsid protein |
| MBSP10G8 | Tape measure | 16 | 44/2e-005 | 24/97 (24) | 16 | 47/2e-006 | 30/114 (26) | Tape measure |
| MBSP3B7 | NSS | 21 | 416/ e-118 | 203/204 (99) | 21 | 319/5e-089 | 154/204 (75) | Tail fiber protein |
| MBSP3B7 | NSS | 22 | 41/2e-010 | 37/38 (97) | 22 | 41/2e-010 | 37/38 (97) | Tail fiber protein |
| MBSP6G6 | NSS | 25 | 44/1e-005 | 36/88 (40) | 25 | 39/4e-004 | 38/130 (29) | Tail fiber protein? |
| SIO51p6B10L | Prophage | 26 | 69/2e-014 | 39/147 (26) | Transcription regulator | |||
| MB61p10D6 | NSS | 26 | 70/8e-014 | 52/162 (32) | Transcription regulator | |||
| SIO51p4H6L | SPO2 polymerase | 38 | 102/5e-023 | 60/197 (30) | 39 | 103/3e-023 | 63/195 (32) | DNA polymerase |
| SIO51p4H6R | NSS | 38 | 92.5/6e-020 | 51/122 (41) | 39 | 47/3e-006 | 25/59 (42) | DNA polymerase |
| MBSP15G9 | Phage polymerase | 39 | 56/5e-009 | 63/234 (26) | DNA polymerase | |||
| SIO51p9E7R | NSS | 42 | 42/4e-005 | 37/146 (25) | ||||
| SIO51p10A1R | Phage replication module | 44 | 173/2e-044 | 88/201 (43) | 45 | 176/4e-045 | 91/208 (43) | Helicase |
| SIO51p4E5L | Helicase | 44 | 89/5e-019 | 50/161 (31) | 45 | 91/2e-019 | 49/141 (34) | Helicase |
| MB61p3F12 | Bacterial chormosome | 44 | 40.5/2e-004 | 24/67 (36) | Helicase | |||
Uncultured marine phage libraries were constructed for the following locations: Scripps Pier, representing an open ocean environment; Mission Bay, representing a shallow, near-coast environment with a lot of human presence; and Mission Bay sediment, a sample taken from the sediment found just underneath the Mission Bay water sample (10; Breitbart and Rohwer, unpublished data). Despite their proximity, the Mission Bay water and sediment libraries are very different both in the apparent types and in the diversity of the phages present (Breitbart and Rohwer, unpublished data). The clones from the uncultured libraries that are similar to both the VP16T and VP16C phages are underlined.
Similarities to either phages, known or putative prophages, parts of phages or known genes, or bacterial chormosomes. NSS, no significant similarity to any GenBank entry.
Number of identical residues/total number of residues (percentage of identical residues).
The closest function suggested by the BLAST results listed in Table 2.
Summary.
We sequenced the genomes of two closely related phages that infect an environmental isolate of V. parahaemolyticus, strain 16. The phage genomes have several features of the genomes of siphophages, including the presence of cos sites and genes encoding several structural and packaging proteins related to those of lambdoid phages. The organization of the genomes most closely resembles that of the dairy phages Sfi11 and Sfi21. However, electron microscopy analysis showed that the tails of our phages are unusual compared to those of other phages and are probably contractile, more like those of myophages. The phages also encode an apparent DNA polymerase, a helicase, and a polypeptide deformylase, the first such protein found to be phage borne. Both phages also carry a gene similar to vapE, which encodes a virulence-associated protein in D. nodosus (14), suggesting the possibility that VP16C and VP16T, like many other phages, contribute to the virulence of their host (8, 28, 40, 62). Interestingly, many proteins encoded by VP16 ORFs are homologous to hypothetical proteins (probably prophage encoded) of Ralstonia solanacearum and Xylella fastidiosa, both of which are plant pathogens. The R. solanacearum chromosome contains at least two putative prophages that exhibit sequence similarity to VP16, one of which is on the megaplasmid present in this bacterium (52). V. parahaemolyticus itself has one probable prophage which exhibits homology to the genes encoding VP16 helicases but to none of the other VP16 genes. Finally, the phages encode two or three putative transcriptional regulators. While we do not yet have clear evidence of lysogeny, we are still investigating this possibility.
