Abstract
In a two-component regulatory system, an important means of signal transduction in microorganisms, a sensor kinase phosphorylates a response regulator protein on an aspartyl residue, resulting in activation. The active site of the response regulator is highly charged (containing a lysine, the phosphorylatable aspartate, two additional aspartates involved in metal binding, and an Mg2+ ion), and introduction of the dianionic phosphoryl group results in the repositioning of charged moieties. Furthermore, substitution of one of the Mg2+-coordinating aspartates with lysine or arginine in the Escherichia coli chemotaxis response regulator CheY results in phosphorylation-independent activation. In order to examine the consequences of altered charge distribution for response regulator activity and to identify possible additional amino acid substitutions that result in phosphorylation-independent activation, we made 61 CheY mutants in which residues close to the site of phosphorylation (Asp57) were replaced by various charged amino acids. Most substitutions (47 of 61) resulted in the complete loss of CheY activity, as measured by the inability to support clockwise flagellar rotation. However, 10 substitutions, all introducing a new positive charge, resulted in the loss of chemotaxis but in the retention of some clockwise flagellar rotation. Of the mutants in this set, only the previously identified CheY13DK and CheY13DR mutants displayed clockwise activity in the absence of the CheA sensor kinase. The absence of negatively charged substitution mutants with residual activity suggests that the introduction of additional negative charges into the active site is particularly deleterious for CheY function. Finally, the spatial distribution of positions at which amino acid substitutions are functionally tolerated or not tolerated is consistent with the presently accepted mechanism of response regulator activation and further suggests a possible role for Met17 in signal transduction by CheY.
Signal transduction in prokaryotes is frequently accomplished by means of two-component regulatory systems (reviewed in reference 42). In a typical manifestation of this scheme, the detection of environmental stimuli by the unique external portion of a transmembrane sensor kinase regulates the autophosphorylation of the conserved internal portion. Transfer of phosphoryl groups from a His residue in the sensor kinase to an Asp residue in the conserved regulatory domain of a response regulator protein regulates the output function of the response regulator (e.g., DNA binding).
Electrostatic charge has long been suspected to play an important role in the activation of response regulators. The highly conserved features of a typical response regulator active site include three negatively charged Asp residues and one positively charged Lys residue (43). The three acidic residues coordinate a divalent cation, typically Mg2+, which is required for phosphorylation and dephosphorylation of the response regulator (28). The usual mechanism of reversible functional activation involves the introduction of a negatively charged phosphoryl group (—PO32−) into the active site, resulting in the repositioning of various charged moieties. In particular, the phosphorus atom of the phosphoryl group forms a covalent bond with a carboxylate oxygen of one of the conserved Asp residues, and the three oxygen atoms of the phosphoryl group interact noncovalently with the divalent cation, with the conserved Lys residue, and with a highly conserved Ser or Thr residue (5, 10, 24, 25). Alternatively, many response regulators can be partially and irreversibly activated in a sensor kinase-independent manner by mutations that cause various charge changes in the vicinity of the active site (7, 15, 21, 23, 30, 40). The active but nonphosphorylated mutant proteins presumably mimic key features of the charge distribution in their phosphorylated wild-type counterparts.
One of the best-characterized response regulators is Escherichia coli CheY. The phosphorylation state of CheY determines the direction of rotation of the flagellar motors and hence the swimming behavior of the cell. Phosphorylated CheY (CheY-P) results in clockwise (CW) flagellar rotation, whereas counterclockwise (CCW) flagellar rotation occurs in the absence of CheY-P. Chemotaxis depends on coordinating rapid activation and inactivation of CheY in response to changes in external chemical concentrations experienced by the cell as it swims through its environment. Replacement of conserved Asp13 with Arg or Lys but no other amino acids among 15 tested resulted in phosphorylation-independent activation of CheY, implying that a positive charge is the critical characteristic leading to the activation of CheY13DK and CheY13DR (7, 8, 32). The phosphorylation-mediated mechanism of activation of a wild-type response regulator outlined above does not offer an obvious explanation for the constitutive activities of these two mutant proteins.
There is evidence that the mechanism of response regulator activation tolerates some variations in the positions of charged moieties, increasing the likelihood that activated structures may be attained by mutation. In particular, (i) replacement of Asp13 with either Arg or Lys activates CheY (7, 8), (ii) mutant FixJ (31) and CheY (2) response regulators lacking the primary Asp phosphorylation site are partially activated by phosphorylation at an adjacent Ser residue, (iii) an engineered —CH2-S-CH2-PO32− side chain at position 57 (cf. —CH2-CO-O-PO32− in wild-type CheY-P) partially activates CheY (16), and (iv) replacement of the aspartyl phosphate side chain with —CH2-S-S-PO32− activates the CheB response regulator (33) but not CheY (36). In order to (i) explore more fully the role of electrostatic charge in response regulator activation, (ii) gain insight into the mechanism underlying the activities of constitutive CheY13DK and CheY13DR mutants, and (iii) identify additional constitutively active CheY mutants, we undertook a large-scale mutagenesis to perturb the distribution of electrostatic charge in the vicinity of the CheY active site. Using the X-ray crystal structure of CheY · Mg2+ (41), we identified the surface residues closest to the site of phosphorylation (Asp57), arbitrarily truncating the list at 13 residues, which represents ∼10% of the amino acids in CheY. Val11, Ser56, and Met85 were excluded from consideration due to their location in the hydrophobic core of CheY, and Gly65 was not analyzed because it has no side chain to participate in structural rearrangements. Each of the chosen 13 residues was then changed to a charged amino acid (Asp, Glu, Arg, His, and Lys), and the phenotypes of the resulting CheY mutants were evaluated.
MATERIALS AND METHODS
Bacterial strains and mutant construction.
