Abstract
Puumala virus (PUUV) is a hantavirus that causes hemorrhagic fever with renal syndrome (HFRS), which is an important public health problem in large parts of Europe. We examined the memory cytolytic T lymphocyte (CTL) responses in 13 Finnish individuals who had HFRS between 1984 and 1995. In seven of these donors, we detected virus-specific CTL responses against the PUUV nucleocapsid (N) protein after in vitro stimulation with PUUV. Six novel CD8+ CTL epitopes were defined on the N protein and were found to be restricted by various HLA alleles including A2, A28, B7, and B8. This is the first demonstration of PUUV-specific CTL responses in humans, and the first identification of CTL epitopes on PUUV. In addition, this study provides one of the few characterizations of a human antiviral memory T cell response, without the complicating issues of virus persistence or reinfection. Interferon (IFN)-γ ELISPOT analysis showed that memory CTL specific for these epitopes were present at high frequency in PUUV-immune individuals many years after acute infection in the absence of detectable viral RNA. The frequencies of PUUV-specific CTL were comparable to or exceeded those found in other viral systems including influenza, EBV and HIV, in which CTL responses may be boosted by periodic reinfection or virus persistence.
Keywords: Puumala virus, hantavirus, immunologic memory, cytotoxic T lymphocytes, T lymphocyte epitopes
Introduction
Puumala virus (PUUV)*, a member of the Hantavirus genus, causes a mild form of hemorrhagic fever with renal syndrome (HFRS), also known as nephropathia epidemica. PUUV is endemic primarily in Northern and central Europe and western parts of Russia and is responsible for ∼4,000–6,000 clinical cases of HFRS annually with a mortality rate of <1%. PUUV infections usually result in a mild form of HFRS, but can also result in a more serious form of the disease characterized by renal failure and circulatory shock. Other hantaviruses that cause more severe forms of HFRS include Hantaan virus (HTNV; Asia) and Dobrava virus (DOBV; Balkans). Related hantaviruses in North and South America (e.g., Sin Nombre [SNV] and Andes [ANDV] virus) cause a frequently fatal pulmonary infection known as hantavirus pulmonary syndrome (HPS). HFRS and HPS are distinct in the primary target organ of virus infection (kidney versus lung) but have important clinical features in common including fever, thrombocytopenia, and a capillary leak syndrome. These common clinical manifestations suggest that the underlying mechanisms of disease may be similar in the two syndromes.
The pathogenesis of hantavirus infection is not well understood. Endothelial cells and monocytes are thought to be the primary cell types infected by the virus, but infection has no direct cytopathic effect on these cells (1–5). Therefore, it is unlikely that direct viral cytotoxicity is the primary cause of pathology in vivo. Several studies have suggested that the clinical syndromes caused by hantavirus infection may be mediated in part by immunopathologic mechanisms, including the action of virus-specific CD8+ and CD4+ T lymphocytes. Evidence to suggest this includes increases in the numbers of activated circulating CD8+ T cells that are seen during HFRS (6), and infiltrating lymphocytes (predominantly CD8+ T cells) found in kidney biopsies from patients with acute PUUV infections (7, 8). In addition, elevated levels of T cell–derived cytokines such as TNF-α and IFN-γ have been observed in both the kidney and peripheral blood of patients with acute hantavirus infections (8–11), and increased numbers of cytokine-producing cells were seen in the lungs of patients with fatal HPS compared with patients with non-HPS acute respiratory distress syndrome (12). Further, a recent study by Mustonen et al. (13) demonstrated a correlation between severity of disease caused by PUUV infection and the HLA haplotype B8, DR3, suggesting that T cells restricted by these HLA alleles may impact the severity of disease.
To begin to understand the role of T lymphocytes in human hantavirus infection, we analyzed memory T cell responses in a group of 13 Finnish individuals who had clinical PUUV infections between the years 1984 and 1995. We were able to demonstrate PUUV-specific T cell responses in seven individuals with the N protein being the most commonly recognized viral protein. We identified six novel CD8+ CTL epitopes on the N protein and found that frequencies of memory CD8+ T cells specific for a single viral epitope were as high as 100–300 per 106 PBMCs for up to 15 y after the acute infection. The inability to detect viral RNA in either PBMCs or urine from these and other PUUV immune individuals suggested that virus-specific memory T cells were maintained in the absence of persistent virus. As such, the PUUV system provides an ideal opportunity to study the development and maintenance of antiviral T cell memory in humans. In addition, the identification of CD8+ T cell epitopes on PUUV will allow us to track specific antiviral T cell responses during acute HFRS, which will provide insight into the role of T cells in the recovery from and/or the immunopathology of disease.
Materials and Methods
Study Subjects.
Donors were Finnish individuals who had HFRS caused by PUUV infection between the years 1984 and 1995. The diagnosis of PUUV infection was confirmed by serological detection of PUUV-specific antibodies (13). PBMCs were isolated from 60 mls of whole blood using CPT vacutainer tubes (Becton Dickinson). Cells were resuspended at 1–2 × 107/ml in RPMI 1640 supplemented with 20% FBS (Sigma-Aldrich), 2 ml l-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, and 10% DMSO and cryopreserved until use.
Synthetic Peptides.
Overlapping peptides that spanned the PUUV strain K27 nucleocapsid protein were synthesized based on the published sequence (14) (GenBank/EMBL/DDBJ accession no. L08804). Peptides were 15 amino acids in length and overlapped by nine amino acids. Peptides were synthesized at the Protein Chemistry Core Facility at the University of Massachusetts Medical School using an automated Rainin Symphony peptide synthesizer.
In Vitro Stimulation of PBMCs.
5–7 × 106 PBMCs were resuspended in 1 ml of AIM-V medium (Life Technologies) supplemented with 10% human AB serum (Advanced Biotechnologies). The PBMCs were stimulated with 20 μl of purified PUUV (2.6 × 106 pfu/ml) (strain K27, provided by Connie S. Schmaljohn, US Amriid, Frederick, MD), grown in Vero E6 cells and purified as described previously (15). Recombinant human IL-2 (5 U/ml) was added on days 5 and 6 of culture in fresh AIM-V/10% human AB serum and the medium was replenished every third day thereafter. 51[Cr]release cytotoxicity assays were performed on days 7 and 8 of culture as described previously (16). Target cells were autologous EBV-transformed B lymphoblastoid cell lines (BLCLs) infected with vaccinia virus recombinants expressing PUUV N, glycoprotein 1 (G1), or glycoprotein 2 (G2). Vaccinia virus recombinants were constructed as described previously (17). Unlabeled BLCLs infected with wild-type vaccinia virus were included in 51[Cr]release assays to reduce levels of vaccinia-specific background lysis (10:1 unlabeled targets:51[Cr] labeled targets). Cultures were maintained and all experiments performed in a biosafety level 3 laboratory according to standard BSL3 guidelines.
