Abstract
We characterized the sae operon, a global regulator for virulence gene expression in Staphylococcus aureus. A Tn917 sae mutant was obtained by screening a Tn917 library of the agr mutant ISP479Mu for clones with altered hemolytic activity. Sequence analysis of the sae operon revealed two additional open reading frames (ORFs) (ORF3 and ORF4) upstream of the two-component regulatory genes saeR and saeS. Four overlapping sae-specific transcripts (T1 to T4) were detected by Northern blot analysis, and the transcriptional initiation points were mapped by primer extension analysis. The T1, T2, and T3 mRNAs are probably terminated at the same stem-loop sequence downstream of saeS. The T1 message (3.1 kb) initiates upstream of ORF4, T2 (2.4 kb) initiates upstream of ORF3, and T3 (2.0 kb) initiates in front of saeR. T4 (0.7 kb) represents a monocistronic mRNA encompassing ORF4 only. sae-specific transcripts were detectable in all of the 40 different clinical S. aureus isolates investigated. Transcript levels were at maximum during the post-exponential growth phase. The sae mutant showed a significantly reduced rate of invasion of human endothelial cells, consistent with diminished transcription and expression of fnbA. The expression of type 5 capsular polysaccharide is activated in the sae mutant of strain Newman, as shown by immunofluorescence and promoter-reporter fusion experiments. In summary, the sae operon constitutes a four-component regulator system which acts on virulence gene expression in S. aureus.
The human pathogen Staphylococcus aureus is the causative agent of a wide spectrum of diseases. This organism's capacity to adapt to different environments in vitro and in vivo is due to a global regulatory network comprising several loci such as agr, sar, sigB, rot, arlRS, svrA, and saeRS (4, 27). Each of these regulators is involved in the control of the expression of virulence factors such as hemolysins, protein A, fibronectin-binding proteins (FnBPA and FnBPB, encoded by fnbA and fnbB, respectively), or capsular polysaccharide (CP, encoded by the cap operon). agr, sar, sigB, svrA, rot, and arlRS constitute a complex interactive regulatory network (3-5, 8, 9, 25). For instance, SarA has been shown to be necessary for full agr activation (5), and SigB (3) and ArlR (8) modulate the transcription of agr and sarA. Mutation of the proposed regulatory locus sae did not affect the transcription of sarA and agr, indicating an independent regulatory circuit (12). A sae transposon mutant was shown to express reduced levels of alpha-hemolysin (Hla, encoded by hla), beta-hemolysin, DNase, coagulase (Coa, encoded by coa), and protein A compared with those expressed by the parental strain, whereas the expression of delta-hemolysin, proteases, and lipase was not altered (15). sae affects target genes on the transcriptional level (12).
The contribution of sae to virulence has been shown after intraperitoneal injection of bacteria into mice (14). In addition, it has been shown that sae, but not agr or sarA, is essential for the transcription of hla during device-related infection in guinea pigs (17). In a different tissue-cage model, it was also shown by microarray analysis that in contrast to that of agr, transcription of sae was not repressed in vivo (42). Sequencing of the transposon insertion site of the sae mutant originally described revealed two open reading frames (ORFs), saeR and saeS, with strong sequence homology to response regulators and histidine kinases of bacterial two-component regulators (11) (GenBank accession number AF129010). According to the general mechanism of these systems (38), SaeS probably functions as a membrane-spanning sensor histidine kinase which, upon sensing the appropriate signal, autophosphorylates and in turn activates the cognate cytosolic response regulator SaeR. The activated response regulator may function as a transcriptional regulator via a specific DNA-binding domain, recognizing motifs near the promoter sequences of target genes.
Since sae seems to be an important modulator of virulence, we decided to further characterize this locus at the molecular level. We show that sae activates the transcription of fnbA, coa, and hla. In contrast, expression of CP5 is inhibited by sae. Transcriptional analysis revealed three overlapping transcripts and two additional putative protein-encoding regions (ORF3 and ORF4) which are cotranscribed with saeS and saeR. sae was found to be differentially expressed depending on the growth phase and the genetic background of the strain studied.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. In vitro growth was performed in CYPG (26) supplemented with the appropriate antibiotics for strains carrying resistance genes (erythromycin at 10 μg/ml; tetracycline at 5 μg/ml). For RNA isolation, cells of an overnight culture were diluted to an initial optical density at 600 nm (OD600) of 0.05 and grown with shaking at 37°C to the mid-exponential (OD600 = 0.8) or post-exponential (OD600 = 8) phase or for 16 h to stationary phase. Clinical isolates were obtained from nose swabs of healthy individuals, from the sputa of patients with cystic fibrosis, and from wound swabs. All isolates were typed by pulsed-field gel electrophoresis (PFGE) as described elsewhere (34).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| Newman | Wild type | 7 |
| ALC355 | Newman Δagr::tetM | 40 |
| ISP479C | Derivative of 8325-4 | 36 |
| ISP479Mu | Spontaneous agrC mutant of ISP479C | Unpublished data |
| AS1 | ISP479Mu sae::Tn917 | This work |
| AS2 | ISP479C sae::Tn917 | This work |
| AS3 | Newman sae::Tn917 | 17 |
| AS4 | Newman Δagr::tetM sae::Tn917 | This work |
| RN6390 | Derivative of NCTC8325 | 31 |
| AS5 | RN6390 sae::Tn917 | This work |
| DU5886 | Newman fnbA::TcrfnbB::Emr | T. J. Foster Dublin |
| Plasmids | ||
| pLT4ts | Tn917 delivery vector | 43 |
| pSN8388 | cap5 promoter fused to the reporter gene xylE in pCL4 | 18 |
| pCWSAE7 | pGEMTeasy with 3.5-kb fragment encompassing ORF4, ORF3, saeS, and saeR of strain ISP479C | This work |
| pCWSAE8 | pCRII with 3.5-kb fragment encompassing ORF4, ORF3, saeS, and saeR of strain Newman | This work |
Tn917 insertional mutagenesis and transduction.