E. coli K-12 strains KO641recA and RBB382, which contain chromosomal deletions of cheY and cheA, respectively, were described previously (8). Plasmid pRBB40, which expresses cheYZ under the control of the Ptrp promoter, also was described previously (8). Note that pRBB40 carries the inadvertent cheZ134EK mutation, which slightly diminishes chemotaxis compared to that in strains expressing wild-type cheZ (6). Site-directed mutations in cheY were made either by the dut ung method (22) or by splicing with overlap extension PCR (18). In either case, restriction fragments carrying a mutant cheY gene and extending from the BstXI site at the 3′ end of cheB to the BsmI site at the 5′ end of cheZ were inserted into a nonmutagenized pRBB40 backbone, and the entire cheY gene was sequenced to confirm that only the intended mutation was present.
Behavioral assays.
To assay chemotaxis, fresh colonies were inoculated by use of a toothpick into motility agar (1% [wt/vol] tryptone, 0.5% [wt/vol] NaCl, 0.3% [wt/vol] Bacto Agar]) and incubated at 30°C for at least 8 h. Each plate included KO641recA/pRBB40 as a positive (Che+) control and KO641recA as a negative (Che−) control. Swarm diameters were measured periodically (five or six time points), and swarm rates (millimeters per hour) were calculated from the slope of linear lines showing the best fit to the data. To correct for variations in individual plate conditions, the swarm rate for each mutant was normalized to the swarm rate for the positive control on the same plate. In 64 individual determinations for KO641recA/pRBB40, the absolute swarm rate ranged from 2.7 to 5.2 mm/h, with a mean of 4.0 mm/h and a standard deviation of 0.60 mm/h. In the same 64 tests, the relative swarm rate for KO641recA ranged from 0.019 to 0.090 the KO641recA/pRBB40 swarm rate, with a mean of 0.056 and a standard deviation of 0.015.
Designation of a mutant phenotype as Che+ or Che− was based on swarm morphology. Che+ swarms on motility agar exhibit two distinct, dense rings of bacteria responding to serine and aspartate, whereas Che− swarms lack rings, exhibit uniform turbidity across the colony, and expand at a slower rate (1, 49). Generally speaking, mutants that swarmed at greater than about one-half the wild-type rate were Che+. To clarify whether mutants that swarmed at approximately one-quarter to one-half the wild-type rate were undergoing genuine chemotaxis or simply migrating randomly through the agar (49), they were inoculated into motility agar alongside Che+ cells, and the swarms were allowed to grow into each other. The rings from two Che+ swarms merged (1), whereas Che+ swarms engulfed Che− swarms (data not shown).
The direction of flagellar rotation was determined by microscopic observation of cells tethered to a coverslip with antiflagellar antibody. Tethered cells were prepared as previously described (9), except that flagella were sometimes sheared off by passage through a 26-gauge needle rather than treatment with a tissue homogenizer, and cells were sometimes prepared at room temperature rather than 0°C.
Biochemical assays.
CheA, CheY, and CheY58WR were purified by previously described methods (17). The kinetics of CheY phosphorylation and dephosphorylation reactions were assessed with a Pi release assay (38). Briefly, 4.5 μM wild-type CheY or CheY58WR was incubated with 3 mM monophosphoimidazole, and the flow of phosphoryl groups from monophosphoimidazole through CheY to Pi in the presence of various concentrations of CheZ was monitored by an enzyme-linked spectroscopic assay (EnzChek Pi kit; Molecular Probes).
CheY autodephosphorylation kinetics were measured essentially as described previously (35). A large molar excess of CheY (280 pmol) was mixed with 32P-labeled CheA-P (28 pmol) in 100 mM Tris (pH 7.5)-10 mM MgCl2. Aliquots were removed from the reaction mixture at various times and quenched with sodium dodecyl sulfate sample buffer, and the reaction products were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Under these conditions, phosphotransfer from CheA to CheY was complete within 5 s. The amount of radioactivity remaining in CheY at various times was quantified by using phosphorimaging analysis.
RESULTS
Behavioral phenotypes of CheY mutants with introduced charge at the active site.
A total of 61 charge substitutions at 13 positions near the CheY active site were constructed, using plasmid pRBB40. For each of the chosen positions, substitutions of Asp, Glu, Arg, His, and Lys were made. Note that 4 of the 13 positions contain charged residues in wild-type CheY—Asp12, Asp13, Asp57, and Lys109. The abilities of the CheY mutants to support chemotaxis on motility agar (indicative of regulatable CheY activity), the swarm rates, and the direction of flagellar rotation (CW rotation being indicative of CheY activity) were assessed (Table 1). Nearly all (57 of 61) alterations of the CheY active site resulted in a loss of chemotactic ability. Only the CheY12DE, CheY14FH, CheY61PK, and CheY61PR proteins supported chemotaxis, indicating that these CheY mutants were capable of environmentally regulated phosphorylation and dephosphorylation and interaction with the flagellar switch similar to those of wild-type CheY. Furthermore, most (47 of 57) of the nonchemotactic mutants exhibited a loss of CheY function, i.e., were incapable of CW flagellar rotation. However, three proteins in this set, CheY61PD, CheY61PE, and CheY61PH, supported relatively high swarm rates, perhaps indicating the ability to produce CW rotation at a frequency not observable with the tethering assay. Because our primary interest was in CheY mutants that retained signaling activity, we did not attempt to ascertain the basis for the loss of function in the 47 CCW Che− mutants.
TABLE 1.