CTL Lines.
PUUV-specific CTL lines were established from bulk cultures by limiting dilution cloning, as described previously (16). In brief, cells were plated at 1, 3, or 10 cells per well in 96-well, round-bottomed plates (Costar) in 0.5 ml AIM-V/10% FBS and stimulated every 14 d with anti-CD3 (12F6; 0.1 μg/ml), recombinant human IL-2 (20 U/ml) and γ-irradiated, allogeneic feeder cells (105 cells per well). Individual wells containing virus-specific CTL were identified by analysis of lytic activity against autologous BLCLs infected with recombinant vaccinia viruses expressing PUUV N, G1, or G2. Wells containing virus-specific CTL were expanded into 48-well plates and restimulated every 14 d as described above. Surface expression of CD4 and CD8 was determined by flow cytometry using FITC-conjugated antibodies (Becton Dickinson). All cell lines were >95% pure CD8 or CD4 as indicated. HLA restriction of CTL lines was determined using partially HLA-matched BLCL targets infected with a vaccinia virus recombinant expressing PUUV N. Fine specificity of CTL lines was determined using overlapping synthetic peptides that span the PUUV N protein and subsequent N- and COOH-terminal peptide truncations.
IFN-γ ELISPOT Assays.
Cryopreserved PBMCs were thawed and incubated for 20–24 h in RPMI 1640/10% FBS. ELISPOT assays were then performed as described previously (18). In brief, 96-well filtration plates (MAIPS45; Millipore) were coated with 15 μg/ml mouse anti–human IFN-γ mAb (clone N1B42; BD PharMingen) overnight. PBMCs were plated at 2–5 × 105 cells per well in RPMI 1640/10% FBS. Peptides were added at 10 μg/ml and plates incubated for 16–18 h at 37°C. Biotinylated mouse anti–human IFN-γ mAb (clone 4S.B3; BD PharMingen) was added for 2 h at room temperature followed by streptavidin alkaline phosphatase (1:400 dilution) for 45 min at room temperature. Substrate (3-amino-9-ethyl-carbazole/0.15%H2O2) (Sigma-Aldrich) was added for 10 min at room temperature. PBMCs stimulated with phytohemagglutinin (2.5 μg/ml) were used as a positive control and unstimulated PBMCs as a negative control. The number of spots in negative control wells (range of 0–5 spots) was subtracted from the number of spots in stimulated wells. T cell frequency was calculated as the number of visible spots/number of total PBMCs per well, and the frequency presented as the number of IFN-γ producing cells/106 PBMCs. Peptide stimulations were performed in triplicate wells unless indicated otherwise. All ELISPOT experiments were performed at least twice and mean values are shown. Responses were considered significant if a minimum of five IFN-γ–producing cells were present in the well and if the number of IFN-γ–producing cells was at least twice that of the negative control (18).
Results
PUUV Nucleocapsid (N) Protein Is a Dominant Target of Memory CTL after Resolution of HFRS.
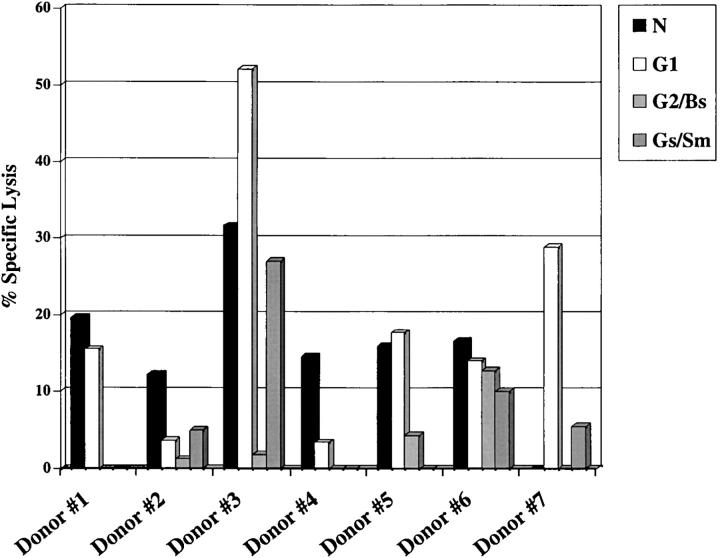
T cell responses against PUUV have not been characterized. To identify PUUV proteins that are targeted by the T cell response during infection, PBMCs from 11 donors who had HFRS caused by PUUV between the years 1984 and 1995 were stimulated in vitro for 7–8 d with live PUUV. After in vitro stimulation, bulk cultures were screened for recognition of autologous BLCLs infected with recombinant vaccinia viruses expressing PUUV proteins. CTL responses against one or more PUUV protein were detectable in PBMC from 7/11 donors tested. Virus-specific CTL recognized N protein in 6 of the cultures (donors 1–6), the G1 glycoprotein in 5 (donors 1, 3, 5, 6, and 7), and the G2 glycoprotein in 2 (donors 3 and 6) of the cultures (Fig. 1) . After restimulation with PUUV and autologous, irradiated PBMCs, we were able to detect N-specific CTL in bulk cultures from two of the donors in which virus-specific responses were undetectable at days 7 and 8 (unpublished data). High levels of background lysis were detected in the bulk cultures from three donors (donors 4, 5, and 7), despite the inclusion of unlabeled vaccinia virus-infected target cells. This background may be due to NK cell activity, although this possibility was not formally tested. The background lysis in these cultures was 26, 18, and 29%, respectively. However, the lysis of target cells infected with recombinant viruses expressing PUUV proteins was significantly higher than control target cells infected with wild-type vaccinia virus. Lysis of vaccinia virus-infected targets in all other cultures was <5%. The specific lysis of targets infected with vaccinia virus recombinants expressing PUUV proteins was considered to be positive if it was >10% above lysis of WT vaccinia virus-infected targets.
Figure 1.
