Abstract
Chronic lymphocytic leukemia (CLL) arises from the clonal expansion of a CD5+ B lymphocyte that is thought not to undergo intraclonal diversification. Using VHDJH cDNA single strand conformation polymorphism analyses, we detected intraclonal mobility variants in 11 of 18 CLL cases. cDNA sequence analyses indicated that these variants represented unique point-mutations (1–35/patient). In nine cases, these mutations were unique to individual submembers of the CLL clone, although in two cases they occurred in a large percentage of the clonal submembers and genealogical trees could be identified. The diversification process responsible for these changes led to single nucleotide changes that favored transitions over transversions, but did not target A nucleotides and did not have the replacement/silent nucleotide change characteristics of antigen-selected B cells. Intraclonal diversification did not correlate with the original mutational load of an individual CLL case in that diversification was as frequent in CLL cells with little or no somatic mutations as in those with considerable mutations. Finally, CLL B cells that did not exhibit intraclonal diversification in vivo could be induced to mutate their VHDJH genes in vitro after stimulation. These data indicate that a somatic mutation mechanism remains functional in CLL cells and could play a role in the evolution of the clone.
Keywords: B lymphocyte, chronic lymphocytic leukemia, somatic hypermutation, Ig gene, V gene diversification
Introduction
Chronic lymphocytic leukemia (CLL)* is the most prevalent adult leukemia in Western countries, accounting for ∼30% of all leukemias (1). It is characterized by the clonal expansion of a CD5+ B cell (2) that was in the past viewed as having a poor propensity to undergo Ig V(D)J gene hypermutation (3–7). This view, however, changed when it was documented that the leukemic cells from ∼60% of CLL patients express Ig VH gene somatic point-mutations (8–13). The frequency of these mutations relates to the VH family expressed, being higher in those cells expressing VH3 genes than in those expressing VH1 and VH4 (13). However, even those studies that described CLL cases with a very high number of somatic mutations reported that these mutations were shared by all the CLL B cells, strongly suggesting that intraclonal diversification does not occur in these leukemic B cells (14–17).
The lack of intraclonal diversification in CLL B cells was considered consistent with the absence or scarcity of somatic mutations in human (2, 18) and murine (19–21) B1a cells. This, however, was not supported by the findings that human CD5+ B cells can undergo somatic hypermutation, antigen selection, and possibly affinity maturation (22–25). Rather, these findings supported the postgerminal center-like features that have been recently suggested to be characteristic of certain CLL cases such as V gene mutations (13), CD38 expression (26), and in vivo isotype class switching (6, 9, 11, 13, 27–29).
Prompted by these discrepancies and the recent advances in the understanding and induction of somatic hypermutation in human B cells (30–33), we decided to determine whether CLL B cells continue to develop V gene mutations after leukemic transformation and, therefore, intraclonally diversify. Using a highly sensitive and ad hoc devised approach (32) based on single-strand DNA conformational polymorphism (SSCP), we determined that intraclonal VHDJH gene diversification and clonal evolution are common features of the in vivo natural history of many CLL B cells. In addition, using appropriate stimuli, we induced somatic mutations in vitro in those leukemic B cells that did not show evidence of in vivo intraclonal diversification. These findings indicate that a hypermutation machinery is functional and active in certain CLL B cells and further point to the importance of a germinal center-like reaction in some of these B lymphocytes.
Materials and Methods
CLL Patients and B Cells.
The 18 patients (12 males and six females) in this study were diagnosed with typical B cell CLL based on clinical criteria and laboratory features. All cases represented clonal expansions of small lymphocytes with high nuclear/cytoplasmic ratios that coexpressed CD19, CD5, CD23, and CD27 along with monotypic H and L chain surface membrane Ig. The median age of the group at the time of analysis was 60 y with a distribution of clinical Rai stages as follows: 10 patients in stages O-II and eight patients in stages III-IV. PBMCs from these patients were obtained from heparinized venous blood by fractionation through Histopaque 1077® (Sigma-Aldrich). B lymphocytes were enriched from PBMCs by depletion of T cells and monocytes as described previously (31). All patients provided informed consent before giving blood samples.
PCR Amplification of V(D)J Transcripts.
RNA was extracted from 2 × 106 B cells using the RNeasy™ Total RNA kit (QIAGEN). mRNA was reverse transcribed using the SuperScript™ Preamplification System for first strand cDNA synthesis (Life Technologies, Inc.). VHDJH-CH cDNAs were amplified using sense primers specific for the sequences of the different VH families together with the antisense primers specific for the CH1-μ sequence (13, 32) and the Pfu Turbo® DNA polymerase (Stratagene). Each reaction consisted of 30 cycles (1 min denaturation at 94°C, 1 min annealing at 58°C, 1 min extension at 72°C, and 10 min extension at 72°C). The PCR cDNA products were purified, ligated into pCR-Blunt II-TOPO vector (Zero Blunt™ TOPO™ Cloning kit; Invitrogen), and transfected in TOP10 One Shot™ competent cells (Invitrogen). Bacterial colonies were screened by PCR and those positive for VHDJH-CH transcripts were selected for SSCP analysis.
Detection of Mutated VHDJH Transcripts by SSCP.
Mutated VHDJH transcripts were identified by SSCP analysis as described previously (32). In brief, cDNAs were amplified with Taq DNA polymerase (Life Technologies, Inc.) by PCR using the cloned cDNA inserted into pCR-Blunt II-TOPO vector as template, in the presence of 1 μCi [α-32P] dCTP (3,000 Ci/mmol; NEN Life Sciences). The internal VH leader sense primer and JH antisense primer (31) were used for VHDJH analysis. Samples were denatured and immediately loaded onto a 6% acrylamide gel (20:1 acrylamide:bis) with 1 × TBE containing 10% Glycerol. Electrophoresis was performed at room temperature for 18 h at 6 W. Gels were autoradiographed on Kodak X-Omat™ AR film (Kodak).
Sequencing Ig V(D)J-C Transcripts.
The Ig VHDJH cDNA clones displaying an altered electrophoretic mobility in SSCP gel as well as at least 5 clones from each patient with typical mobility were analyzed by sequencing to confirm and characterize the nature of the mutations (13, 32). Sequences were compared with the germline counterpart (34) and with the original CLL VHDJH sequence using MacVector v. 5.0 software (International Biotechnologies).
Mutational Analysis.
The census of the somatic point-mutations was determined by counting identical mutations in more than one transcript only once. Comparison of the observed with the expected frequency of replacement (R) and silent (S) point-mutations was performed using the inherent mutation rate of the CLL VHDJH sequences, calculated using the Inh. Sus. Calc. Program, version 1.0 for the Macintosh as reported by B. Chang and P. Casali (35). The expected frequency of mutations was calculated by taking into account the base composition of the unmutated CLL V(D)J sequence, i.e., it was corrected by the frequency of occurrence of the individual nucleotides, or di-, tri-, tetra-nucleotides considered within the CLL B cells V(D)J sequence assuming randomness. In the absence of negative or positive selective pressure on a gene product, nucleotide changes yielding amino acid R or S mutations are randomly distributed throughout the coding sequences. If a DNA segment displays a number of R mutations higher than that expected by chance alone, a positive selective pressure for variability is the likely cause. Conversely, if a DNA segment displays a number of R mutations lower than that expected by chance, it is likely that a negative pressure was exerted on the gene product to select against mutations, such that the protein structure is preserved.
T Cells.
CD4+ T cells were positively selected from PBMCs by fractionation through Histopaque 1077® (Sigma-Aldrich) using CD4-conjugated magnetic beads® (Miltenyi Biotec). Selected cells were cultured in FCS-RPMI 1640, and expanded by weekly stimulation with a feeder cell mixture containing irradiated (1,200 rads) PBMCs, 100 μg/ml of phytohemagglutinin (Life Technologies Inc.), and 100 U/ml of human recombinant IL-2 (Genzyme). For T/B cell coculture experiments, CD4+ T cells were used at least 2 wk after their last activation, and were incubated for 6 h with 20 ng/ml of 13-phorbol 12-myristate acetate (Sigma-Aldrich), and 500 ng/ml of ionomycin (Calbiochem-Novabiochem) before culture with B cells.
B/T Cell Cocultures.