Why did the mixture of two phages originally escape notice? After the original isolation of plaques of VP16 on V. parahaemolyticus strain 16, several rounds of plaque purification were performed on marine agar. Because the two phage plaques cannot be distinguished on this medium, one possibility is that some contamination may have occurred. This is very unlikely based on repeated plaque purification and the inoculation of lysates with single plaques (J. Paul, personal communication). Alternatively, the original phage may have recombined with a related defective prophage in a lysogenic host, generating two different recombinant types. However, we have yet to detect VP16-related homology in our V. parahaemolyticus strain 16 by Southern analysis or by PCR (data not shown; I. Feng, Y. Xu, and A. Segall, unpublished results), and the phages are very strain specific among Vibrio strains which we and other workers have tested (26; Xu and Segall, unpublished results).
A related question is how these two closely related phages coexist without losing their genome identities. The two phages should have ample opportunity to recombine if they coinfect the same host cell, although recombination should not be very efficient since most of the exact and nearly exact homologies that they have are unlikely to permit frequent recombination. Such recombination would be expected to generate a cohort of phages with different arrangements of the same family of DNA segments, making sequencing of the phages very hard or impossible. The original lysate which we used as a source of DNA contained a mixture of the two phages, but we have no evidence of hybrid phage genomes; the sequencing reactions performed with the mixed lysate yielded contigs that matched only one or the other of the two phage genomes, and there was no evidence of chimeras between the two. In addition, the degree of sequence divergence between the two phages suggests that the phages may have been separated for as long as 100 million years. A reasonable explanation for this apparent reproductive isolation is that each VP16 phage encodes superinfection exclusion functions like those encoded by phage P22; such exclusion functions would allow coexistence of the two phages within a population of bacteria, but coinfection would occur only rarely. Reproductive isolation enforced by low frequencies of recombination and/or by superinfection exclusion probably limits the extent of modular phage evolution. We are examining the effect of coinfection and successive infection on the yields and genome structures of VP16T and VP16C phage genomes to test this hypothesis. If the superinfection exclusion hypothesis is correct, evolution of new recombinant types should be rare.
The vibriophage genomes add to the body of sequence data available for marine phages. Like the roseophage SIO1 genes, up to ∼75% of the genes of the two vibriophages is not significantly similar to genes with known functions (51). A single gene (ORF16) was found to be similar to a gene found in V. harveyi phage VHML, the only other marine siphophage that has been sequenced (43). No other similarities (e value < 0.001) were found between the VP16 phages and any of the other vibriophages whose sequences have been deposited in the GenBank database. Analyses of bulk phage DNA from marine and nonmarine environments have revealed the same extensive diversity (10; Breitbart et al., submitted). In fact, we have glimpsed only a very small fraction of the phage metagenome, and the diversity is not limited to marine phages; about the same fraction, 75%, of the genes of a recently described set of 10 mycophages is not significantly similar to genes with known functions (47, 48). Whether marine phages are more closely related to other marine phages than to nonmarine phages is still an open question and one which can only be answered as more marine phage genomes are entered into the databases.
Acknowledgments
We thank John Paul (University of South Florida) for providing the original VP16 phage lysate and its host strain, as well as for his continued interest and encouragement. We thank David Mead (Lucigen Corporation) for providing the LASL libraries. We are particularly grateful to Rob Edwards (University of Tennessee) for his cheerful advice and gracious help provided to A.M.S. with some of the bioinformatic analyses and for his careful reading of the manuscript. Mya Breitbart (San Diego State University) performed the BLAST analyses of the vibriophage genomes and the uncultured libraries.
This project was supported by funding from the San Diego State University RSCA program and by NSF Biocomplexity grant OCE0221763. Victor Seguritan received funding from NIH MBRS program grant R25 GM58906, and I-Wei Feng received funding from the San Diego State University RSCA program.
Footnotes
Dedicated to Gisela Mosig, whose support of young scientists, excitement, and rigorous approach to science affected, inspired, and infected so many. We will miss her.