Bulk culture recognition of PUUV proteins. PBMCs (4–5 × 106) were stimulated with 20 μl of infectious PUUV (strain K27; 2.6 × 106 pfu/ml) and cytotoxicity assays were performed using autologous BLCLs infected with recombinant vaccinia viruses expressing PUUV N, G1, G2/Bs or G2/Sm protein as targets. The recombinant vaccinia virus designated vac-G2/Bs contains the first half of the G2 cDNA (amino acids 1 to 256), and the recombinant designated vac-G2/Sm contains the second half of the G2 protein (amino acids 227 to 490). E/T ratios were either 60:1 or 80:1. Unlabeled wild-type vaccinia virus infected BLCL were included in all wells at a ratio of 10:1 unlabeled: labeled targets, to reduce levels of vaccinia virus-specific lysis. Data shown represent the percent specific lysis after subtraction of wild-type vaccinia virus background lysis. Responses >10% above background were considered positive.
PUUV-specific CTL cell lines were identified and characterized by in vitro expansion and cloning of bulk cultures from donors 1 and 2. 43 PUUV-specific cell lines were isolated from donor 1, of these 42 were N-specific and 1 was G1-specific. 20 cell lines were isolated from donor 2, of these 19 were N-specific and 1 was G2-specific. The predominance of cell lines specific for N suggests that epitopes on the N protein may be the major target of the CTL response during PUUV infection.
PUUV Infection Induces a Broad CTL Response to N Protein in Two Individuals.
N-specific CTL lines isolated from donors 1 and 2 were characterized using a panel of overlapping 15-mer peptides that span the PUUV N sequence. Optimal CTL epitopes were then defined by testing each CTL line against sequential N- and COOH-terminal truncations of the appropriate15-mer peptide (unpublished data). The restricting HLA alleles required for peptide recognition by these CTL lines was determined using a panel of partially HLA-matched BLCLs as targets in 51[Cr]release assays. A summary of the epitopes and representative CTL lines identified from these donors is shown in Table I.
Table I.
PUUV N-specific CD8+ CTL Lines
| Donor | CTL line | Epitope | HLA restrict. | PUUV | % Specific lysisa HTNV V | SNV N |
|---|---|---|---|---|---|---|
| 1 | 1A-C11 | N204-12 | A2 | 44.7 | −1.1 | −1.1 |
| 1A-D10 | N204-12 | A2 | 41.5 | −1.2 | −3.3 | |
| 1A-E5 | N204-12 | A2 | 49.6 | −5.6 | −3.8 | |
| 1B-C3 | N204-12 | A2 | 50.6 | −1.7 | −1.4 | |
| 3-C3 | N204-12 | A2 | 36.4 | −2.8 | −3.9 | |
| 3-D10 | N204-12 | A2 | 56.3 | −4.6 | −6.6 | |
| 3-E4 | N204-12 | A2 | 41.4 | −6.9 | −8.2 | |
| 3-E11 | N204-12 | A2 | 43.4 | −3.0 | −6.7 | |
| 10-B10 | N204-12 | A2 | 37.6 | −2.8 | −3.9 | |
| 1 | 3-C8 | N173-81 | B7 | 18.0 | −7.8 | −3.7 |
| 10-F5 | N173-81 | B7 | 33.6 | 25.5 | 0.5 | |
| 10-G4 | N173-81 | B8 | 22.4 | 2.7 | −1.8 | |
| 1 | 1A-G6 | N243-51 | B8 | 76.7 | ND | ND |
| 2 | 1A-D11 | N236-50 | ND | 19.1 | −1.0 | 2.2 |
| 1B-G9 | N236-50 | ND | 20.3 | −1.8 | 22.0 | |
| 3-B2 | N236-50 | ND | 19.3 | −0.3 | 3.2 | |
| 3-E9 | N236-50 | ND | 24.0 | 3.1 | 2.2 | |
| 10-B4 | N236-50 | ND | 18.2 | 2.0 | 2.7 | |
| 10-B5 | N236-50 | ND | 17.9 | 0.9 | 3.8 | |
| 10-E11 | N236-50 | ND | 20.8 | 2.6 | 1.7 | |
| 2 | 3-D3 | N212-20 | ND | 27.3 | −1.7 | −3.5 |
| 3-D3 | N212-20 | ND | 27.6 | 0.4 | 1.9 | |
| 2 | 3-B8 | N164-78 | A28 | 27.4 | 25.9 | −3.0 |
E/T = 10.
CD8+ CTL lines from donor 1 were found to recognize one of four epitopes: 1 restricted by HLA-A2 (N204–12), 1 restricted by B7 (N173–81), and two restricted by B8 (N173–81, N243–51). CD8+ CTL lines from donor 2 also recognized one of four epitopes: 1 restricted by HLA-A2 (N204–12; not shown), 1 restricted by A28 (N164–78) and 2 (N236–50, N212–20) for which the restricting HLA allele has yet to be determined (Table I). Fig. 2 demonstrates the cytolytic activity of the CD8+ CTL lines using target cells pulsed with increasing peptide dilutions.
Figure 2.

CTL activity of PUUV N-specific CD8+ T cell clones isolated from donor 1 against autologous BLCL target cells pulsed with increasing dilutions of the optimal peptide. E/T = 10. (A) CTL lines specific for the A2-restricted epitope N204–12. (B) CTL lines specific for the B7,B8-restricted epitope N173–81. (C) CTL line specific for the epitope N236–50. Lysis of targets not pulsed with peptide was <5% in all assays.
The CTL lines specific for N204–212 appeared to have a range of affinities for the peptide/MHC complex, as reflected by the wide range of peptide concentrations required for sensitization of target cells (Fig. 2 A). This suggests that the CTL response to this epitope is polyclonal. This peptide (GLFPTQIQV) conforms to the defined HLA binding motif for A2, with the preferred leucine at position 2 and valine at position 9 (19).
The CTL lines specific for N236–50 and N173–181 displayed more uniform peptide dose–response curves, suggesting that their TCRs have similar affinities for the peptide–MHC complex (Fig. 2, B and C). It is interesting to note that we identified both B7-restricted (3-C8 and 10-F5) and B8-restricted (10-G4) CTL lines that recognized the same epitope (Table I). This peptide (RPKHLYVSM) conforms to the defined HLA B7 motif reasonably well, with the preferred proline at position 2 and a methionine at position 9 (L or F preferred); and to the B8 motif less well, with the preferred lysine at position 3 and a leucine at position 5 (K or R preferred; reference 19). The second B8-restricted CTL epitope (N243–51: ECPFIKPEV) conforms to the B8 motif very poorly. This highlights the fact that natural CD8+ T cell epitopes do not necessarily conform to published HLA-binding motifs. Overall, these data show that individuals who had past PUUV infections developed broad CTL cell responses to at least four epitopes on the N protein.