B/T cell cocultures were performed as described previously (31). In brief, CLL B cells were cultured at 0.5 × 105 cells per well in the presence of 2.5 × 105 irradiated (4,000 rads) CD4+ T cells, 106 irradiated (4,000 rads) human CD40L-transfected 293 cells (CD40L-293 cells) and cytokines, including IL-4 (100 U/ml) and IL-2 (100 U/ml) in a U-bottomed, 96-well plate (200 μl FCS-RPMI 1640 vol). To cross-link the BCR, CLL B cells were incubated for 2 h at 4°C with Sepharose®-conjugated rabbit Abs to human IgM and Ig (H + L) chain (2 μg/ml; Irvine Scientific), and then washed with cold PBS. After 7 d of culture, CLL B cells were collected, freed of dead cells and debris by fractionation through Histopaque® 1077 (Sigma-Aldrich), exposed again to anti-BCR Abs, washed, and reseeded over a new layer of irradiated T cells and CD40L-293 cells, in the presence of cytokines. At day 14 of culture, CLL cells were harvested for total RNA extraction.
Results
CLL VHDJH Gene Diversification Determined by SSCP Analyses.
The VH, D, and JH genes expressed by the 18 CLL cases included in this study are listed in Table I. Of the VH genes identified, VH3 family members were the most frequent, and JH4 was the most represented JH gene, as reported for other CLL cases (13). The origin of two D gene segments could not be determined.
Table I.
Ig VHDJH Genes and Somatic Point-Mutations Expressed in CLL B Cells
| Point mutations
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CLL case |
GenBank accession number |
VH gene | D gene | JH gene | Number of transcripts analyzed |
Total | Shareda | Partiallysharedb | Uniquec | Number of mutations expected by PCR error |
| 63 | AF021974 | 1-46 | D6-6 | JH6b | 29 | 0 | 0 | 0 | 0 | 0.29 |
| 67 | AF021990 | 4-34 | D2-15 | JH4b | 24 | 0 | 0 | 0 | 0 | 0.24 |
| 270 | AY055487 | 1-02 | D6-19 + D3-9 | JH4b | 28 | 1 | 0 | 0 | 1 | 0.28 |
| 141 | AF022005 | 4-34 | D2-2 | JH5b | 29 | 2 | 0 | 0 | 2 | 0.29 |
| 156 | AY055478 | 1-45 | D2-2 + D3-10 | JH5b | 27 | 2 | 0 | 0 | 2 | 0.27 |
| 7 | AY055477 | 3-49 | D3-3 | JH5a | 29 | 2 | 2 | 0 | 0 | 0.29 |
| 258 | AY05485 | 1-69 | D3-16 | JH3b | 30 | 3 | 0 | 0 | 3 | 0.30 |
| 216 | AY055483 | 3-49 | D5-12 | JH6b | 40 | 3 | 3 | 0 | 0 | 0.40 |
| 48 | AF0211969 | 1-02 | D6-13 | JH3b | 26 | 8 | 8 | 0 | 0 | 0.26 |
| 175 | AY055484 | 3 (HHG4) | D3-16 | JH6b | 28 | 9 | 2 | 0 | 7 | 0.28 |
| 249 | AY055479 | 3-23 | D2-21 | JH4b | 30 | 10 | 4 | 0 | 6 | 0.30 |
| 165 | AY055482 | 3-15 | D1-26 | JH6b | 30 | 14 | 14 | 0 | 0 | 0.30 |
| 178 | AY055482 | 2-05 | ND | JH4b | 30 | 17 | 13 | 0 | 4 | 0.30 |
| 136 | AF022002 | 4-34 | D2-2 | JH6b | 40 | 18 | 18 | 0 | 0 | 0.40 |
| 113 | AF021989 | 2-05 | ND | JH4b | 57 | 24 | 21 | 0 | 3 | 0.57 |
| 169 | AY055480 | 3-33 | D3-9 | JH4b | 50 | 26 | 24 | 0 | 2 | 0.50 |
| 261 | AY055486 | 3-33 | D5-12 | JH4b | 80 | 36 | 14 | 8 | 14 | 0.80 |
| 105 | AF021986 | 3-23 | D2-21 | JH4b | 60 | 40 | 5 | 11 | 24 | 0.60 |
Point-mutations in the Ig VHDJH gene transcripts of CLL B cells.
Shared, mutations shared by all the Ig VHDJH gene transcripts analyzed.
Partially shared, mutations shared but some but not all the Ig VHDJH gene transcripts analyzed.
Unique, mutations unique to distinct Ig VHDJH gene transcripts analyzed.
Based on error rate of 10−6 change/base/PCR cycle.
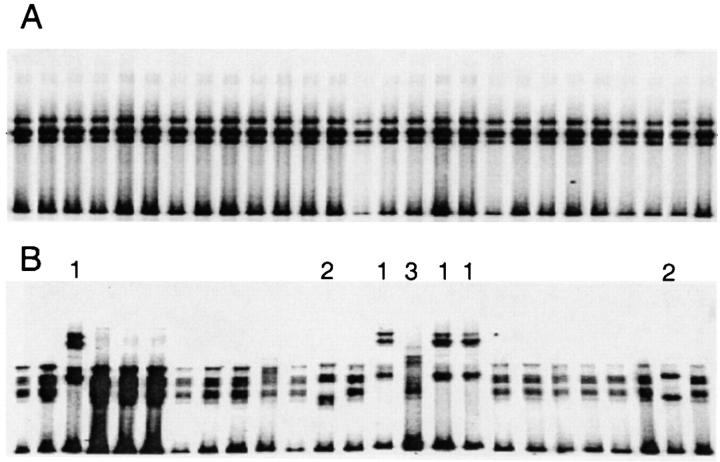
The Ig VHDJH cDNAs amplified by PCR using high-fidelity Pfu Turbo® DNA polymerase were cloned into appropriate vectors for nested PCR amplification in the presence of [α-32P] dCTP. The amplified [α-32P] labeled VHDJH cDNAs were then subjected to SSCP analysis. The cDNAs from seven cases were homogeneous in electrophoretic mobility, as exemplified by case 216 (Fig. 1 A). In the remaining 11 cases, the VHDJH cDNA transcripts displayed inconstant patterns of electrophoretic mobility, suggesting a variable degree of intraclonal diversification, as exemplified by case 105 (Fig. 1 B).
Figure 1.
SSCP analysis from two CLL cases displaying either the absence or presence of intraclonal diversification. All 27 VHDJH transcripts from CLL 216 showed identical mobility (A). In CLL 105, seven of the 25 VHDJH transcripts showed a mobility pattern different from that displayed by the remaining 18 VHDJH transcripts (B). Sequence analysis showed that the VHDJH cDNAs 1–3 were all collinear and collinear with the most represented transcripts confirming their monoclonality. However, these transcripts displayed nucleotide variations distributed randomly throughout the VH segment. VHDJH transcripts labeled 1 were identical among themselves but different, though collinear, from the VHDJH transcripts 2 and 3, and the most represented transcripts. VHDJH transcripts 2 were all identical but different, though collinear, from the VHDJH transcripts 1, 3, and the most represented transcripts. VHDJH transcripts 3 were identical but different, though collinear, from the VHDJH transcripts 1, 2 and the most represented transcripts.
DNA Sequence Analyses Confirm that CLL B Cells Can Accumulate New Ig V Gene Mutations and Thereby Intraclonally Diversify.
The sequences of the VHDJH cDNAs that differed in their SSCP mobility profiles from the majority of the VHDJH cDNAs and the sequences of at least five cDNAs representative of the dominant mobility profile were determined. In each case, all the VHDJH cDNAs were collinear, strengthening the monoclonality of the leukemic cells. However, as expected, intraclonal nucleotide differences were detected in those VHDJH cDNAs that displayed altered electrophoretic mobility (Table I). These findings were consistent, regardless of how many transcripts were analyzed. This suggests that the data provided by our SSCP analyses accurately reflected the nature of the in vivo process.
The nucleotide differences detected in these sequence analyses were distributed randomly throughout VH. In most instances, these differences were single base substitutions, resulting in both S and R mutations. The frequency of R mutations in the complementarity determining regions (CDR) and framework regions (FR) of the VH gene segments were not different from those expected by chance alone, suggesting the lack of a selective pressure applied to these Ig VH gene products.