CheY active-site charge substitution mutants
| CheY mutanta | Chemotaxis phenotype | Swarm rateb | Rotational biasc | Reference or source |
|---|---|---|---|---|
| None (wild type) | Che+ | 1.0 | ND | |
| No CheY | Che− | 0.056 (64) | CCW | 8 |
| 12DE | Che+ | 0.80 (4) | ND | 8 |
| 12DH | Che− | 0.075 (2) | CCW | This work |
| 12DK | Che− | 0.096 (3) | CCW | 8 |
| 12DR | Che− | 0.055 (3) | CCW | This work |
| 13DE | Che− | 0.044 (2) | CCW | 8 |
| 13DH | Che− | 0.052 (3) | CCW | 7 |
| 13DK | Che− | 0.098 (2) | CW | 8 |
| 13DR | Che− | 0.11 (4) | CW | 7 |
| 14FD | Che− | 0.058 (2) | CCW | This work |
| 14FE | Che− | 0.057 (3) | CCW | This work |
| 14FH | Che+ | 0.50 (2) | ND | This work |
| 14FK | Che− | 0.059 (2) | CCW | This work |
| 14FR | Che− | 0.45 (5) | REV | This work |
| 17MD | Che− | 0.037 (2) | CCW | This work |
| 17ME | Che− | 0.056 (2) | CCW | This work |
| 17MH | Che− | 0.052 (4) | CCW | This work |
| 17MK | Che− | 0.042 (4) | CCW | This work |
| 17MR | Che− | 0.052 (2) | CCW | This work |
| 57DE | Che− | 0.046 (2) | CCW | 8 |
| 57DH | Che− | 0.052 (3) | CCW | This work |
| 57DK | Che− | 0.023 (2) | CCW | 8 |
| 57DR | Che− | 0.047 (2) | CCW | 2 |
| 58WD | Che− | 0.052 (3) | CCW | This work |
| 58WE | Che− | 0.076 (2) | CCW | This work |
| 58WH | Che− | 0.20 (2) | REV | This work |
| 58WK | Che− | 0.12 (2) | REV | This work |
| 58WR | Che− | 0.19 (2) | REV | This work |
| 59ND | Che− | 0.047 (4) | CCW | 38 |
| 59NE | Che− | 0.084 (5) | CCW | 38 |
| 59NH | Che− | 0.16 (2) | CW | 38 |
| 59NK | Che− | 0.20 (3) | REV | 38 |
| 59NR | Che− | 0.23 (3) | CW | 38 |
| 60MD | Che− | 0.030 (2) | CCW | This work |
| 60ME | Che− | 0.052 (3) | CCW | This work |
| 60MH | Che− | 0.095 (2) | REV | This work |
| 60MK | Che− | 0.052 (3) | CCW | This work |
| 60MR | Che− | 0.044 (2) | CCW | This work |
| 61PD | Che− | 0.26 (2) | CCW | This work |
| 61PE | Che− | 0.30 (2) | CCW | This work |
| 61PH | Che− | 0.45 (2) | CCW | This work |
| 61PK | Che+ | 0.70 (2) | ND | This work |
| 61PR | Che+ | 0.81 (2) | ND | This work |
| 86VD | Che− | 0.054 (2) | CCW | This work |
| 86VE | Che− | 0.047 (2) | CCW | This work |
| 86VH | Che− | 0.045 (2) | CCW | This work |
| 86VK | Che− | 0.056 (2) | CCW | This work |
| 86VR | Che− | 0.067 (2) | CCW | This work |
| 87TD | Che− | 0.029 (2) | CCW | This work |
| 87TE | Che− | 0.081 (4) | CCW | 3 |
| 87TH | Che− | 0.051 (2) | CCW | This work |
| 87TK | Che− | 0.073 (4) | CCW | 3 |
| 87TR | Che− | 0.041 (3) | CCW | This work |
| 88AD | Che− | 0.057 (3) | CCW | This work |
| 88AE | Che− | 0.052 (4) | CCW | This work |
| 88AH | Che− | 0.049 (2) | CCW | This work |
| 88AK | Che− | 0.055 (4) | CCW | This work |
| 88AR | Che− | 0.074 (2) | CCW | This work |
| 109KD | Che− | 0.097 (2) | CCW | This work |
| 109KE | Che− | 0.027 (2) | CCW | 7 |
| 109KH | Che− | 0.033 (2) | CCW | 7 |
| 109KR | Che− | 0.040 (2) | CCW | 7 |
All cheY mutants were assayed in a KO641recA/pRBB40 strain background.
Results are the means of two to five measurements (number of measurements), normalized in each case to the value for the wild-type control on the same motility agar plate.
ND, flagellar rotational bias was not determined for Che+ mutants; CCW, predominantly or exclusively CCW; CW, predominantly or exclusively CW; REV, both CCW and CW episodes were frequently observed.
Of particular interest were mutants that were nonchemotactic but that retained some CW flagellar rotation. In these mutants, the CW rotation could have been the result of constitutive activity. Such strains would not be expected to respond to a chemical gradient or show rings in a swarm assay. Ten CheY proteins in our set (CheY13DK, CheY13DR, CheY14FR, CheY58WH, CheY58WK, CheY58WR, CheY59NH, CheY59NK, CheY59NR, and CheY60MH) did not support chemotaxis but retained the ability to sustain at least some CW flagellar rotation (Table 1), as judged by direct observation of rotational direction. It is noteworthy that all of these proteins incorporated positively charged substitutions (Arg, His, or Lys) and involved just 5 of the 13 positions tested (residues 13, 14, 58, 59, and 60).
Wild-type CheY normally receives phosphoryl groups from the CheA sensor kinase, which results in CW flagellar rotation. Therefore, plasmid pRBB40 containing each of the 10 cheY mutations was transformed into the ΔcheA strain RBB382, and the direction of flagellar rotation was observed in order to assess the CheA dependence of the mutant phenotypes. As previously reported, the CheY13DK and CheY13DR proteins supported strong CW flagellar rotation (i.e., retain activity) in the absence of CheA and even were active in the absence of phosphorylation from any source (7, 8). However, none of the other eight proteins (CheY14FR, CheY58WH, CheY58WK, CheY58WR, CheY59NH, CheY59NK, CheY59NR, orCheY60MH) supported any CW flagellar rotation in the absence of CheA (data not shown). Therefore, this set of substitutions, designed to redistribute electrostatic charge at the CheY active site, did not yield additional constitutively active CheY mutants.