CTL Lines Isolated from Donors 1 and 2 Are Primarily PUUV-specific.
Next, we tested whether the CTL lines isolated from donors 1 and 2 were cross-reactive with other human hantaviruses. To do this, we tested whether these CTL lines could recognize autologous BLCLs infected with recombinant vaccinia viruses expressing either PUUV, HTNV, or SNV N proteins (Table I). The CTL lines specific for N202–12 failed to recognize the N protein from either HTNV or SNV virus. The sequences of HTNV and SNV N proteins both differ from the PUUV sequence at this epitope by four amino acids, including a V to A change at the anchor position 9 (Table II). Of the two B7-restricted and one B8-restricted CTL lines that recognize PUUV N173–81, only clone 10-F5 displayed cross-reactive recognition of the HTNV N protein. This epitope is well conserved among the viruses, with the HTNV and SNV epitopes containing two conservative amino acid changes, and with both expressing the preferred P at position 2 (Table II).
Table II.
Sequence Comparison of Epitopes between PUUV, HTNV, and SNV Viruses
| N204-12 | PUUV | G | L | F | P | T | Q | I | Q | V | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HTNV | G | L | Y | P | A | Q | I | K | A | |||||||
| SNV | G | L | F | P | A | Q | V | K | A | |||||||
| N173-81 | PUUV | R | P | K | H | L | Y | V | S | M | ||||||
| HTNV | K | P | K | H | L | Y | V | S | L | |||||||
| SNV | K | P | R | H | L | Y | V | S | M | |||||||
| N243-51 | PUUV | E | C | P | F | I | K | P | E | V | ||||||
| HTNV | P | C | K | L | L | P | D | T | A | |||||||
| SNV | R | C | P | F | L | P | E | Q | K | |||||||
| N236-50 | PUUV | I | R | E | F | M | E | K | E | C | P | F | I | K | P | E |
| HTNV | I | E | Q | W | L | I | E | P | C | K | L | L | P | D | T | |
| SNV | I | D | D | F | L | A | A | R | C | P | F | L | P | E | Q | |
| N212-20 | PUUV | V | R | N | I | M | S | P | V | M | ||||||
| HTNV | A | R | Q | M | I | S | P | V | M | |||||||
| SNV | A | R | N | I | I | S | P | V | M | |||||||
| N164-78 | PUUV | T | S | F | E | D | I | N | G | I | R | R | P | K | H | L |
| HTNV | S | S | F | E | D | V | N | G | I | R | K | P | K | H | L | |
| SNV | S | S | Y | E | E | V | N | G | I | R | K | P | R | H | L |
The CD8+ CTL lines that recognize PUUV N236–250 are also primarily PUUV-specific with most displaying no recognition of targets expressing either the HTNV or SNV N protein. However, one CTL line (1B-G9) consistently recognized targets expressing the SNV N protein as well as those expressing the PUUV N protein (Table I). The sequence at this epitope is extremely variable among different hantaviruses (Table II), and it is surprising that this CTL line was able to recognize the corresponding sequence from the SNV N protein.
Neither of the cell lines that recognize PUUV N212–20 was cross-reactive with HTNV or SNV N protein, despite a relatively high level of sequence conservation at this epitope (Table II). The N212–20 peptides from the PUUV, HTNV, and SNV N proteins share the same amino acids at the putative anchor positions 2 and 9 (R and M, respectively). However, since we have not yet determined the HLA restriction of these CTL lines, we cannot accurately predict which residues within the epitope might be critical for recognition of the peptide-HLA complex by the TCR or for binding of this peptide to the HLA molecule. Finally, the CTL line that recognizes N164–178 (3-B8) is cross-reactive with the HTNV N protein, but does not recognize the SNV N protein. This epitope is relatively well conserved between PUUV and HTNV, with the corresponding epitope on HTNV N differing by three amino acids and less well conserved between PUUV and SNV, which differ by six amino acids at this epitope.
In summary, of 22 CTL lines tested against HTNV and SNV viruses only three CTL lines showed cross-reactive lysis, suggesting that the CTL responses identified in this study were highly PUUV-specific. None of these cross-reactive CTL lines recognized the N protein from both HTNV and SNV viruses, despite the fact that some of the epitopes share considerable sequence identity among the different hantaviruses.
PUUV-specific Memory CD8+ T Cells Circulate at High Frequencies in Immune Individuals.
To determine whether the epitopes identified in this study are commonly targeted in infected individuals, we studied a panel of PBMCs from 13 PUUV-immune donors, including the seven donors in which PUUV-specific responses were detected in bulk cultures (Fig. 1). Using IFN-γ ELISPOT assays, we determined the frequency of epitope-specific CD8+ T cells in the memory pool.
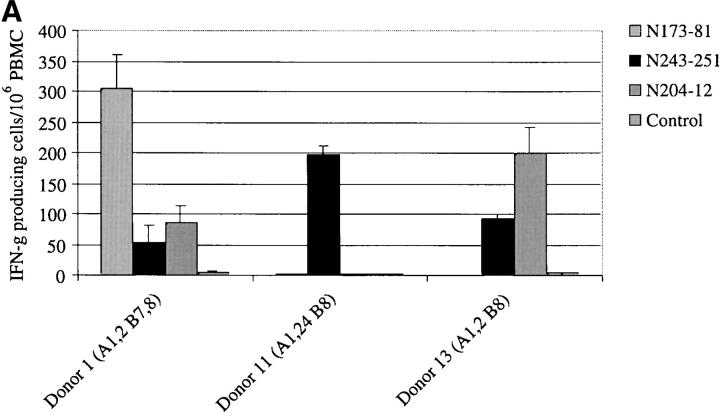
Of the 13 individuals tested, eight had detectable PUUV-specific T cell responses to one or more PUUV N epitope (Fig. 3) . Three individuals had high frequencies of circulating PUUV N-specific memory T cells (donors 1, 11, and 13; Fig. 3 A), with some epitope-specific T cells present at frequencies as high as 300 per 106 PBMCs. The hierarchy of epitope recognition varied from donor to donor, with different epitope-specific populations dominating in different individuals.
Figure 3.