Table I lists the number and characteristics of the cDNAs analyzed in each CLL case. These transcripts are noted as containing either “shared” (mutations present in all the Ig VHDJH gene transcripts analyzed), “partially shared” (mutations exhibited by some but not all VHDJH transcripts), or “unique” point-mutations (mutations found only in distinct VHDJH transcripts) as compared with the respective germline template. The number of partially shared and unique point-mutations differed between the different cases analyzed, ranging from 1 (case 270) to 35 (case 105).
The Presence of Intraclonal Diversification Is Not Related to the Initial Load of Shared Mutations.
CLL cases can be divided into two subgroups based on the presence of Ig V gene mutations (13). To determine whether the degree of leukemic B cell intraclonal diversification was related to this original mutational load, we analyzed by linear regression the relationship of the number of partially shared and unique mutations to the total number of mutations (Table I). When all the cases were included in this analysis, the degree of intraclonal diversification was found to be dependent on the overall load of point-mutations (P < 0.05). However, when we excluded from the analysis the two most intraclonally diversified cases (105 and 261) that were atypical in their extent and patterns of diversification (vide infra), the relationship was no longer significant (P > 0.1). Thus, intraclonal diversity appears to occur in CLL B cells regardless of their original Ig V gene mutational load.
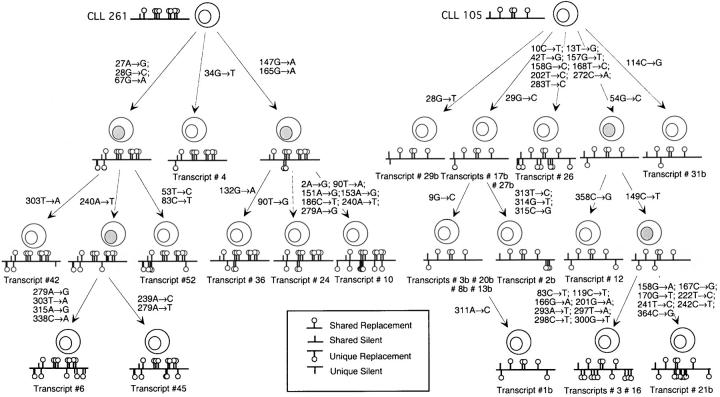
The High Degree of Intraclonal Diversification in Certain CLL Cases Allows the Construction of Genealogical Trees.
To outline the evolution of the changes that occurred within each CLL clone, we aligned all the VHDJH cDNAs to the closest germline gene sequence and analyzed them on the assumption that the shared mutations occurred due to single rather than independent events. This allowed us to identify in each case the putative progenitor VHDJH gene sequence and to assess the level of intraclonal complexity. Of the 11 cases that exhibited intraclonal diversification, nine (four originally unmutated CLL 141, 156, 258, and 270, and five originally mutated CLL 113, 169, 175, 178, and 249) displayed only one level of diversification, i.e., they expressed only unique mutations and no partially shared mutations. However, two cases, CLL 105 and 261, displayed different levels of complexity with several partially shared mutations that allowed us to construct genealogical trees, as reported previously (36, 37).
CLL 261 showed 14 unique and eight partially shared point-mutations, compared with the progenitor VHDJH gene sequence (Fig. 2 A). These mutations identified a genealogical tree with three branches that spanned 1–3 generations. CLL 105 showed 24 unique point-mutations and 11 partially shared mutations, compared with the progenitor VHDJH gene sequence (Fig. 2). There were five branches to this genealogical tree, spanning 1–3 generations.
Figure 2.
Genealogical tree constructed using VHDJH sequences of CLL nos. 261 and 105. Point-mutations are indicated by their codon number and the nature of the base change. Shared point-mutations and acquired unique point-mutations are indicated above and below the line, respectively. Vertical bars depict S mutations, and lollipops depict R mutations. The putative intermediate elements are depicted with gray nuclei.
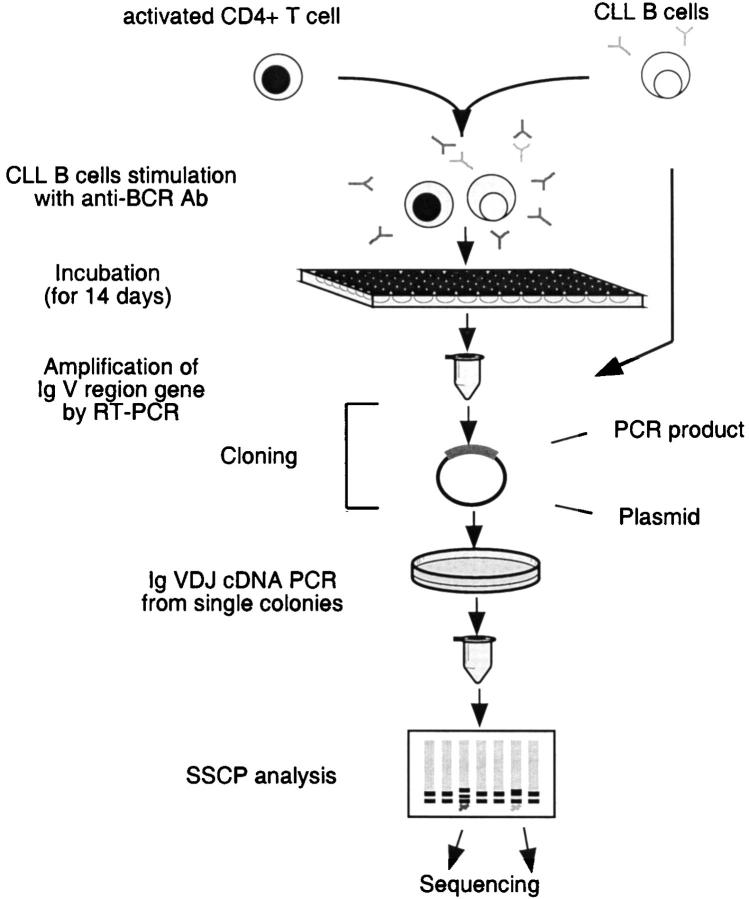
Induction of Somatic Mutation in CLL B Cells In Vitro.
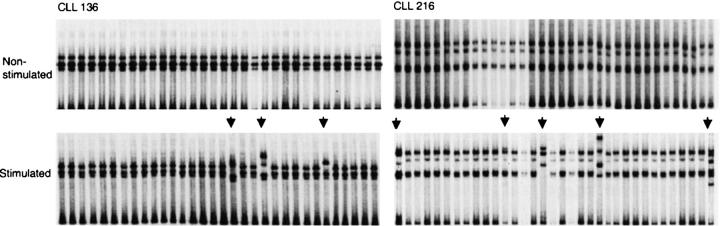
To determine whether CLL B cells could be induced to mutate the expressed VHDJH genes in vitro, the leukemic cells from CLL cases 136 and 216 were reacted with immobilized Abs to human Ig, and then cultured in the presence of activated normal allogeneic human CD4+ T cells and IL-2 and IL-4. After 14 d of culture, VHDJH-CH cDNAs were analyzed as illustrated in Fig. 3 . These cases were chosen for these studies because their B cells did not exhibit evidence for in vivo intraclonal variability (40 independent bacterial clones containing VHDJH cDNAs screened by SSCP and cDNA sequencing revealed that all cDNAs were identical; Table I).
Figure 3.
Schematic representation of the steps involved in the in vitro induction of somatic hypermutation in CLL B cells.
After in vitro stimulation, 3 of 32 (9%) VHDJH cDNAs in CLL 136 and 5 of 34 (14.7%) in CLL 216 displayed a gel mobility different from that of the corresponding cDNAs from the unstimulated CLL cells (Fig. 4) . All of the clones with a different gel mobility pattern in SSCP contained new point-mutations. In CLL 136, we detected three mutations. These were independent point-mutations confined to the VHDJH sequence (375 bp) and comprised three transversions (clone 11: 4 G > C; clone 20: 23 G > T; clone 9: 120 T > G). Thus, CLL 136 was induced to undergo mutation with an overall frequency of 2.7 × 10−4 changes/base, >sixfold the PCR amplification error rate with high-fidelity Pfu Turbo® DNA polymerase mentioned above. In CLL 216, a total of nine mutations were found in VHDJH transcripts. These were also independent point-mutations that were confined to the VHDJH sequence and consisted of nine transitions (clone 2: 25T > C, 36T > C, 88G > A; clone 4: 75T > C; clone 23: 87G > A, 21G > A; clone 32: 21T > C, 157G > A; clone 18: 75A > G). These mutations occurred at a frequency of 6.86 × 10−4 changes/base. Thus, these CLL cells were triggered to mutate their expressed VHDJH genes with the same modalities that induce mutations in normal B cells and monoclonal B cell lines (30, 31).