REFERENCES
- 1.Abremski, K. E., and R. H. Hoess. 1992. Evidence for a second conserved arginine residue in the integrase family of recombination proteins. Protein Eng. 5:87-91. [DOI] [PubMed] [Google Scholar]
- 2.Agatonovic-Kustrin, S., and R. Beresford. 2000. Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J. Pharm. Biomed. Anal. 22:717-727. [DOI] [PubMed] [Google Scholar]
- 3.Almeida, J. S. 2002. Predictive non-linear modeling of complex data by artificial neural networks. Curr. Opin. Biotechnol. 13:72-76. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 7.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beutin, L., D. Geier, S. Zimmermann, and H. Karch. 1995. Virulence markers of Shiga-like toxin-producing Escherichia coli strains originating from healthy domestic animals of different species. J. Clin. Microbiol. 33:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloomfield, G. A., G. Whittle, M. B. McDonagh, M. E. Katz, and B. F. Cheetham. 1997. Analysis of sequences flanking the vap regions of Dichelobacter nodosus: evidence for multiple integration events, a killer system, and a new genetic element. Microbiology 143:553-562. [DOI] [PubMed] [Google Scholar]
- 10.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine phage communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruessow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 12.Carniel, E. 1999. The Yersinia high-pathogenicity island. Int. Microbiol. 2:161-167. [PubMed] [Google Scholar]
- 13.Catalano, C. E., D. Cue, and M. Feiss. 1996. Virus DNA packaging: the strategy used by phage λ. Mol. Microbiol. 16:1075-1086. [DOI] [PubMed] [Google Scholar]
- 14.Cheetham, B. F., and M. E. Katz. 1995. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol. Microbiol. 18:201-208. [DOI] [PubMed] [Google Scholar]
- 15.Chen, F., and J. Lu. 2002. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl. Envrion. Microbiol. 68:2589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, F., C. A. Suttle, and S. M. Short. 1996. Genetic diversity of marine algal virus communities as revealed by sequence analysis of DNA polymerase genes. Appl. Environ. Microbiol. 62:2869-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Feng, I. W. 2003. M.S. thesis. San Diego State University, San Diego, Calif.
- 18.Figueroa-Bossi, N., and L. Bossi. 1999. Inducible prophages contribute to Salmonella virulence in mice. Mol. Microbiol. 33:167-176. [DOI] [PubMed] [Google Scholar]
- 19.Fuhrman, J. A. 1999. Marine viruses: biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 20.Fuhrman, J. A., and L. Campbell. 1998. Microbial microdiversity. Nature 393:410-411. [Google Scholar]
- 21.Geourjon, C., and G. Deleage. 1995. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Applic. Biosci. 11:681-684. [DOI] [PubMed] [Google Scholar]
- 22.Haggard-Ljungquist, E., C. Halling, and R. Calendar. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardies, S. C., A. M. Comeau, P. Serwer, and C. A. Suttle. 2003. The complete sequence of marine bacteriophage VpV262 infecting Vibrio parahaemolyticus indicates that an ancestral component of a T7 viral supergroup is widespread in the marine environment. Virology 310:359-371. [DOI] [PubMed] [Google Scholar]
- 24.Heldal, M., and G. Bratbak. 1991. Production and decay of viruses in aquatic environments. Mar. Ecol. Prog. Ser. 72:205-212. [Google Scholar]
- 25.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345-369. [DOI] [PubMed] [Google Scholar]
- 26.Kellogg, C. A., J. B. Rose, S. C. Jiang, J. M. Turmond, and J. H. Paul. 1995. Genetic diversity of related vibriophages isolated from marine environments around Florida and Hawaii, USA. Mar. Ecol. Prog. Ser. 120:89-98. [Google Scholar]
- 27.Lawrence, J. G., and H. Ochman. 1997. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44:383-397. [DOI] [PubMed] [Google Scholar]
- 28.Lindsay, J. A., A. Ruzin, H. F. Ross, N. Kurepina, and R. P. Novick. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29:527-543. [DOI] [PubMed] [Google Scholar]
- 29.Lindsey, D. F., D. A. Mullin, and J. R. Walker. 1989. Characterization of the crytpic lambdoid prophage DLP12 of Escherichia coli and overlap of the DLP12 integrase gene with the tRNA gene, argU. J. Bacteriol. 171:6197-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luccchini, S., F. Desiere, and H. Brussow. 1999. The genetic relationship between virulent and temperate Streptococcus thermophilus bacteriophages: whole genome comparison of cos-site phages Sfi19 and Sfi21. Virology 260:232-243. [DOI] [PubMed] [Google Scholar]
- 32.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagamori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 33.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Mannisto, R. H., H. M. Kivela, H. Paulin, D. H. Bamford, and J. K. Bamford. 1999. The complete genome sequence of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262:355-363. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Hackert, E., and A. M. Stock. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269:301-312. [DOI] [PubMed] [Google Scholar]