Detection of virus-specific CD8+ memory T cells by IFN-γ ELISPOT. PBMCs were stimulated with 10 μg/ml of the indicated peptides in a 16–18 h assay. Input cell numbers ranged from 2–4 × 106 cells per well. Peptide stimulations were performed in triplicate wells. PBMCs stimulated with a B8-restricted epitope from HCV NS3 (NS31402–1411) was included for comparison with PUUV-specific B8-restricted epitopes. PBMCs incubated with media alone was included as negative controls. The data is presented as the number of IFN-γ producing cells/106 PBMCs, with media control values subtracted. The number of spots in the media control wells ranged from 0 to 5. All ELISPOT experiments were performed at least twice and mean values are shown. (A) PUUV-immune individuals who had high precursor frequencies (>100 specific T cells/106 PBMC) of CD8+ T cells specific for one or more CD8+ epitopes on PUUV N. (B) PUUV-immune individuals who had lower precursor frequencies of CD8+ T cells specific for one or more CD8+ epitopes on PUUV N.
Five additional donors (donors 2, 6, 7, 9, and 10) had somewhat lower frequencies of memory cells specific for these epitopes (Fig. 3 B), again with a variable hierarchy of recognition. The frequency of individual epitope-specific T cells seen in these donors was between 13 and 61/106 PBMCs. These responses are comparable to frequencies of many commonly recognized influenza virus epitopes (18, 20). In fact, when the three A2+ donors in this study were stimulated with the influenza A virus peptide M158–66 in ELISPOT assays, none of the donors had detectable responses to this epitope.
A summary of the range of precursor frequencies seen in these donors is shown in Table III. Of the three epitopes for which T cell responses were detected, the A2-restricted epitope (N204–12) and the B8-restricted epitope (N243–51) were recognized most consistently, with detectable responses to the former seen in all A2+ donor tested (3/3) and to the latter in 8 of 12 B8+ donors tested. The N173–81 (B7 or B8 restricted) was recognized in only 2 of 12 donors (Table III). We did not detect IFN-γ responses to the N164–78 epitope in any of the A28+ donors tested, including the donor from which the N164–78-specific clone was isolated (donor 2). Since we used a 15-mer peptide to stimulate the PBMCs in the ELISPOT assays, the conditions for stimulation may have been suboptimal. It is also possible that the frequencies of T cells specific for this epitope were below the limit of detection. These data demonstrate that PUUV N-specific memory CTL are abundant in the peripheral blood of immune individuals for many years after acute infection, and that detection of these memory CTL populations does not require prolonged in vitro restimulation.
Table III.
Recognition of CD8+ PUUV Epitopes in PBMCs from Immune Donors by IFN-γ ELISPOT
| Epitope | HLA-restriction | Number of donors testeda | Number of responders | Range of IFN-γ–producingT cells/106 PBMCs |
|---|---|---|---|---|
| N 173-181 | B7, B8 | 12 | 2 | 52-304 |
| N 243-251 | B8 | 12 | 8 | 13-188 |
| N 204-212 | A2 | 3 | 3 | 16-200 |
| N 164-178 | A28 | 3 | 0 | NDb |
| Flu M158-66 | A2 | 3 | 0 | ND |
Tested only donors possessing the indicated HLA allele.
Below the limit of detection.
We did not detect responses to any of the four CD8+ epitopes tested in the PBMCs of the other five donors (donors 3, 4, 5, 8, and 12). It is clear that virus-specific memory T cells are present in the PBMCs of these donors, since N-specific CTL activity is detectable in at least three of these donors PBMCs (donors 3–5) after 7 d of in vitro stimulation with infectious PUUV (Fig. 1). Undoubtedly, there are other CD8+ epitopes on the N protein that have not yet been identified, and that CTL with distinct specificities contribute to the CTL response seen in these donors. We have also demonstrated proliferation in response to inactivated PUUV in PBMCs from these donors (unpublished data), further demonstrating that these donors possess virus-specific memory T cells.
To determine whether the maintenance of PUUV-specific memory T cells was due to the persistence of virus, we performed nested RT-PCR on PBMCs and urine samples from PUUV immune donors, including 9 donors enrolled in the present study and seven donors from previous studies (21, 22). We were unable to detect viral RNA in convalescent PBMC or urine samples from any individual (unpublished data). In vitro, this assay is capable of detecting as few as 10–100 copies of viral RNA (unpublished data).
Discussion
In this study, we analyzed the memory CD8+ T cell responses in PUUV immune individuals. The majority of PBMCs from immune individuals stimulated with live PUUV recognized the viral nucleocapsid, and the N protein was the target of the majority of CTL lines isolated from two immune individuals. In contrast, the PUUV G1 glycoprotein was recognized at the bulk culture level in 5 of 7 donors, but we were able to isolate only a single G1-specific CTL line (17). These data, as well as the high precursor frequency of N-specific CD8+ T cells observed in many of the immune donors, suggest that the N protein may be the dominant target of CD8+ CTL during PUUV infection. Six CD8+ CTL epitopes on N protein were identified in this study (Table I). The hierarchy of the epitope-specific CD8+ T cell populations differed between individuals, suggesting that no single epitope was immunodominant. The CTL response in two donors was simultaneously targeted to at least four epitopes on PUUV N and was restricted by a variety of HLA alleles, thus demonstrating a diverse PUUV-specific T cell repertoire. The identification of PUUV-specific CD8+ T cell epitopes provides framework for future studies of PUUV-specific CTL responses during acute HFRS, allowing us to evaluate the kinetics and magnitude of the virus-specific T cell response and the potential role of T cell activation in the pathology of disease.
There are at least 20 distinct hantaviruses, approximately half of which are known to cause human disease. We were interested in examining whether protective immunity against one hantavirus confers immunity against other strains of hantavirus. This is an important consideration in geographic regions where more than one hantavirus cocirculates (23, 24). In a previous study, CTL responses directed against a conserved epitope on HTNV N protein were found to be highly cross-reactive, while CTL specific for another, more variable, epitope were HTNV-specific (16). The epitopes identified in this study are clustered near the center of the PUUV N protein in a region that shares relatively little sequence identity among the different hantaviruses. It was not surprising, therefore, to find that the majority of the CTL lines isolated in this study failed to recognize target cells expressing the N protein from either HTNV or SNV. In previous studies, B cell epitopes in humans and protective epitopes in bank voles were all mapped to the amino-terminal third of the N protein (25, 26). This suggests that distinct regions of the N protein are targeted by B and T cells during the immune response to PUUV. This may be an important consideration in the design of effective vaccines capable of eliciting both neutralizing antibodies and antiviral CTL.