Figure 4.
In vitro induction of somatic hypermutation. CLL 136 and CLL 216 did not exhibit evidence of in vivo intraclonal diversity (see Table I). After in vitro stimulation, 3 of 32 VHDJH transcripts in CLL 136 and 5 of 34 in CLL 216 displayed gel mobilities different from that of corresponding transcripts obtained from the unstimulated CLL cells. Transcripts were sequenced and each contained at least one nucleotide change.
Comparison of the Mutations Induced In Vitro with Those Occurring In Vivo.
Randomly occurring somatic point-mutations are expected to be one-third transitions and two-thirds transversions (38). Among the 128 original in vivo shared VHDJH point-mutations, transitions exceeded transversions, 72:56 (Table II). As in normal B cells, the original shared mutations targeted A nucleotides at a frequency (46/128 total mutations) that was ∼48% higher than expected by chance alone, after correcting for base composition, i.e., normalizing for the relative occurrence of A in the unmutated VHDJH sequence. A > G mutations accounted for 59% of the total A mutations and 38% of the total transitions observed (Table II).
Table II.
Nature of the Base Substitutions in the Ig VHDJH Gene Segment of CLL B Cells In Vivo and In Vitro
| In vivo shared point-mutations | In vivo partially shared and unique point-mutations | In vivo induced point-mutations | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transitions | G > A | A > G | C > T | T > C | Transitions | G > A | A > G | C > T | T > C | Transitions | G > A | A > G | C > T | T > C |
| 72 | 24 | 27 | 17 | 4 | 39 | 9 | 8 | 12 | 10 | 9 | ||||
| [12.8] | [10.6] | [9.8] | [10.2] | [7.4] | [5.9] | [5.4] | [5.6] | |||||||
| Transversions | G > C | A > C | C > A | T > A | Transversions | G > C | A > C | C >A | T > A | Transversions | G > C | A > C | C >A | T > A |
| 27 | 11 | 9 | 4 | 3 | 12 | 4 | 2 | 3 | 3 | 1 | 1 | 0 | 0 | 0 |
| [12.8] | [10.6] | [9.8] | [10.2] | [7.4] | [5.9] | [5.4] | [5.6] | [1.2] | [0.92] | [0.92] | [0.92] | |||
| G > T | A > T | C > G | T > G | G > T | A > T | C > G | T > G | G > T | A > T | C > G | T > G | |||
| 29 | 5 | 10 | 5 | 9 | 23 | 7 | 5 | 7 | 4 | 2 | 1 | 0 | 01 | |
| [1.2] | [0.92] | [0.92] | [0.92] | |||||||||||
| Total | G > N | A > N | C > N | T > N | Total | G > N | A > N | C > N | T > N | Total | G > N | A > N | C > N | T > N |
| 128 | 40 | 46 | 26 | 16 | 74 | 20 | 15 | 22 | 17 | 12 | 6 | 1 | 0 | 5 |
| [38.4] | [31.8] | [29.4] | [30.7] | [22.2] | [17.7] | [16.2] | [17] | [3.6] | [2.76] | [2.76] | [2.76] | |||
Shared, partially shared, and unique point-mutations in the Ig VHDJH gene segment of CLL B cells were analyzed in vivo and after in vitro induction.
Identical mutations in different transcripts of the same and different isotypes were assumed not to be independent and were counted only once.
The [expected] number of mutations (from a given nucleotide residue to another given nucleotide residue) was normalized for the base composition of the unmutated VHDJH sequence. It was calculated by multiplying the frequency of occurrence of the nucleotide target of mutation in the unmutated sequence by the total number of observed mutations, and dividing this product by three. For instance, the expected number of G > A mutations was calculated by multiplying 0.30 (G frequency of occurrences the unmutated VHDJH sequence) by 128 = 38.4, divided by 3 (as G > A, G > C, and G > T mutations have all the same theoretical probability to occur) = 12.8.
Shared point-mutations in the Ig VHDJH gene segment of CLL 7, 216, 48, 175, 249, 165, 178, 136, 113, 169, and 105.
Partially shared and unique point-mutations in the Ig VHDJH gene segment of CLL 270, 141, 156, 175, 113, 169, 261, and 105.
The 74 in vivo partially shared and unique point-mutations showed a similar, albeit lesser bias for transitions over transversions (39:35). However, these mutations lacked an A base preference since only 15/74 mutations (20%) targeted A nucleotides. Among the two cases with the more extensive partially shared and unique mutations, CLL 261 demonstrated some evidence for A targeting (9/22 mutations involving A), whereas CLL 105 exhibited minimal evidence for this tendency (2/35 mutations involving A). These two cases also differed in the ratio of transitions to transversions detected among these mutations (CLL 261–14:8 and CLL 105–15:20).
The in vitro–induced mutations also favored transitions over transversions (9:3). However, the small number of these mutations did not allow us to draw firm conclusions regarding their nature.
Discussion
The generation of high affinity–specific antibodies usually occurs in the germinal centers of organized lymphoid tissues and depends on a complex series of interactions involving antigen activated B cells, specific T cells, and follicular dendritic cells (39). This process usually introduces point-mutations into Ig V genes and thus increases receptor diversity (40, 41). Subsequently, certain B cells are selected for survival and expansion based on the affinity for antigen of the BCR encoded by the newly mutated V genes (42, 43). Recently, it has been suggested that somatic mutations may occur via another pathway that does not require the participation of T cells and may occur outside of classical germinal center structures (44, 45).
CLL B cells have been viewed as B lymphocytes that cannot sustain effective somatic hypermutation of their Ig VHDJH genes. However, analyses of the expressed Ig V genes in CLL indicate that as many as 50% of IgM+ CLL and 75% of non-IgM+ (IgG and IgA) CLL cases show evidence of somatic mutations with a subset of these displaying R mutations in a pattern consistent with antigen selection (13). Thus, although the traditional views about the presence of Ig V gene mutations in the leukemic cells of this disease have been modified, the literature has remained consistent in characterizing these V gene mutations as static, without evidence for intraclonal instability and the accumulation of diversity. Occasional studies have reported the detection of somatic variants over time in typical CLL cases and in a CD5− case (11, 46, 47). However, these and other studies did not demonstrate detailed evidence that significant intraclonal diversification occurred in the leukemic cells of many CLL B cases, leading to the conclusion that somatic mutation ceases as a consequence of leukemic transformation (15–17). In this report, we document that intraclonal VHDJH gene diversification and clonal evolution do occur, albeit at variable degrees, in >50% (11/18) of CLL B cases. Additionally, in those cases in which this phenomenon is not detected in vivo, diversity can be induced in vitro after providing physiologically relevant signals.
It is unlikely that these findings represent PCR errors. First, the frequency of unique nucleotide changes ranged from 3.5-fold (case 270) to 58-fold (case 105) higher than the expected frequency of misinsertions calculated based on the error rate of the high-fidelity Pfu Turbo® DNA polymerase provided by the manufacturer (10−6 base/cycle, i.e., 0.01 base changes in a 375 bp DNA sequence after 30 cycles). This published error rate is actually somewhat higher than that calculated by us using the sequence data generated for the Ig CH region from the exact same cDNA used for the VH data presented above and from other cDNA prepared in an identical manner (0.6 × 10−6 base/cycle). Nevertheless, we have used the manufacturer's standard in order to be conservative about our estimations. However, even if we are extremely conservative in our calculations and exclude those cases with ≤2 point-mutations, a significant number of the remaining cases (7/18; ∼35%) still exhibit from 3–21 mutations, which is inconsistent with these PCR error rates. Second, when we subjected a subclone of the Burkitt's lymphoma cell line RAMOS (clone 1; references 48 and 49) to the same RT-PCR-SSCP cloning and sequencing procedure that we used for the CLL cases, a mutation frequency of 1.3 × 10−4/bp was identified, which compares favorably with the published mutation frequencies determined for this RAMOS subclone (0.8–0.93 × 10−4/bp; references 48 and 49). These two comparisons also suggest that the reverse transcription step was not a major source of error in our analyses, since these calculations of potential error are very similar despite the fact that our analyses were based on RNA as the starting material and the others on DNA.