Characterization of CheY58WR.
Of the mutant proteins that did not sustain chemotaxis but that did support some CW flagellar rotation, CheY59NR had the strongest phenotype (Table 1) and was previously characterized (37, 38). CheY59NR is highly resistant to CheZ-mediated dephosphorylation, a fact which accounts for the strong phosphorylation-dependent activity of this mutant. Resistance to CheZ-mediated dephosphorylation likely contributes to the CW rotation phenotypes associated with the CheY59NH and CheY59NK proteins as well (38).
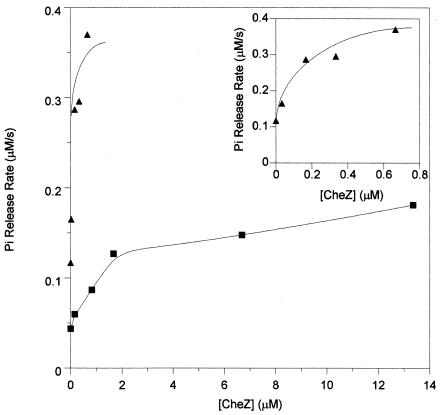
Of the five remaining CheY charge substitution mutant proteins with an unexplained partially CW rotation phenotype, CheY58WR supported the greatest degree of CW flagellar rotation (data not shown) and therefore was chosen for further analysis. Because this mutant does not contain the useful Trp58 fluorophore, we were not able to exploit the intrinsic fluorescence methods commonly used to monitor phosphorylation events. Instead, we measured the rate of Pi release to determine several kinetic parameters. In the presence of 3 mM monophosphoimidazole, CheY58WR catalyzed Pi release (the consequence of CheY autophosphorylation, followed by autodephosphorylation) at ∼40% the rate of wild-type CheY (Fig. 1). This rate reflects either autophosphorylation or autodephosphorylation, whichever is rate limiting. Under these conditions, autodephosphorylation is rate limiting for wild-type CheY (38). The addition of CheZ increased the rate of Pi release for both wild-type CheY-P and CheY58WR-P (Fig. 1), suggesting that autodephosphorylation was also rate limiting for CheY58WR. This result implies that CheY58WR catalyzes autodephosphorylation at about 40% the rate of wild-type CheY. This conclusion was confirmed by direct determination of autodephosphorylation rates by measuring the rate of loss of radioactivity from 32P-labeled CheY-P. The mean autodephosphorylation rate constants determined from duplicate experiments were 0.051 s−1 for wild-type CheY-P, a value similar to previous measurements (3, 38), and 0.021 s−1 for CheY58WR-P (data not shown).
FIG. 1.
Phosphate release from CheY. Wild-type CheY (triangles) or CheY58WR (squares) was incubated with the phosphodonor monophosphoimidazole in the presence of various concentrations of CheZ. The resulting rate of Pi release was monitored as described in Materials and Methods. The inset plots the data for wild-type CheY with a different x-axis scale.
Titration of the Pi release rate with CheZ gave additional information about the phosphorylation properties of CheY58WR. First, CheY58WR was significantly less sensitive to CheZ than was wild-type CheY (Fig. 1). Whereas about 80 to 100 nM CheZ was needed to double the rate of Pi release from wild-type CheY, about 700 to 900 nM CheZ was required to produce a similar effect on CheY58WR. Second, the rate of Pi release eventually saturates with increasing CheZ concentrations, because CheY autophosphorylation becomes rate limiting (38). The lower plateau level of Pi release from CheY58WR-P than from wild-type CheY-P at high CheZ concentrations (Fig. 1) indicates that the autophosphorylation rate of CheY58WR is about half that of wild-type CheY.
In summary, the data in Fig. 1 indicate that CheY58WR is modestly (∼2- to 3-fold) deficient in autophosphorylation and autodephosphorylation and moderately (∼10-fold) deficient in sensitivity to CheZ. In the absence of any other effects, decreased susceptibility to dephosphorylation by CheZ should result in marked CW flagellar rotation, which was not the observed result. Therefore, there are likely compensatory defects, for example, in interactions with CheA or FliM, that result in the observed behavior (Table 1). The Che− phenotype of the CheY58WR mutant may reflect an inability of the cells to change the concentration of CheY-P sufficiently rapidly in response to environmental conditions because of resistance to CheZ (Fig. 1) or retarded phosphotransfer from CheA.
DISCUSSION
The spatial distribution of charge substitution effects on CheY activity supports the present model of response regulator activation.
The 13 positions in CheY investigated during the present study of charge substitutions can be divided into three groups, based on the ability of mutant CheY proteins bearing amino acid replacements at those positions to generate CW flagellar rotation (Table 2). “Tolerant” positions (residues 13, 14, and 58 to 61) could undergo substitution by chemically unrelated amino acids and still permit the mutant protein to support at least some CW flagellar rotation. “Partially tolerant” positions (Asp12 and Thr87) could undergo substitution only by closely related amino acids for the mutant protein to retain any function. Finally, substitution of “intolerant” positions (17, 57, 86, 88, and 109) by any of a large variety of residues yielded proteins that lost all ability to support CW flagellar rotation. This classification of residues, derived from mutant phenotypes, correlates extremely well with the changes in nuclear magnetic resonance (NMR) signals that these residues exhibit upon phosphorylation of wild-type CheY (26). All (seven of seven) of the intolerant and partially tolerant positions underwent significant changes in their NMR chemical shifts in the presence of phosphorylation, which presumably reflected structural changes. In contrast, nearly all (five of six) of the tolerant residues had NMR chemical shifts that were not affected by phosphorylation.