- 36.McInerney, J. O. 1998. GCUA (General Codon Usage Analysis). Bioinformatics 14:372-373. [DOI] [PubMed] [Google Scholar]
- 37.Morgan, G. J., G. F. Hatfull, S. Casjens, and R. W. Hendrix. 2002. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 317:337-359. [DOI] [PubMed] [Google Scholar]
- 38.Mosig, G. 1998. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu. Rev. Genet. 32:379-413. [DOI] [PubMed] [Google Scholar]
- 39.Mosig, G., J. Gewin, A. Luder, N. Colowick, and D. Vo. 2001. Two recombination-dependent DNA replication pathways of bacteriophage T4, and their roles in mutagenesis and horizontal gene transfer. Proc. Natl. Acad. Sci. USA 98:8306-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newland, J. W., and R. J. Neill. 1988. DNA probes for Shiga-like toxins I and II and for toxin-converting bacteriophages. J. Clin. Microbiol. 26:1292-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. A neural network method for the identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8:581-599. [DOI] [PubMed] [Google Scholar]
- 42.Noble, R. T., and J. A. Fuhrman. 2000. Rapid virus production and removal as measured with fluorescently labeled viruses as tracers. Appl. Environ. Microbiol. 66:3790-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oakey, H. J., B. R. Cullen, and L. Owens. 2002. The complete nucleotide sequence of the Vibrio harveyi bacteriophage VHML. J. Appl. Microbiol. 93:1089-1098. [DOI] [PubMed] [Google Scholar]
- 44.Ochman, H., and A. C. Wilson. 1987. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 26:74-87. [DOI] [PubMed] [Google Scholar]
- 45.Ortmann, A. C., J. E. Lawrence, and C. A. Suttle. 2002. Lysogeny and lytic viral production during a bloom of the cyanobacterium Synechococcus spp. Microb. Ecol. 43:225-231. [DOI] [PubMed] [Google Scholar]
- 46.Paul, J. H., M. B. Sullivan, A. M. Segall, and F. Rohwer. 2002. Marine phage genomics. Comp. Biochem. Physiol. 133:463-476. [DOI] [PubMed] [Google Scholar]
- 47.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 48.Rohwer, F. 2003. Global phage diversity. Cell 113:141. [DOI] [PubMed] [Google Scholar]
- 49.Rohwer, F., and R. A. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohwer, F., V. Seguritan, D. H. Choi, A. M. Segall, and F. Azam. 2001. Production of shotgun libraries using random amplification. BioTechniques 31:108-118. [DOI] [PubMed] [Google Scholar]
- 51.Rohwer, F., A. Segall, G. Steward, V. Seguritan, F., Wolven, M. Breitbart, and F. Azam. 2000. The complete genome sequence of the marine roseophage SIO1 shares homology with nonmarine phages. Limnol. Oceanogr. 45:408-418. [Google Scholar]
- 52.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Biliaut, P. Brottier, J. C. Camus, L. Cattolico, et al. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:197-202. [DOI] [PubMed] [Google Scholar]
- 53.Sandmeier, H. 1994. Acquisition and rearrangement of sequence motifs in the evolution of bacteriophage tail fibres. Mol. Microbiol. 12:343-350. [DOI] [PubMed] [Google Scholar]
- 54.Semsey, S., B. Blaha, K. Koles, L. Orosz, and P. P. Papp. 2002. Site-specific integrative elements of rhizobiophage 16-3 can integrate into proline tRNA (CGG) genes in different bacterial genera. J. Bacteriol. 184:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen, P., and H. V. Huang. 1986. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112:441-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Short, S. M., and C. A. Suttle. 2002. Sequence analysis of marine virus communities reveals that groups of related algal viruses are widely distributed in nature. Appl. Environ. Microbiol. 68:1290-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steward, G. F., J. L. Montiel, and F. Azam. 2000. Genome size distributions indicate variability and similarities among marine viral assemblages from diverse environments. Limnol. Oceanog. 45:1697-1706. [Google Scholar]
- 58.Suttle, C. A. 1993. Enumeration and isolation of viruses, p. 121-134. In P. F. Kemp, B. F. Sherr, E. F. Sherr, and J. J. Cole, (ed.), Current methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 59.Suttle, C. A., and F. Chen. 1992. Mechanisms and rates of decay of marine viruses in seawater. Appl. Environ. Microbiol. 58:3721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tatusova, T. A., and T. L. Madden. 1999. Blast 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 61.Tettelin, H., D. Radune, S. Kasif, H. Khouri, and S. L. Salzberg. 1999. Optimized multiplex PCR: efficiently closing a whole-genome shotgun sequencing project. Genomics 62:500-507. [DOI] [PubMed] [Google Scholar]
- 62.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 63.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wommack, K. E., R. T. Hill, T. A. Muller, and R. R. Colwell. 1996. Effects of sunlight on bacteriophage viability and structure. Appl. Environ. Microbiol. 62:1336-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xue, Q., and J. B. Egan. 1995. DNA sequences of tail fiber genes of coliphage 186 and evidence for a common ancestor shared by dsDNA phage fiber genes. Virology 212:128-133. [DOI] [PubMed] [Google Scholar]