Perhaps the most provocative finding of this study is that PUUV-specific memory CTL were maintained at high frequencies in individuals who had clinical infections with PUUV 6–15 y earlier, without evidence of continued antigenic stimulation. There is no evidence of persistent hantavirus infection, and clinical reinfections with PUUV or other human hantaviruses have never been reported. Viral RNA can be easily detected during within the first 9 d of symptomatic PUUV infection, but convalescent samples (>10 d after the onset of symptoms) are consistently PCR negative (21, 22, 27, 28). Similarly, patients with acute HPS have been shown to have high plasma levels of SNV RNA as detected by competitive RT-PCR, but viral RNA is rapidly cleared and becomes undetectable during convalescence (29, 30). To further address the question of virus persistence, we performed nested RT-PCR analysis on PBMCs and urine samples taken from 17 PUUV-immune individuals, including nine donors from this study. For ten of these donors, PCR analysis was performed during the acute phase of illness, and five had detectable PUUV RNA in their PBMCs or urine at that time. In agreement with published results, PUUV RNA was undetectable in all convalescent PBMCs and urine samples tested, suggesting that high levels of memory T cells persist in the absence of detectable virus.
There has been considerable debate regarding the role of antigen in the maintenance of memory CD8+ T cells. The definitive studies were performed in murine models in which LCMV-specific CD8+ T cells were transferred into naive wild-type or class I–deficient mice. These studies clearly demonstrated that the survival of memory CD8+ T cells requires neither persistent antigen nor the expression of MHC class I molecules on host cells (31, 32). Similar studies performed in B cell–deficient mice also ruled out the possibility that the survival of memory T cells relies on depots of antigen trapped on follicular dendritic cells in the form of antigen–antibody complexes (33). In humans, vaccinia virus-specific memory CTL responses have been detected in individuals who were vaccinated up to three decades ago. Since vaccinia virus does not cause a chronic infection in humans and reexposure to the virus is unlikely, the maintenance of T cell memory in this system also appears to be antigen independent (34).
The frequencies of PUUV-specific memory CTL reported here are comparable or higher than frequencies of CTL specific for many commonly recognized epitopes on influenza A virus. (18). In one study, the highest precursor frequencies of memory CD8+ T cells specific for dominant influenza A virus epitopes on the M1 and NP proteins ranged from 9–286/106 PBMCs and 15–67/106 PBMCs, respectively (18, 20). The highest frequencies of PUUV N-specific T cells measured in this study were 52–304/106 and 16–200/106 PBMCs. Interestingly, none of the three A2+ donors in this study had detectable responses to the influenza A virus M1 epitope (Table III). Like PUUV, influenza virus causes an acute infection that is rapidly cleared by the immune system. However, influenza virus is able to periodically reinfect individuals with existing influenza virus-specific T cell memory, and thus boost T cell responses. While periodic exposures to PUUV may occur, secondary infections have never been reported for this or any other human hantavirus. High levels of PUUV-specific neutralizing antibodies circulate after resolution of infection and these antibodies can be detected for many years (35–37). These neutralizing antibodies probably prevent or minimize the ability of the virus to establish a productive infection in host cells. As a result, the memory T cell response is unlikely to be effectively boosted since viral antigens are unlikely to gain access to the MHC class I processing pathway and be presented by a significant number of host cells. Indeed, the passive transfer of hantavirus-specific neutralizing antibodies in hamsters has been shown to provide protection against subsequent virus challenge, suggesting that the presence of neutralizing antibodies prevented the virus from establishing a productive infection (38).
Frequency analysis of measles virus-specific CTL in healthy adult donors who had childhood infections with measles reported that 100–960 cells per 106 PBMCs were virus-specific (39). In this study measles-infected BLCLs, rather than defined peptide epitopes, were used as stimulator cells in IFN-γ ELISPOT assays. Therefore, these numbers reflect the total of all measles virus-specific T cells capable of producing IFN-γ in these donors. The total frequency of the PUUV-specific T cells analyzed in the present study was between 13 and 444/106 PBMCs. Since we measured the responses to only four CD8+ CTL epitopes, restricted by only four possible HLA alleles (A2, A28, B7, B8), we are probably underestimating the total PUUV-specific T cells response in these donors. The frequencies of virus-specific memory CD8+ T cells measured in the present study are also comparable to the frequencies of some virus-specific CTL in adult donors with chronic virus infections such as EBV and HIV (40–43). In these systems, CTL precursor frequencies are often very high, probably due to periodic reactivation as a result of the persisting virus infection.
Overall, the data show that the frequencies of epitope-specific CD8+ T cells in PUUV-immune individuals are remarkably high and are comparable to or exceed those measured in other acute virus infections such as influenza and measles. The reported frequencies are also comparable to those measured in chronic virus infections such as EBV and HIV. This is notable given the fact that the primary PUUV infections in these individuals occurred between 6 and 15 y earlier, and that PUUV RNA is no longer detectable as early as 9 d after the onset of acute illness. This system provides a rare opportunity to study the development and maintenance of antiviral CD8+ T cell memory in humans after an acute infection in the absence of detectable persisting virus.
Acknowledgments
We would like to sincerely thank the Finnish individuals who agreed to donate blood for this study, and the Medical Research Fund of Tampere University Hospital. We also thank C.S. Schmaljohn for providing purified PUUV, A. Plyusnin and T. Sironen for performing the PCR analysis, J. Partanen for determining the HLA haplotypes of the donors, M. Mangada for intracellular cytokine staining, A.L. Rothman for his critical review of the manuscript, and A. Leporati for propagation of vaccinia virus recombinants.
This work was supported by EU contract QLK2-CT-1999-0119, Sigrid Juselius Foundation, and NIH-NIAID training grant T32-AIO7272.
H.L. Van Epps current address is Division of Infectious Diseases, Memorial Sloan-Kettering Cancer Center, 1275 York Ave., New York, NY 10021.
Footnotes
Abbreviations used in this paper: BLCL, B lymphoblastoid cell line; HFRS, hemorrhagic fever with renal syndrome; HPS, hantavirus pulmonary syndrome; HTNV, Hantaan virus; PUUV, Puumala virus; SNV, Sin Nombre virus.