TABLE 2.
Classification of CheY active-site positions according to functional tolerance of amino acid substitutions
| Level of tolerance | CheY position | Substitutions giving
|
|
|---|---|---|---|
| Some CW activity (reference or source) | No CW activity (reference or source) | ||
| Tolerant | Asp13 | Arg (7) | Ala, Cys, Gln, Gly, His, Leu, Phe, Tyr (7) |
| Lys (8) | Glu (8) | ||
| Asn, Met, Ser, Thr (32) | |||
| Phe14 | His, Arg (Table 1) | Asp, Glu, Lys (Table 1) | |
| Tyr (7) | |||
| Leua | |||
| Trp58 | Arg, His, Lys (Table 1) | Asp, Glu (Table 1) | |
| Ile, Pheb | |||
| Asn59 | Arg, His, Lys (Table 1) | Asp, Glu (Table 1) | |
| Ala (38) | |||
| Gln, Sera | |||
| Met60 | His (Table 1) | Asp, Glu, Lys, Arg (Table 1) | |
| Pro61 | Arg, Lys (Table 1) | Asp, Glu, His (Table 1) | |
| Ser, Thra | |||
| Glnb | |||
| Partially tolerant | Asp12 | Glu (8) | Arg, His (Table 1) |
| Gly (7) | |||
| Asn, Lys (8) | |||
| Alab | |||
| Thr87 | Ala, Cys, Ser (3) | Arg, Asp, His (Table 1) | |
| Glu, Lys (3) | |||
| Ile (13) | |||
| Glya | |||
| Intolerant | Met17 | Arg, Asp, Glu, His, Lys (Table 1) | |
| Ser, Thra | |||
| Glnb | |||
| Asp57 | Arg, His (Table 1) | ||
| Ala, Gln, Leu, Ser (7) | |||
| Asn, Glu, Lys (8) | |||
| Cys (36) | |||
| Val86 | Arg, Asp, Glu, His, Lys (Table 1) | ||
| Ala88 | Arg, Asp, Glu, His, Lys (Table 1) | ||
| Met, Vala | |||
| Glnb | |||
| Lys109 | Asp (Table 1) | ||
| Cys, Gln, Glu, Gly, His, Leu, Met, Phe, Pro, Ser, Thr, Tyr, Val (7) | |||
| Arg (27) | |||
J. G. Smith et al., unpublished data.
R. B. Bourret, unpublished data.
Plotting of the functional tolerance of amino acid substitutions on the three-dimensional structure of CheY reveals a remarkable distribution (Fig. 2). The members of the tolerant and intolerant groups form contiguous and nonoverlapping clusters, with the members of the partially tolerant group sandwiched in between. A parsimonious interpretation of this highly nonrandom pattern is that the amino acid residues at partially tolerant and intolerant positions are critical for signaling by CheY, whereas the amino acid residues at tolerant positions are dispensable for function. This interpretation is generally consistent with the presently accepted model of phosphorylation-mediated conformational change in response regulators. The conserved Thr87 and Lys109 residues interact directly with the phosphoryl group on Asp57 and are thought to convey conformational changes toward the α4-β5-α5 surface, where signaling interactions with other proteins or domains presumably occur. Conversely, the retention of the ability of mutant CheY proteins bearing a variety of amino acid substitutions at residues 13, 14, 58, 59, 60, and 61 to generate CW flagellar rotation suggests that functionally important conformational changes are not propagated from the phosphoryl group toward the α2 and α3 helices (Fig. 2). Most of the detailed information available concerning the mechanism of response regulator activation is derived from comparisons of high-resolution atomic structures of response regulators crystallized in active and inactive conformations (5, 24, 25). The direct, but static, views of the dynamic process of conformational change that are provided by crystal structures can be usefully supplemented by the less direct, but functionally relevant, views of signal transduction that are provided by genetic studies such as that reported here.
FIG. 2.
Functional tolerance of amino acid substitutions plotted on the three-dimensional structure of CheY · Mg2+. Residues shown in green (13 and 14; 58 to 61) permit CW flagellar rotation following substitution with amino acids that are chemically different from the wild-type amino acid. Residues shown in yellow (12 and 87) permit CW flagellar rotation only when substituted with amino acids chemically similar to the wild-type amino acid. Residues shown in red (17, 57, 86, 88, and 109) have no substitutions that permit CW flagellar rotation. The Mg2+ ion is shown in blue, and the CheY backbone (41) is shown in gray. Table 2 shows the substitutions tested.
A possible role for Met17 in signal transduction by CheY.
The functional intolerance of CheY to replacement of Met17 by a variety of amino acids highlights the lack of a known role for this residue. Met17 could play a role in propagation and/or coupling of the concerted movement of central residues Thr87 and Tyr106 upon phosphorylation of Asp57. Conformational flexibility of the αl N terminus (the location of Met17) has been observed for the Spo0F (12), FixJ (5, 14), and CheY (20) response regulators but has not yet been demonstrated to be a part of the activation mechanism. Another possibility is that the importance of the α1 N terminus may arise from direct involvement in interactions with CheA and/or CheZ. The α1 helix of Spo0F (including Ile15, which correponds to CheY Met17) is part of a hydrophobic patch at the interface with the four-helix bundle of the Spo0B phosphotransferase (50), and Met17 is conserved as a hydrophobic residue in CheY proteins from different species. Similarly, the αl helix of CheY (including Met17) interacts extensively with the four-helix bundle of the CheZ phosphatase (51). The location of sensor kinase phosphorylation sites within helical bundle structures (29, 44) suggests a conserved recognition strategy in which the α1 helix of the response regulator mediates interactions with sensor kinases as well. Replacement of CheY Met17 with hydrophobic amino acids, rather than the polar substitutions constructed to date, as well as biochemical characterization of mutant CheY proteins bearing various amino acids at residue 17 may distinguish between the two hypotheses proposed above for the role of Met17 (conformational change or protein-protein interactions).