Thus, the calculated rate of IgV gene mutations in these B cells is clearly greater than that occurring spontaneously in vivo elsewhere in the genome or to that attributable to in vitro PCR error with high-fidelity Pfu Turbo® DNA polymerase (Table I). It is also considerably lower than that seen in other B cell lymphomas thought to originate in the germinal center such as follicular lymphoma and Burkitt's lymphoma (50–54). These mutation frequency differences are in line with the in vivo proliferative differences of the malignant cells in CLL and these two subtypes of B cell lymphoproliferative disorders.
Our findings indicate that a somatic mutation process is active in at least certain members of the CLL clone, indicating that the leukemic cells are not functionally inert, but are in this regard functionally active. What is unclear is the extent to which this mutational activity is mediated by the classic IgV gene hypermutation mechanism, and also whether it is induced by external stimuli or is inherent to the B cells due to genetic alterations that occurred as a consequence of the leukemic transformation. Although our data do not directly indicate that the observed in vivo intraclonal diversification in CLL B cells is externally mediated, there is evidence from these studies to suggest that this could occur. In vitro stimulation through the BCR and other critical costimulatory molecules expressed on activated T cells induced mutations in those CLL B cells that did not display intraclonal diversity in vivo (Fig. 4). These mutations occurred at similar frequencies to those identified in vivo. We cannot formally exclude that mutated VHDJH transcripts predated the in vitro induction and were, in some way, positively selected in our culture conditions. However, the complete absence of mutating Ig VHDJH transcripts among the large number of B cells analyzed before in vitro stimulation (Table 1) makes this possibility unlikely. Thus, in these cases the BCR was still effective in inducing somatic mutation in vitro implying that in vivo external triggering could result in the observed Ig V gene mutational heterogeneity and suggesting a potential role for ongoing receptor stimulation in the evolution of the CLL clone. The identification of mutations in only a small subset of the CLL B cells might be consistent with this view, since presumably not all members of the clone would have access to the relevant antigen(s) or other BCR cross-linkers. Furthermore, the occurrence of intraclonal variation in most, but not all, CLL B cells would be consistent with differences in BCR-mediated signaling among CLL cases, as has been suggested (13, 55–59). Alternatively, these point-mutations could occur spontaneously or result from ongoing stimulation via other receptors in leukemic subclones made competent to receive them by genetic alterations. The apparent lack of selection for R mutations in the CDR and against R mutations in the framework regions among the partially shared and unique new mutations may support this interpretation.
In addition, the lack of targeting of R mutations to the CDR and the apparent different nature of the originally shared (A preference as in normal B cells) versus unique point-mutations (non-A preference) favor the idea that the unique point-mutations detected in the leukemic cells might occur via mechanisms different from the canonical Ig V gene hypermutation machinery. However, other features (Tables I and II) are compatible with a canonical process of somatic hypermutation (individual point-mutations resulting in more transitions than would be predicted for a stochastic event; reference 38). Further studies will be necessary to identify more precisely the mechanisms responsible for this intraclonal V gene diversity. In this regard, we have begun to study the relationship between the intraclonal VHDJH gene diversification and the downregulation of DNA pol η/pol ζ expression ratio. BCR engagement and subsequent Ig V(D)J gene mutation can be associated with downregulation of the translesion DNA pol η, and the maintenance or upregulation of the translesion DNA pol ζ (33), although studies in mice differ in this regard (60). In preliminary studies, we have analyzed the expression of DNA pol η and pol ζ in B cells of six CLL cases with and four without intraclonal diversification. DNA pol η was expressed at a normal level in all four cases without in vivo intraclonal diversification, but was significantly downregulated in all but one of the cases with intraclonal diversification (unpublished data). DNA pol ζ was expressed at comparable levels in all cases, yielding normal and low pol η/pol ζ expression ratios in the nondiversified and diversified CLL cases, respectively. Furthermore, preliminary studies suggest that activation-induced cytidine deaminase, that appears to play an essential role in somatic hypermutation (61, 62), can be detected by RT-PCR in some CLL cases, although a correlation between enzyme mRNA expression and intraclonal diversification is not clear at this point (unpublished data).
The CLL cases studied differed in the extent of intraclonal diversification identified. In most instances, the Ig V gene mutations detected were unique to individual clonal members within a specific CLL patient. However, in two cases (nos. 105 and 261) mutations were shared among clonal members (Table I), making it possible to construct genealogical trees outlining the clonal evolution of the CLL B cells (Fig. 2). The reason(s) for the occurrence of such sublineages among certain, but not all, patients is not clear. Several possibilities should be considered. First, if the mutational machinery is not active at all points throughout the “life” of the leukemic clone, then those cases in which the machinery was active earlier and for a longer time interval would be more likely to have shared mutations than those cases in which the machinery was active later and for a shorter time interval. In addition, differences in the inherent proliferative rates of CLL cases or in individual members of the clone could affect the number and frequency of point mutations, and therefore the sharing of point-mutations. Alternatively, additional genetic damage, induced by this or other mutational processes, could have affected the potential for somatic hypermutation, causing its acceleration or termination in individual subclones. This latter issue could be especially important not only for the diversification of the Ig V gene repertoire, but also for the potential level of “malignancy” of an individual subclone since in normal B cells and in other B cell lymphoproliferative disorders a somatic hypermutation process can target non-Ig genes (32, 63–67). Finally, it is conceivable that the two patients with the extensive mutational lineages (CLL nos. 261 and 105) differ from the others that exhibit unique mutations but do not display this feature. For instance, the somatic mutation processes could have occurred by different mechanisms and/or at different anatomic sites in these two patients than those cases with solely unique point mutations or even possibly with the original shared mutations. As mentioned earlier, recent data suggest that human B cells can accumulate Ig V gene mutations outside of classical germinal center structures and adequate T cell help (44, 45, 68).
Consistent with its heterogeneity in terms of incidence and magnitude, the occurrence of ongoing somatic hypermutation in CLL B cells could also depend on the microenvironment. This dependency could lead either to an induction or abolishment of the mutational process during the course of the disease. It is becoming increasingly clear that the natural history and behavior of CLL B cells reflect not only intrinsic defects of the leukemic cells, but also extrinsic factors. For instance, bidirectional lymphocyte–nonlymphocyte cell interactions may lead to the inhibition of apoptosis in neoplastic B cells (69–72). Similarly, the presence or absence of proliferation centers that resemble germinal centers and alterations in the B/T cell network could affect the ability of CLL B cells to undergo somatic hypermutation (73, 74). Our finding that CLL B cells from cases 136 and 216, in which no intraclonal diversification was found, effectively mutated the VHDJH genes in vitro upon application of appropriate stimuli supports the notion that external factors can overcome the putative “differentiation block” of these leukemic cells. This is consistent with previous studies indicating that activated normal T cells or polyclonal B cell activators can induce terminal differentiation of CLL B cells (75, 76).