References
- 1.Pensiero, M.N., J.B. Sharefkin, C.W. Dieffenbach, and J. Hay. 1992. Hantaan virus infection of human endothelial cells. J. Virol. 66:5929–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Temonen, M., O. Vapalahti, H. Holthofer, M. Brummer-Korvenkontio, A. Vaheri, and H. Lankinen. 1993. Susceptibility of human cells to Puumala virus infection. J. Gen. Virol. 74:515–518. [DOI] [PubMed] [Google Scholar]
- 3.Zaki, S.R., P.W. Greer, L.M. Coffield, C.S. Goldsmith, K.B. Nolte, K. Foucar, R.M. Feddersen, R.E. Zumwalt, G.L. Miller, and A.S. Khan. 1995. Hantavirus pulmonary syndrome. Pathogenesis of an emerging infectious disease. Am. J. Pathol. 146:552–579. [PMC free article] [PubMed] [Google Scholar]
- 4.Yao, Z.O., W.S. Yang, W.B. Zhang, and X.F. Bai. 1989. The distribution and duration of hantaan virus in the body fluids of patients with hemorrhagic fever with renal syndrome. J. Infect. Dis. 160:218–224. [DOI] [PubMed] [Google Scholar]
- 5.Nagai, T., O. Tanishita, Y. Takahashi, T. Yamanouchi, K. Domae, K. Kondo, J.R. Dantas, Jr., M. Takahashi, and K. Yamanishi. 1985. Isolation of haemorrhagic fever with renal syndrome virus from leukocytes of rats and virus replication in cultures of rat and human macrophages. J. Gen. Virol. 66:1271–1278. [DOI] [PubMed] [Google Scholar]
- 6.Huang, C., B. Jin, M. Wang, E. Li, and C. Sun. 1994. Hemorrhagic fever with renal syndrome: relationship between pathogenesis and cellular immunity. J. Infect. Dis. 169:868–870. [DOI] [PubMed] [Google Scholar]
- 7.Mustonen, J., H. Helin, K. Pietila, M. Brummer-Korvenkontio, K. Hedman, A. Vaheri, and A. Pasternack. 1994. Renal biopsy findings and clinicopathologic correlations in nephropathia epidemica. Clin. Nephrol. 41:121–126. [PubMed] [Google Scholar]
- 8.Temonen, M., J. Mustonen, H. Helin, A. Pasternack, A. Vaheri, and H. Holthofer. 1996. Cytokines, adhesion molecules, and cellular infiltration in nephropathia epidemica kidneys: an immunohistochemical study. Clin. Immunol. Immunopathol. 78:47–55. [DOI] [PubMed] [Google Scholar]
- 9.Krakauer, T., J.W. Leduc, J.C. Morrill, A.O. Anderson, and H. Krakauer. 1994. Serum levels of α and γ interferons in hemorrhagic fever with renal syndrome. Viral Immunol. 7:97–101. [DOI] [PubMed] [Google Scholar]
- 10.Krakauer, T., J.W. Leduc, and H. Krakauer. 1995. Serum levels of tumor necrosis factor-α, interleukin-1, and interleukin-6 in hemorrhagic fever with renal syndrome. Viral Immunol. 8:75–79. [DOI] [PubMed] [Google Scholar]
- 11.Linderholm, M., C. Ahlm, B. Settergren, A. Waage, and A. Tarnvik. 1996. Elevated plasma levels of tumor necrosis factor (TNF)-α, soluble TNF receptors, interleukin (IL)-6, and IL-10 in patients with hemorrhagic fever with renal syndrome. J. Infect. Dis. 173:38–43. [DOI] [PubMed] [Google Scholar]
- 12.Mori, M., A.L. Rothman, I. Kurane, J.M. Montoya, K.B. Nolte, J.E. Norman, D.C. Waite, F.T. Koster, and F.A. Ennis. 1999. High levels of cytokine-producing cells in the lung tissues of patients with fatal hantavirus pulmonary syndrome. J. Infect. Dis. 179:295–302. [DOI] [PubMed] [Google Scholar]
- 13.Mustonen, J., J. Partanen, M. Kanerva, K. Pietila, O. Vapalahti, A. Pasternack, and A. Vaheri. 1996. Genetic susceptibility to severe course of nephropathia epidemica caused by Puumala hantavirus. Kidney Int. 49:217–221. [DOI] [PubMed] [Google Scholar]
- 14.Xiao, S.Y., K.W. Spik, D. Li, and C.S. Schmaljohn. 1993. Nucleotide and deduced amino acid sequences of the M and S genome segments of two Puumala virus isolates from Russia. Virus Res. 30:97–103. [DOI] [PubMed] [Google Scholar]
- 15.Schmaljohn, C.S., S.E. Hasty, S.A. Harrison, and J.M. Dalrymple. 1983. Characterization of Hantaan virions, the prototype virus of hemorrhagic fever with renal syndrome. J. Infect. Dis. 148:1005–1012. [DOI] [PubMed] [Google Scholar]
- 16.Van Epps, H.L., C.S. Schmaljohn, and F.A. Ennis. 1999. Human memory cytotoxic T-lymphocyte (CTL) responses to Hantaan virus infection: identification of virus-specific and cross-reactive CD8+ CTL epitopes on nucleocapsid protein. J. Virol. 73:5301–5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terajima, M., H.L. Van Epps, D. Li, A. Leporati, S.E. Juhlin, J. Mustonen, A. Vaheri, and F.A. Ennis. 2002. Generation of recombinant vaccinia viruses expressing Puumala virus proteins and their use in isolating cytotoxic T cells specific for Puumala virus. Virus Res. 84:67–77. [DOI] [PubMed] [Google Scholar]
- 18.Lalvani, A., R. Brookes, S. Hambleton, W.J. Britton, A.V. Hill, and A.J. McMichael. 1997. Rapid effector function in CD8+ memory T cells. J. Exp. Med. 186:859–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rammensee, H.G., T. Friede, and S. Stevanoviic. 1995. MHC ligands and peptide motifs: first listing. Immunogenetics. 41:178–228. [DOI] [PubMed] [Google Scholar]
- 20.Jameson, J., J. Cruz, and F.A. Ennis. 1998. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J. Virol. 72:8682–8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plyusnin, A., J. Horling, M. Kanerva, J. Mustonen, Y. Cheng, J. Partanen, O. Vapalahti, S.K. Kukkonen, J. Niemimaa, H. Henttonen, et al. 1997. Puumala hantavirus genome in patients with nephropathia epidemica: correlation of PCR positivity with HLA haplotype and link to viral sequences in local rodents. J. Clin. Microbiol. 35:1090–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plyusnin, A., J. Mustonen, K. Asikainen, A. Plyusnina, J. Niemimaa, H. Henttonen, and A. Vaheri. 1999. Analysis of Puumala hantavirus genome in patients with nephropathia epidemica and rodent carriers from the sites of infection. J. Med. Virol. 59:397–405. [DOI] [PubMed] [Google Scholar]