Negatively charged substitutions in the CheY active site are not functionally tolerated.
Every amino acid substitution examined in this study that introduced a new negative charge into the active site resulted in a loss of CheY function (Table 1), including substitutions at residue 14, which is an Asp or a Glu residue in many other wild-type response regulators (46). Although the basis for the loss of function in negatively charged substitution mutants was not investigated here, a plausible explanation includes charge-charge repulsion that interferes with (i) the negatively charged Asp residues coordinating the Mg2+ ion, (ii) the introduction or position of the negatively charged phosphoryl group, or (iii) interactions of CheY with the FliM protein at the flagellar switch.
Positively charged substitutions in the CheY active site are compatible with CW rotation-generating activity but have different consequences.
All 10 Che− mutants listed in Table 1 that retained at least partial CW rotation-generating function contained positively charged replacements, as did 3 of the 4 Che+ mutants. This striking common characteristic of mutants with partial or full activity appears to be a consequence of the deleterious nature of negatively charged substitutions in the CheY active site discussed above, combined with the circumstance that only positively or negatively charged substitutions were used in this study. An alternative interpretation is that the presence of a new positive charge confers partial function. If the insertion of a positive charge at the active site leads to CW rotation-generating function, then a common mechanism of CheY activation (e.g., a direct interaction of the new CheY positive charge with FliM) might be expected. However, the observation that substitutions of positive charges at various positions within the CheY active site are associated with demonstrably different phenotypes argues against this hypothesis. For example, CheY13DK and CheY13DR exhibited phosphorylation-independent activity (7, 8), the activity of CheY59NR resulted from resistance to dephosphorylation by CheZ (37, 38), and CheY58WR exhibited decreased phosphorylation and dephosphorylation (Fig. 1).
Why do positively charged substitutions in the CheY active site result in constitutive activity only when made at residue 13?
It has long been mystifying that the introduction of either a cationic Arg or Lys at CheY residue 13 or a dianionic phosphoryl group at residue 57 has similar functional consequences. Genetic characterization of CheY87TX/13DK (3) and CheY109KX/13DK (7) double mutants, together with an understanding of the structural basis for stabilization of the active conformation by Thr87 and Lys109, suggests that Lys or Arg at position 13 imposes conformational changes similar to those induced by the phosphorylation of Asp57. Nevertheless, in the X-ray crystal structure of CheY13DK, the α4-β4 and α5-β5 loops (which contain Thr87 and Lys109, respectively) are in inactive conformations, and Tyr106 significantly populates both its interior and its exterior conformations (19). Presumably, the active and inactive conformations of CheY are close enough in energy that the equilibrium between states can be shifted by intermolecular crystalline forces (19). The ability of NMR analysis to detect both the active and the inactive conformations of the analogous response regulator NtrC in solution (45) is consistent with similar energies for the two conformations.
The molecular basis for the increased tendencies of CheY13DK and CheY13DR to acquire an active conformation still is not entirely apparent. Positive charge appears to be important because only Arg and Lys substitutions (out of 15 tested) activate CheY (7, 8, 32). The present study revealed that the location of the positive charge is also critical. The introduction of a positive charge at numerous nearby residues in the active site did not result in constitutive activation (Table 1), suggesting that residue 13 possesses special features. In this regard, Asp13 is one of only two CheY side chains (the other is Asp57) that directly coordinate Mg2+ (41), and CheY13DK does not detectably bind Mg2+ (11, 48). There is some evidence suggesting that apo-CheY (CheY lacking Mg2+ at the active site) acquires the active conformation in the absence of phosphorylation more readily than does CheY · Mg2+. Mg2+ participates in multiple interactions with CheY, thus altering and constraining the structure of the active site (41). In contrast, a high-resolution crystal structure of apo-CheY shows two distinct conformations of the β4-α4 loop and Tyr106, one of which displays features of activated CheY (39). Furthermore, in the apo forms of wild-type CheY (47), CheY13DK (19), and CheY95IV (34), the solvent-exposed positions of the Tyr106 side chains are less extreme than those observed in either of the reported CheY · Mg2+ crystal structures (4, 41). Although there is no phosphoryl group to stabilize the activated orientations of the Thr87, Tyr106, and Lys109 side chains in CheY13DK, the presence of a positively charged side chain specifically at position 13 prevents Mg2+ binding and may accentuate the natural ability of the resulting apo-CheY molecule to assume the active conformation in the absence of phosphorylation.
Acknowledgments
We thank Kristin Boesch, Daniel Pennington, and Martin Schuster for technical assistance. We thank the anonymous reviewers for extensive suggestions that improved the clarity of this document.
This work was supported by Public Health Service grant GM50860 from the National Institute of General Medical Sciences and grant 44594 from the University of North Carolina Research Council.