CLL cases can be segregated into two subgroups based on the mutational load of the expressed VH sequence (13), as defined by the number of shared mutations, and these subgroups differ very significantly in clinical course and outcome (26, 77). It is surprising that the occurrence and frequency of intraclonal Ig V gene variants is independent of the original mutation status of the individual CLL cases. This suggests several points about the relatedness of these two subgroups of CLL cases. First, these data indicate that all CLL cells, regardless of their initial V gene mutation status, retain the capacity to develop V gene mutations. If these mutations occur via the normal Ig V gene hypermutation process, then it is unlikely that the differences in V gene mutations between the two subgroups is inherent and a consequence of the leukemic process. Indeed, this might support the notion that all CLL cases derived from antigen-experienced (78) or memory (79) B cells. Second, these data may provide some insight into whether the differences in the load of shared mutations reflect distinct maturation stages of mature B cells at which these CLL cells arose, e.g., pregerminal center B cells versus post-germinal center B cells (10, 13, 14, 26, 77), or activation pathways that the precursor B cells followed, e.g., T cell–dependent versus T cell–independent triggers (13, 26). Finally, the occurrence of significant intraclonal diversification in a subset of CLL patients (nos. 261 and 105; Fig. 2) may occasionally impact on the utility of IgV gene mutation status as a prognostic marker in CLL. Recent studies indicate that the presence of significant numbers of V gene mutations (≥2% VH difference from the most similar germline gene) correlates with a relatively benign clinical course, whereas the absence of such mutations is associated with a more accelerated and unfavorable clinical course and outcome (26, 77). It remains to be seen whether the extent of V gene changes that converted the VH gene mutation status of CLL 105 from the “unmutated” to the “mutated” subgroup, will impact on the predictive value of this marker in such cases, although this seems unlikely since this type of “conversion” appears to be relatively infrequent.
Acknowledgments
We thank Dr. Kozaburo Yamaji and Dr. Andras Schaffer for their most helpful discussion. We thank Shefali Shah and Patricia Dramitinos for their help with tissue culture and SSCP analysis.
National Institutes of Health (NIH) grants AG13910, AR40908, AI45011, and AI07621 to P. Casali, and NIH grants CA81554, CA87956, and AI10811 to N. Chiorazzi supported this work. Support was also provided by the Joseph Eletto Leukemia Research Fund, the Jean Walton Fund for Lymphoma & Myeloma Research, the Jerry and Cecile Shore Fund for Immunologic Research, and the Richard and Nancy Leeds Fund.
P. Casali and N. Chiorazzi contributed equally to this work.
Footnotes
Abbreviations used in this paper: CDR, complementarity determining region; CLL, chronic lymphocytic leukemia; SSCP, single-strand conformation polymorphism.
References
- 1.Rai, K., and D. Patel. 1995. Chronic lymphocytic leukemia. Hematology: Basic Principles and Practice. 2nd edition. R. Hoffman, E. Benz, S. Shattil, B. Furie, H. Cohen, and L. Silberstein, editors. Churchill Livingstone, New York. pp. 1308–1321.
- 2.Kipps, T.J. 1989. The CD5 B cell. Adv. Immunol. 47:117–185. [DOI] [PubMed] [Google Scholar]
- 3.Meeker, T.C., J.C. Grimaldi, R. O'Rourke, J. Loeb, G. Juliusson, and S. Einhorn. 1988. Lack of detectable somatic hypermutation in the V region of the Ig H chain gene of a human chronic B lymphocytic leukemia. J. Immunol. 141:3994–3998. [PubMed] [Google Scholar]
- 4.Pratt, L.F., L. Rassenti, J. Larrick, B. Robbins, P.M. Banks, and T.J. Kipps. 1989. Ig V region gene expression in small lymphocytic lymphoma with little or no somatic hypermutation. J. Immunol. 143:699–705. [PubMed] [Google Scholar]
- 5.Kuppers, R., A. Gause, and K. Rajewsky. 1991. B cells of chronic lymphatic leukemia express V genes in unmutated form. Leuk. Res. 15:487–496. [DOI] [PubMed] [Google Scholar]
- 6.Friedman, D.F., J.S. Moore, J. Erikson, J. Manz, J. Goldman, P.C. Nowell, and L.E. Silberstein. 1992. Variable region gene analysis of an isotype-switched (IgA) variant of chronic lymphocytic leukemia. Blood. 80:2287–2297. [PubMed] [Google Scholar]
- 7.Rassenti, L.Z., and T.J. Kipps. 1993. Lack of extensive mutations in the VH5 genes used in common B cell chronic lymphocytic leukemia. J. Exp. Med. 177:1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, J., C. Humphries, A. Richardson, and P.W. Tucker. 1992. Extensive and selective mutation of a rearranged VH5 gene in human B cell chronic lymphocytic leukemia. J. Exp. Med. 176:1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto, S., M. Wakai, J. Silver, and N. Chiorazzi. 1992. Biased usage of variable and constant-region Ig genes by IgG+, CD5+ human leukemic B cells. Ann. NY Acad. Sci. 651:477–479. [DOI] [PubMed] [Google Scholar]
- 10.Schroeder, H.W., Jr., and G. Dighiero. 1994. The pathogenesis of chronic lymphocytic leukemia: analysis of the antibody repertoire. Immunol. Today. 15:288–294. [DOI] [PubMed] [Google Scholar]
- 11.Hashimoto, S., M. Dono, M. Wakai, S.L. Allen, S.M. Lichtman, P. Schulman, V.P. Vinciguerra, M. Ferrarini, J. Silver, and N. Chiorazzi. 1995. Somatic diversification and selection of immunoglobulin heavy and light chain variable region genes in IgG+ CD5+ chronic lymphocytic leukemia B cells. J. Exp. Med. 181:1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oscier, D.G., A. Thompsett, D. Zhu, and F.K. Stevenson. 1997. Differential rates of somatic hypermutation in V(H) genes among subsets of chronic lymphocytic leukemia defined by chromosomal abnormalities. Blood. 89:4153–4160. [PubMed] [Google Scholar]
- 13.Fais, F., F. Ghiotto, S. Hashimoto, B. Sellars, A. Valetto, S.L. Allen, P. Schulman, V.P. Vinciguerra, K. Rai, L.Z. Rassenti, et al. 1998. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J. Clin. Invest. 102:1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silberstein, L.E., S. Litwin, and C.E. Carmack. 1989. Relationship of variable region genes expressed by a human B cell lymphoma secreting pathologic anti-Pr2 erythrocyte autoantibodies. J. Exp. Med. 169:1631–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dono, M., S. Hashimoto, F. Fais, V. Trejo, S.L. Allen, S.M. Lichtman, P. Schulman, V.P. Vinciguerra, B. Sellars, P.K. Gregersen, et al. 1996. Evidence for progenitors of chronic lymphocytic leukemia B cells that undergo intraclonal differentiation and diversification. Blood. 87:1586–1594. [PubMed] [Google Scholar]
- 16.Schettino, E.W., A. Cerutti, N. Chiorazzi, and P. Casali. 1998. Lack of intraclonal diversification in Ig heavy and light chain V region genes expressed by CD5+IgM+ chronic lymphocytic leukemia B cells: a multiple time point analysis. J. Immunol. 160:820–830. [PMC free article] [PubMed] [Google Scholar]
- 17.Garand, R., S.S. Sahota, H. Avet-Loiseau, P. Talmant, N. Robillard, A. Moreau, F. Gaillard, F.K. Stevenson, and R. Bataille. 2000. IgG-secreting lymphoplasmacytoid leukaemia: a B-cell disorder with extensively mutated VH genes undergoing Ig isotype-switching frequently associated with trisomy 12. Br. J. Haematol. 109:71–80. [DOI] [PubMed] [Google Scholar]