- 23.Sibold, C., H. Meisel, A. Lundkvist, A. Schulz, F. Cifire, R. Ulrich, O. Kozuch, M. Labuda, and D.H. Kruger. 1999. Short report: simultaneous occurrence of Dobrava, Puumala, and Tula Hantaviruses in Slovakia. Am. J. Trop. Med. Hyg. 61:409–411. [DOI] [PubMed] [Google Scholar]
- 24.Avsic-Zupanc, T., K. Nemirov, M. Petrovec, T. Trilar, M. Poljak, A. Vaheri, and A. Plyusnin. 2000. Genetic analysis of wild-type Dobrava hantavirus in Slovenia: co-existence of two distinct genetic lineages within the same natural focus. J. Gen. Virol. 81:1747–1755. [DOI] [PubMed] [Google Scholar]
- 25.Vapalahti, O., H. Kallio-Kokko, A. Narvanen, I. Julkunen, A. Lundkvist, A. Plyusnin, H. Lehvaslaiho, M. Brummer-Korvenkontio, A. Vaheri, and H. Lankinen. 1995. Human B-cell epitopes of Puumala virus nucleocapsid protein, the major antigen in early serological response. J. Med. Virol. 46:293–303. [DOI] [PubMed] [Google Scholar]
- 26.Lundkvist, A., and B. Niklasson. 1992. Bank vole monoclonal antibodies against Puumala virus envelope glycoproteins: identification of epitopes involved in neutralization. Arch. Virol. 126:93–105. [DOI] [PubMed] [Google Scholar]
- 27.Horling, J., A. Lundkvist, K. Persson, M. Mullaart, T. Dzagurova, A. Dekonenko, E. Tkachenko, and B. Niklasson. 1995. Detection and subsequent sequencing of Puumala virus from human specimens by PCR. J. Clin. Microbiol. 33:277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grankvist, O., P. Juto, B. Settergren, C. Ahlm, L. Bjermer, M. Linderholm, A. Tarnvik, and G. Wadell. 1992. Detection of nephropathia epidemica virus RNA in patient samples using a nested primer-based polymerase chain reaction. J. Infect. Dis. 165:934–937. [DOI] [PubMed] [Google Scholar]
- 29.Terajima, M., J.D. Hendershot III, H. Kariwa, F.T. Koster, B. Hjelle, D. Goade, M.C. DeFronzo, and F.A. Ennis. 1999. High levels of viremia in patients with the Hantavirus pulmonary syndrome. J. Infect. Dis. 180:2030–2034. [DOI] [PubMed] [Google Scholar]
- 30.Hjelle, B., C.F. Spiropoulou, N. Torrez-Martinez, S. Morzunov, C.J. Peters, and S.T. Nichol. 1994. Detection of Muerto Canyon virus RNA in peripheral blood mononuclear cells from patients with hantavirus pulmonary syndrome. J. Infect. Dis. 170:1013–1017. [DOI] [PubMed] [Google Scholar]
- 31.Lau, L.L., B.D. Jamieson, T. Somasundaram, and R. Ahmed. 1994. Cytotoxic T-cell memory without antigen. Nature. 369:648–652. [DOI] [PubMed] [Google Scholar]
- 32.Murali-Krishna, K., L.L. Lau, S. Sambhara, F. Lemonnier, J. Altman, and R. Ahmed. 1999. Persistence of memory CD8 T cells in MHC class I-deficient mice. Science. 286:1377–1381. [DOI] [PubMed] [Google Scholar]
- 33.Asano, M.S., and R. Ahmed. 1996. CD8 T cell memory in B cell-deficient mice. J. Exp. Med. 183:2165–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demkowicz, W.E., Jr., R.A. Littaua, J. Wang, and F.A. Ennis. 1996. Human cytotoxic T-cell memory: long-lived responses to vaccinia virus. J. Virol. 70:2627–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lundkvist, A., J. Horling, and B. Niklasson. 1993. The humoral response to Puumala virus infection (nephropathia epidemica) investigated by viral protein specific immunoassays. Arch. Virol. 130:121–130. [DOI] [PubMed] [Google Scholar]
- 36.Lundkvist, A., S. Bjorsten, and B. Niklasson. 1993. Immunoglobulin G subclass responses against the structural components of Puumala virus. J. Clin. Microbiol. 31:368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horling, J., A. Lundkvist, J.W. Huggins, and B. Niklasson. 1992. Antibodies to Puumala virus in humans determined by neutralization test. J. Virol. Methods. 39:139–147. [DOI] [PubMed] [Google Scholar]
- 38.Schmaljohn, C.S., Y.K. Chu, A.L. Schmaljohn, and J.M. Dalrymple. 1990. Antigenic subunits of Hantaan virus expressed by baculovirus and vaccinia virus recombinants. J. Virol. 64:3162–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nanan, R., A. Rauch, E. Kampgen, S. Niewiesk, and H.W. Kreth. 2000. A novel sensitive approach for frequency analysis of measles virus-specific memory T-lymphocytes in healthy adults with a childhood history of natural measles. J. Gen. Virol. 81:1313–1319. [DOI] [PubMed] [Google Scholar]
- 40.Benninger-Doring, G., S. Pepperl, L. Deml, S. Modrow, H. Wolf, and W. Jilg. 1999. Frequency of CD8+ T lymphocytes specific for lytic and latent antigens of Epstein-Barr virus in healthy virus carriers. Virology. 264:289–297. [DOI] [PubMed] [Google Scholar]
- 41.Dalod, M., M. Harzic, I. Pellegrin, B. Dumon, B. Hoen, D. Sereni, J.C. Deschemin, J.P. Levy, A. Venet, and E. Gomard. 1998. Evolution of cytotoxic T lymphocyte responses to human immunodeficiency virus type 1 in patients with symptomatic primary infection receiving antiretroviral triple therapy. J. Infect. Dis. 178:61–69. [DOI] [PubMed] [Google Scholar]
- 42.Dalod, M., M. Dupuis, J.C. Deschemin, C. Goujard, C. Deveau, L. Meyer, N. Ngo, C. Rouzioux, J.G. Guillet, J.F. Delfraissy, et al. 1999. Weak anti-HIV CD8+ T-cell effector activity in HIV primary infection. J. Clin. Invest. 104:1431–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan, L.C., N. Gudgeon, N.E. Annels, P. Hansasuta, C.A. O'Callaghan, S. Rowland-Jones, A.J. McMichael, A.B. Rickinson, and M.F. Callan. 1999. A re-evaluation of the frequency of CD8+ T cells specific for EBV in healthy virus carriers. J. Immunol. 162:1827–1835. [PubMed] [Google Scholar]