REFERENCES
- 1.Adler, J. 1966. Chemotaxis in bacteria. Science 153:708-716. [DOI] [PubMed] [Google Scholar]
- 2.Appleby, J. L., and R. B. Bourret. 1999. Activation of CheY mutant D57N by phosphorylation at an alternate site, Ser56. Mol. Microbiol. 34:915-925. [DOI] [PubMed] [Google Scholar]
- 3.Appleby, J. L., and R. B. Bourret. 1998. Proposed signal transduction role for conserved CheY residue Thr87, a member of the response regulator active site quintet. J. Bacteriol. 180:3563-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellsollel, L., J. Prieto, L. Serrano, and M. Coll. 1994. Magnesium binding to the bacterial chemotaxis protein CheY results in large conformational changes involving its functional surface. J. Mol. Biol. 238:489-495. [DOI] [PubMed] [Google Scholar]
- 5.Birck, C., L. Mourey, P. Gouet, B. Fabry, J. Schumacher, P. Rousseau, D. Kahn, and J.-P. Samama. 1999. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure 7:1505-1515. [DOI] [PubMed] [Google Scholar]
- 6.Boesch, K. C., R. E. Silversmith, and R. B. Bourret. 2000. Isolation and characterization of nonchemotactic CheZ mutants of Escherichia coli. J. Bacteriol. 182:3544-3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourret, R. B., S. K. Drake, S. A. Chervitz, M. I. Simon, and J. J. Falke. 1993. Activation of the phosphosignaling protein CheY. II. Analysis of activated mutants by 19F NMR and protein engineering. J. Biol. Chem. 268:13089-13096. [PMC free article] [PubMed] [Google Scholar]
- 8.Bourret, R. B., J. F. Hess, and M. I. Simon. 1990. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc. Natl. Acad. Sci. USA 87:41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray, D., R. B. Bourret, and M. I. Simon. 1993. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. Mol. Biol. Cell 4:469-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, H. S., S.-Y. Lee, D. Yan, X. Pan, J. S. Parkinson, S. Kustu, D. S. Wemmer, and J. G. Pelton. 2000. NMR structure of activated CheY. J. Mol. Biol. 297:543-551. [DOI] [PubMed] [Google Scholar]
- 11.Drake, S. K., R. B. Bourret, L. A. Luck, M. I. Simon, and J. J. Falke. 1993. Activation of the phosphosignaling protein CheY. I. Analysis of the phosphorylated conformation by 19F NMR and protein engineering. J. Biol. Chem. 268:13081-13088. [PMC free article] [PubMed] [Google Scholar]
- 12.Feher, V. A., and J. Cavanagh. 1999. Millisecond-timescale motions contribute to the function of the bacterial response regulator protein Spo0F. Nature 400:289-293. [DOI] [PubMed] [Google Scholar]
- 13.Ganguli, S., H. Wang, P. Matsumura, and K. Volz. 1995. Uncoupled phosphorylation and activation in bacterial chemotaxis. The 2.1Å structure of a threonine to isoleucine mutant at position 87 of CheY. J. Biol. Chem. 270:17386-17393. [PubMed] [Google Scholar]
- 14.Gouet, P., B. Fabry, V. Guillet, C. Birck, L. Mourey, D. Kahn, and J.-P. Samama. 1999. Structural transitions in the FixJ receiver domain. Structure 7:1517-1526. [DOI] [PubMed] [Google Scholar]
- 15.Gupte, G., C. Woodward, and V. Stout. 1997. Isolation and characterization of rcsB mutations that affect colanic acid capsule synthesis in Escherichia coli K-12. J. Bacteriol. 179:4328-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halkides, C. J., X. Zhu, D. P. Phillion, P. Matsumura, and F. W. Dahlquist. 1998. Synthesis and biochemical characterization of an analogue of CheY-phosphate, a signal transduction protein in bacterial chemotaxis. Biochemistry 37:13674-13680. [DOI] [PubMed] [Google Scholar]
- 17.Hess, J. F., R. B. Bourret, and M. I. Simon. 1991. Phosphorylation assays for proteins of the two-component regulatory system controlling chemotaxis in E. coli. Methods Enzymol. 200:188-204. [DOI] [PubMed] [Google Scholar]
- 18.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Mol. Biotechnol. 3:93-99. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, M., R. B. Bourret, M. I. Simon, and K. Volz. 1997. Uncoupled phosphorylation and activation in bacterial chemotaxis: the 2.3-Å structure of an aspartate to lysine mutant at position 13 of CheY. J. Biol. Chem. 272:11850-11855. [DOI] [PubMed] [Google Scholar]
- 20.Kato, M., T. Shimizu, T. Mizuno, and T. Hakoshima. 1999. Structure of the histidine-containing phosphotransfer (HPt) domain of the anaerobic sensor kinase ArcB complexed with the chemotaxis response regulator CheY. Acta Crystallogr. Sect. D 55:1257-1263. [DOI] [PubMed] [Google Scholar]
- 21.Klose, K. E., D. S. Weiss, and S. Kustu. 1993. Glutamate at the site of phosphorylation of nitrogen-regulatory protein NTRC mimics aspartyl-phosphate and activates the protein. J. Mol. Biol. 232:67-78. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel, T. A., J. D. Roberts, and R. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 23.Lan, C.-Y., and M. M. Igo. 1998. Differential expression of the OmpF and OmpC porin proteins in Escherichia coli K-12 depends upon the level of active OmpR. J. Bacteriol. 180:171-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, S.-Y., H. S. Cho, J. G. Pelton, D. Yan, E. A. Berry, and D. E. Wemmer. 2001. Crystal structure of activated CheY: comparison with other activated receiver domains. J. Biol. Chem. 276:16425-16431. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, R. J., J. A. Brannigan, K. Muchova, I. Barak, and A. J. Wilkinson. 1999. Phosphorylated aspartate in the structure of a response regulator protein. J. Mol. Biol. 294:9-15. [DOI] [PubMed] [Google Scholar]