- 18.Fischer, M., U. Klein, and R. Kuppers. 1997. Molecular single-cell analysis reveals that CD5-positive peripheral blood B cells in healthy humans are characterized by rearranged Vκ genes lacking somatic mutation. J. Clin. Invest. 100:1667–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herzenberg, L.A., A.M. Stall, P.A. Lalor, C. Sidman, W.A. Moore, D.R. Parks, and L.A. Herzenberg. 1986. The Ly-1 B cell lineage. Immunol. Rev. 93:81–102. [DOI] [PubMed] [Google Scholar]
- 20.Hardy, R.R., and K. Hayakawa. 1992. Developmental origins, specificities and immunoglobulin gene biases of murine Ly-1 B cells. Int. Rev. Immunol. 8:189–207. [DOI] [PubMed] [Google Scholar]
- 21.Herzenberg, L.A., and A.B. Kantor. 1993. B-cell lineages exist in the mouse. Immunol. Today. 14:79–83. [DOI] [PubMed] [Google Scholar]
- 22.Harindranath, N., I.S. Goldfarb, H. Ikematsu, S.E. Burastero, R.L. Wilder, A.L. Notkins, and P. Casali. 1991. Complete sequence of the genes encoding the VH and VL regions of low- and high-affinity monoclonal IgM and IgA1 rheumatoid factors produced by CD5+ B cells from a rheumatoid arthritis patient. Int. Immunol. 3:865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mantovani, L., R.L. Wilder, and P. Casali. 1993. Human rheumatoid B-1a (CD5+ B) cells make somatically hypermutated high affinity IgM rheumatoid factors. J. Immunol. 151:473–488. [PMC free article] [PubMed] [Google Scholar]
- 24.Kasaian, M.T., and P. Casali. 1995. B-1 cellular origin and VH segment structure of IgG, IgA, and IgM anti-DNA autoantibodies in patients with systemic lupus erythematosus. Ann. NY Acad. Sci. 764:410–423. [DOI] [PubMed] [Google Scholar]
- 25.Schettino, E.W., S.K. Chai, M.T. Kasaian, H.W. Schroeder, Jr., and P. Casali. 1997. VHDJH gene sequences and antigen reactivity of monoclonal antibodies produced by human B-1 cells: evidence for somatic selection. J. Immunol. 158:2477–2489. [PMC free article] [PubMed] [Google Scholar]
- 26.Damle, R.N., T. Wasil, F. Fais, F. Ghiotto, A. Valetto, S.L. Allen, A. Buchbinder, D. Budman, K. Dittmar, J. Kolitz, et al. 1999. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 94:1840–1847. [PubMed] [Google Scholar]
- 27.Malisan, F., A.C. Fluckiger, S. Ho, C. Guret, J. Banchereau, and H. Martinez-Valdez. 1996. B-chronic lymphocytic leukemias can undergo isotype switching in vivo and can be induced to differentiate and switch in vitro. Blood. 87:717–724. [PubMed] [Google Scholar]
- 28.Efremov, D.G., M. Ivanovski, F.D. Batista, G. Pozzato, and O.R. Burrone. 1996. IgM-producing chronic lymphocytic leukemia cells undergo immunoglobulin isotype-switching without acquiring somatic mutations. J. Clin. Invest. 98:290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fais, F., B. Sellars, F. Ghiotto, X.J. Yan, M. Dono, S.L. Allen, D. Budman, K. Dittmar, J. Kolitz, S.M. Lichtman, et al. 1996. Examples of in vivo isotype class switching in IgM+ chronic lymphocytic leukemia B cells. J. Clin. Invest. 98:1659–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denepoux, S., D. Razanajaona, D. Blanchard, G. Meffre, J.D. Capra, J. Banchereau, and S. Lebecque. 1997. Induction of somatic mutation in a human B cell line in vitro. Immunity. 6:35–46. [DOI] [PubMed] [Google Scholar]
- 31.Zan, H., A. Cerutti, P. Dramitinos, A. Schaffer, Z. Li, and P. Casali. 1999. Induction of Ig somatic hypermutation and class switching in a human monoclonal IgM+ IgD+ B cell line in vitro: definition of the requirements and modalities of hypermutation. J. Immunol. 162:3437–3447. [PMC free article] [PubMed] [Google Scholar]
- 32.Zan, H., Z. Li, K. Yamaji, P. Dramitinos, A. Cerutti, and P. Casali. 2000. B cell receptor engagement and T cell contact induce Bcl-6 somatic hypermutation in human B cells: identity with Ig hypermutation. J. Immunol. 165:830–839. [DOI] [PubMed] [Google Scholar]
- 33.Zan, H., A. Komori, Z. Li, A. Cerutti, A. Schaffer, M.F. Flajnik, M. Diaz, and P. Casali. 2001. The translesion DNA polymerase ζ plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 14:643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomlinson, I., S. Williams, S. Corbett, J. Cox, and G. Winter. 1996. V BASE sequence directory. MRC Centre for Protein Engineering, Cambridge, UK. http://www.mrc-cpe.cam.ac.uk-Vbase.
- 35.Chang, B., and P. Casali. 1994. The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol. Today. 1:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shlomchik, M.J., A. Marshak-Rothstein, C.B. Wolfowicz, T.L. Rothstein, and M.G. Weigert. 1987. The role of clonal selection and somatic mutation in autoimmunity. Nature. 328:805–811. [DOI] [PubMed] [Google Scholar]
- 37.Matolcsy, A., E.J. Schattner, D.M. Knowles, and P. Casali. 1999. Clonal evolution of B cells in transformation from low- to high-grade lymphoma. Eur. J. Immunol. 29:1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yelamos, J., N. Klix, B. Goyenechea, F. Lozano, Y.L. Chui, A. Gonzalez Fernandez, R. Pannell, M.S. Neuberger, and C. Milstein. 1995. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature. 376:225–229. [DOI] [PubMed] [Google Scholar]
- 39.MacLennan, I.C. 1994. Germinal centers. Annu. Rev. Immunol. 12:117–139. [DOI] [PubMed] [Google Scholar]
- 40.Jacob, J., R. Kassir, and G. Kelsoe. 1991. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl) acetyl. I. The architecture and dynamics of responding cell populations. J. Exp. Med. 173:1165–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berek, C., A. Berger, and M. Apel. 1991. Maturation of the immune response in germinal centers. Cell. 67:1121–1129. [DOI] [PubMed] [Google Scholar]
- 42.Jacob, J., G. Kelsoe, K. Rajewsky, and U. Weiss. 1991. Intraclonal generation of antibody mutants in germinal centres. Nature. 354:389-392. [DOI] [PubMed] [Google Scholar]
- 43.Kuppers, R., M. Zhao, M.L. Hansmann, and K. Rajewsky. 1993. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO J. 12:4955–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weller, S., A. Faili, C. Garcia, M.C. Braun, F.F. Le Deist, G.G. de Saint Basile, O. Hermine, A. Fischer, C.A. Reynaud, and J.C. Weill. 2001. CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc. Natl. Acad. Sci. USA. 98:1166–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Vinuesa, C.G., M.C. Cook, J. Ball, M. Drew, Y. Sunners, M. Cascalho, M. Wabl, G.G. Klaus, and I.C. MacLennan. 2000. Germinal centers without T cells. J. Exp. Med. 191:485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roudier, J., G.J. Silverman, P.P. Chen, D.A. Carson, and T.J. Kipps. 1990. Intraclonal diversity in the VH genes expressed by CD5-chronic lymphocytic leukemia-producing pathologic IgM rheumatoid factor. J. Immunol. 144:1526–1530. [PubMed] [Google Scholar]
- 47.Korganow, A.S., T. Martin, J.C. Weber, B. Lioure, P. Lutz, A.M. Knapp, and J.L. Pasquali. 1994. Molecular analysis of rearranged VH genes during B cell chronic lymphocytic leukemia: intraclonal stability is frequent but not constant. Leuk. Lymphoma 14:55–69. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, W., P.D. Bardwell, C.J. Woo, V. Poltoratsky, M.D. Scharff, and A. Martin. 2001. Clonal instability of V region hypermutation in the Ramos Burkitt's lymphoma cell line. Int. Immunol. 13:1175–1184. [DOI] [PubMed] [Google Scholar]
- 49.Martin, A., P.D. Bardwell, C.J. Woo, M. Fan, M.J. Shulman, and M.D. Scharff. 2002. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature. 415:802–806. [DOI] [PubMed] [Google Scholar]