- 26.Lowry, D. F., A. F. Roth, P. B. Rupert, F. W. Dahkquist, F. J. Moy, P. J. Domaille, and P. Matsumura. 1994. Signal transduction in chemotaxis; a propagating conformational change upon phosphorylation of CheY. J. Biol. Chem. 269:26358-26362. [PubMed] [Google Scholar]
- 27.Lukat, G. S., B. H. Lee, J. M. Mottonen, A. M. Stock, and J. B. Stock. 1991. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J. Biol. Chem. 266:8348-8354. [PubMed] [Google Scholar]
- 28.Lukat, G. S., A. M. Stock, and J. B. Stock. 1990. Divalent metal ion binding to the CheY protein and its significance to phosphotransfer in bacterial chemotaxis. Biochemistry 29:5436-5442. [DOI] [PubMed] [Google Scholar]
- 29.Mourey, L., S. De Re, J.-D. Pedelacq, T. Tolstykh, C. Faurie, V. Guillet, J. B. Stock, and J.-P. Samama. 2001. Crystal structure of the CheA histidine phosphotransfer domain that mediates response regulator phosphorylation in bacterial chemotaxis. J. Biol. Chem. 276:31074-31082. [DOI] [PubMed] [Google Scholar]
- 30.Pazour, G. J., C. N. Ta, and A. Das. 1992. Constitutive mutations of Agrobacterium tumefaciens transcriptional activator virG. J. Bacteriol. 174:4169-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyrat, J.-M., M. David, J. Batut, and P. Boistard. 1994. FixL of Rhizobium meliloti enhances the transcriptional activity of a mutant FixJD54N protein by phosphorylation of an alternate residue. J. Bacteriol. 176:1969-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders, D. A., B. L. Gillece-Castro, A. M. Stock, A. L. Burlingame, and D. E. Koshland, Jr. 1989. Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J. Biol. Chem. 264:21770-21778. [PubMed] [Google Scholar]
- 33.Saxl, R. L., G. S. Anand, and A. M. Stock. 2001. Synthesis and characterization of a phosphorylated analogue of the response regulator CheB. Biochemistry 40:12896-12903. [DOI] [PubMed] [Google Scholar]
- 34.Schuster, M., R. Zhao, R. B. Bourret, and E. J. Collins. 2000. Correlated switch binding and signaling in bacterial chemotaxis. J. Biol. Chem. 275:19752-19758. [DOI] [PubMed] [Google Scholar]
- 35.Silversmith, R. E., J. L. Appleby, and R. B. Bourret. 1997. Catalytic mechanism of phosphorylation and dephosphorylation of CheY: kinetic characterization of imidazole phosphates as phosphodonors and the role of acid catalysis. Biochemistry 36:14965-14974. [DOI] [PubMed] [Google Scholar]
- 36.Silversmith, R. E., and R. B. Bourret. 1998. Synthesis and characterization of a stable analogue of the phosphorylated form of the chemotaxis protein CheY. Protein Eng. 11:205-212. [DOI] [PubMed] [Google Scholar]
- 37.Silversmith, R. E., G. P. Guanga, L. Betts, C. Chu, R. Zhao, and R. B. Bourret. 2003. CheZ-mediated dephosphorylation of the Escherichia coli response regulator CheY: a role for CheY glutamate 89. J. Bacteriol. 185:1495-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silversmith, R. E., J. G. Smith, G. P. Guanga, J. T. Les, and R. B. Bourret. 2001. Alteration of a nonconserved active site residue in the chemotaxis response regulator CheY affects phosphorylation and interaction with CheZ. J. Biol. Chem. 276:18478-18484. [DOI] [PubMed] [Google Scholar]
- 39.Simonovic, M., and K. Volz. 2001. A distinct meta-active conformation in the 1.1 Å resolution structure of wild-type apoCheY. J. Biol. Chem. 276:28637-28640. [DOI] [PubMed] [Google Scholar]
- 40.Stewart, R. C. 1993. Activating and inhibitory mutations in the regulatory domain of CheB, the methylesterase in bacterial chemotaxis. J. Biol. Chem. 268:1921-1930. [PubMed] [Google Scholar]
- 41.Stock, A. M., E. Martinez-Hackert, B. F. Rasmussen, A. H. West, J. B. Stock, D. Ringe, and G. A. Petsko. 1993. Structure of the Mg2+-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry 32:13375-13380. [DOI] [PubMed] [Google Scholar]
- 42.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 43.Stock, J. B., A. J. Ninfa, and A. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomomori, C., T. Tanaka, R. Dutta, H. Park, S. K. Saha, Y. Zhu, R. Ishima, D. Liu, K. I. Tong, H. Kurokawa, H. Qian, M. Inouye, and M. Ikura. 1999. Solution structure of the homodimeric core domain of Esherichia coli histidine kinase EnvZ. Nat. Struct. Biol. 6:729-734. [DOI] [PubMed] [Google Scholar]
- 45.Volkman, B. F., D. Lipson, D. E. Wemmer, and D. Kern. 2001. Two-state allosteric behavior in a single-domain signaling protein. Science 291:2329-2330. [DOI] [PubMed] [Google Scholar]
- 46.Volz, K. 1993. Structural conservation in the CheY superfamily. Biochemistry 32:11741-11753. [DOI] [PubMed] [Google Scholar]
- 47.Volz, K., and P. Matsumura. 1991. Crystal structure of Escherichia coli CheY refined at 1.7-Å resolution. J. Biol. Chem. 266:15511-15519. [DOI] [PubMed] [Google Scholar]
- 48.Welch, M., K. Oosawa, S.-I. Aizawa, and M. Eisenbach. 1994. Effects of phosphorylation, Mg2+, and conformation of the chemotaxis protein CheY on its binding to the flagellar switch protein FliM. Biochemistry 33:10470-10476. [DOI] [PubMed] [Google Scholar]
- 49.Wolfe, A. J., and H. C. Berg. 1989. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. USA 86:6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zapf, J., U. Sen, Madhusudan, J. A. Hoch, and K. I. Varughese. 2000. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Struct. Fold Des. 8:851-862. [DOI] [PubMed] [Google Scholar]
- 51.Zhao, R., E. J. Collins, R. B. Bourret, and R. E. Silversmith. 2002. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat. Struct. Biol. 9:570-575. [DOI] [PubMed] [Google Scholar]