- 50.Zelenetz, A.D., T.T. Chen, and R. Levy. 1992. Clonal expansion in follicular lymphoma occurs subsequent to antigenic selection. J. Exp. Med. 176:1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chapman, C.J., C.I. Mockridge, M. Rowe, A.B. Rickinson, and F.K. Stevenson. 1995. Analysis of VH genes used by neoplastic B cells in endemic Burkitt's lymphoma shows somatic hypermutation and intraclonal heterogeneity. Blood. 85:2176–2181. [PubMed] [Google Scholar]
- 52.Chapman, C.J., J.X. Zhou, C. Gregory, A.B. Rickinson, and F.K. Stevenson. 1996. VH and VL gene analysis in sporadic Burkitt's lymphoma shows somatic hypermutation, intraclonal heterogeneity, and a role for antigen selection. Blood. 88:3562–3568. [PubMed] [Google Scholar]
- 53.Aarts, W.M., R. Willemze, R.J. Bende, C.J. Meijer, S.T. Pals, and C.J. van Noesel. 1998. VH gene analysis of primary cutaneous B-cell lymphomas: evidence for ongoing somatic hypermutation and isotype switching. Blood. 92:3857–3864. [PubMed] [Google Scholar]
- 54.Lossos, I.S., A.A. Alizadeh, M.B. Eisen, W.C. Chan, P.O. Brown, D. Botstein, L.M. Staudt, and R. Levy. 2000. Ongoing immunoglobulin somatic mutation in germinal center B cell-like but not in activated B cell-like diffuse large cell lymphomas. Proc. Natl. Acad. Sci. USA. 97:10209–10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zupo, S., L. Isnardi, M. Megna, R. Massara, F. Malavasi, M. Dono, E. Cosulich, and M. Ferrarini. 1996. CD38 expression distinguishes two groups of B-cell chronic lymphocytic leukemias with different responses to anti-IgM antibodies and propensity to apoptosis. Blood. 88:1365–1374. [PubMed] [Google Scholar]
- 56.Lankester, A.C., G.M. van Schijndel, C.E. van der Schoot, M.H. van Oers, C.J. van Noesel, and R.A. van Lier. 1995. Antigen receptor nonresponsiveness in chronic lymphocytic leukemia B cells. Blood. 86:1090–1071. [PubMed] [Google Scholar]
- 57.Alfarano, A., S. Indraccolo, P. Circosta, S. Minuzzo, A. Vallario, R. Zamarchi, A. Fregonese, F. Calderazzo, A. Faldella, M. Aragno, et al. 1999. An alternatively spliced form of CD79b gene may account for altered B-cell receptor expression in B-chronic lymphocytic leukemia. Blood. 93:2327–2335. [PubMed] [Google Scholar]
- 58.Gordon, M.S., R.M. Kato, F. Lansigan, A.A. Thompson, R. Wall, and D.J. Rawlings. 2000. Aberrant B cell receptor signaling from B29 (Igβ, CD79b) gene mutations of chronic lymphocytic leukemia B cells. Proc. Natl. Acad. Sci. USA. 97:5504–5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zupo, S., R. Massara, M. Dono, E. Rossi, F. Malavasi, M.E. Cosulich, and M. Ferrarini. 2000. Apoptosis or plasma cell differentiation of CD38-positive B-chronic lymphocytic leukemia cells induced by cross-linking of surface IgM or IgD. Blood. 95:1199–1206. [PubMed] [Google Scholar]
- 60.Zeng, X., D.B. Winter, C. Kasmer, K.H. Kraemer, A.R. Lehmann, and P.J. Gearhart. 2001. DNA polymerase ζ is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2:537–541. [DOI] [PubMed] [Google Scholar]
- 61.Muramatsu, M., V.S. Sankaranand, S. Anant, M. Sugai, K. Kinoshita, N.O. Davidson, and T. Honjo. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470–18476. [DOI] [PubMed] [Google Scholar]
- 62.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 63.Pasqualucci, L., A. Migliazza, N. Fracchiolla, C. William, A. Neri, L. Baldini, R.S.K. Chaganti, U. Klein, R. Kuppers, K. Rajewsky, and R. Dalla-Favera. 1998. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc. Natl. Acad. Sci. USA. 95:11816–11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peng, H.Z., M.Q. Du, A. Koulis, A. Aiello, A. Dogan, L.X. Pan, and P.G. Isaacson. 1999. Nonimmunoglobulin gene hypermutation in germinal center B cells. Blood. 93:2167–2172. [PubMed] [Google Scholar]
- 65.Sahota, S.S., Z. Davis, T.J. Hamblin, and F.K. Stevenson. 2000. Somatic mutation of bcl-6 genes can occur in the absence of V(H) mutations in chronic lymphocytic leukemia. Blood. 95:3534–3540. [PubMed] [Google Scholar]
- 66.Capello, D., F. Fais, D. Vivenza, G. Migliaretti, N. Chiorazzi, G. Gaidano, and M. Ferrarini. 2000. Identification of three subgroups of B cell chronic lymphocytic leukemia based upon mutations of BCL-6 and IgV genes. Leukemia. 14:811–815. [DOI] [PubMed] [Google Scholar]
- 67.Pasqualucci, L., A. Neri, L. Baldini, R. Dalla-Favera, and A. Migliazza. 2000. BCL-6 mutations are associated with immunoglobulin variable heavy chain mutations in B-cell chronic lymphocytic leukemia. Cancer Res. 60:5644–5648. [PubMed] [Google Scholar]
- 68.Toellner, K.M., W.E. Jenkinson, D.R. Taylor, M. Khan, D.M. Sze, D.M. Sansom, C.G. Vinuesa, and I.C. MacLennan. 2002. Low-level hypermutation in T cell-independent germinal centers compared with high mutation rates associated with T cell-dependent germinal centers. J. Exp. Med. 195:383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chilosi, M., G. Pizzolo, F. Caligaris-Cappio, A. Ambrosetti, F. Vinante, L. Morittu, F. Bonetti, L. Fiore-Donati, and G. Janossy. 1985. Immunohistochemical demonstration of follicular dendritic cells in bone marrow involvement of B-cell chronic lymphocytic leukemia. Cancer. 56:328–332. [DOI] [PubMed] [Google Scholar]
- 70.Panayiotidis, P., D. Jones, K. Ganeshaguru, L. Foroni, and A.V. Hoffbrand. 1996. Human bone marrow stromal cells prevent apoptosis and support the survival of chronic lymphocytic leukaemia cells in vitro. Br. J. Haematol. 92:97–103. [DOI] [PubMed] [Google Scholar]
- 71.Lagneaux, L., A. Delforge, D. Bron, C. De Bruyn, and P. Stryckmans. 1998. Chronic lymphocytic leukemic B cells but not normal B cells are rescued from apoptosis by contact with normal bone marrow stromal cells. Blood. 91:2387–2396. [PubMed] [Google Scholar]
- 72.Lagneaux, L., A. Delforge, C. De Bruyn, M. Bernier, and D. Bron. 1999. Adhesion to bone marrow stroma inhibits apoptosis of chronic lymphocytic leukemia cells. Leuk. Lymphoma. 35:445–453. [DOI] [PubMed] [Google Scholar]
- 73.Pizzolo, G., M. Chilosi, A. Ambrosetti, G. Semenzato, L. Fiore-Donati, and G. Perona. 1983. Immunohistologic study of bone marrow involvement in B-chronic lymphocytic leukemia. Blood. 62:1289–1296. [PubMed] [Google Scholar]
- 74.Cerutti, A., E.C. Kim, S. Shah, E.J. Schattner, H. Zan, A. Schaffer, and P. Casali. 2001. Dysregulation of CD30+ T cells by leukemia impairs isotype switching in normal B cells. Nat. Immunol. 2:150–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fu, S.M., N. Chiorazzi, H.G. Kunkel, J.P. Halper, and S.R. Harris. 1978. Induction of in vitro differentiation and immunoglobulin synthesis of human leukemic B lymphocytes. J. Exp. Med. 148:1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chiorazzi, N., S. Fu, G. Montazeri, H. Kunkel, K. Rai, and T. Gee. 1979. T cell helper defect in patients with chronic lymphocytic leukemia. J. Immunol. 122:1087–1090. [PubMed] [Google Scholar]
- 77.Hamblin, T.J., Z. Davis, A. Gardiner, D.G. Oscier, and F.K. Stevenson. 1999. Unmutated Ig VH genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 94:1848–1854. [PubMed] [Google Scholar]
- 78.Damle, R.N., F. Ghiotto, A. Valetto, E. Albesiano, F. Fais, X.J. Yan, C.P. Sison, S.L. Allen, J. Kolitz, P. Schulman, et al. 2002. B-cell chronic lymphocytic leukemia cells express a surface membrane phenotype of activated, antigen-experienced B lymphocytes. Blood. 99:4087–4093. [DOI] [PubMed] [Google Scholar]
- 79.Klein, U., Y. Tu, G.A. Stolovitzky, M. Mattioli, G. Cattoretti, H. Husson, A. Freedman, G. Inghirami, L. Cro, L. Baldini, et al. 2001. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J. Exp. Med. 194:1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]