The identification of a sae::Tn917 mutant was described previously (17). Briefly, a Tn917 library of the spontaneous agrC mutant strain ISP479Mu (ISP479Mu pTV1ts) was screened for differences in the hemolytic pattern observed after growth in air alone versus air supplemented with 5% CO2. One colony which remained nonhemolytic under elevated CO2 was chosen for further investigation. In order to determine the Tn917 insertion site, we digested chromosomal DNA with EcoRI, separated it by electrophoresis, blotted it onto a nylon membrane, and hybridized it with a Tn917-specific probe. The reactive fragment was gel eluted, cloned into pUC18, and sequenced. The mutation was transduced into strains ISP479Mu, ISP479C, RN6390, and Newman and the agr mutant of strain Newman (ALC355 [40]) by using a φ11 lysate of the original transposon mutant.
RNA isolation and Northern blot hybridization.
RNA isolation and Northern blot analysis were performed as described previously (16). Briefly, approximately 109 S. aureus cells were lysed in 1 ml of Trizol reagent (Invitrogen Life Technologies, Karlsruhe, Germany) with 0.5 ml of zirconia-silica beads (diameter, 0.1 mm) in a high-speed homogenizer (Savant Instruments, Farmingdale, N.Y.). RNA was isolated as described in the instructions provided by the manufacturer of Trizol.
Several digoxigenin (DIG)-labeled probes for the detection of specific transcripts were generated by using the DIG-labeling PCR kit according to the manufacturer's instructions (Roche Biochemicals, Mannheim, Germany). Oligonucleotides are listed in Table 2.
TABLE 2.
Oligonucleotides used for PCR and primer extension analysis
| Description | Basisa | Nameb | Primer sequence |
|---|---|---|---|
| Probe 1 | AJ556795 | sae285U | CAAATTGAAGAAATGAGGAGTTA |
| sae564L | ACCTTTTGATGATTTGTAGTTAG | ||
| Probe 2 | AJ556795 | sae727U | TTCGGCGGCGCTAAATTAAA |
| sae981L | TGCTTAAGCTAAACAAACCTC | ||
| Probe 3 | AJ556795 | sae992U | TCAAACACTTCCTGTTCACA |
| sae1283L | TGACGTCGTATGTGCAACTA | ||
| Probe 4 | AJ556795 | sae1980U | TGGTCACGAAGTCCCTATGC |
| sae2458L | TGCTTGCGTAATTTCCGTTAG | ||
| Probe 5 | AJ556795 | sae3364U | AAACTTTATGGGTATCCTTCTCA |
| csbL | TTGGCTTGTTTATCTTTTCAATA | ||
| fnbA probe | J04151 | fnbAU | TGCAAATACGACAGATACTT |
| fnbAL | TTGGCCACCTTCATAACCTA | ||
| coa probe | X16457 | coaU | CGAGACCAAGATTCAACAAG |
| coaL | AAAGAAAACCACTCACATCA | ||
| agr probe | X52543 | RNAIIIU | TATATTTTAACGGCGGGTCTCA |
| RNAIIIL | TTAATTAAGGAAGGAGTGATTT | ||
| hla probe | X01645 | hlaU | AGAAAATGGCATGCACAAAAA |
| hlal | TGTAGCGAAGTCTGGTGAAAA | ||
| Tn917 probe | M11180 | Tn917U | TAAGAGTGTGTTGATAG |
| Tn917L | ACGATAAGTTGAATAGATAG | ||
| sae cloning | AJ556795 | sae1U | TTATTGTGGCAAAAGGTTTAT |
| sae3530L | ATTATTAGGCGGCATACAG | ||
| Primer extension 1 (P1) | AJ556795 | sae337, CY5 | ACCTAAAGCTAATGTTGTGATAACAGCACC |
| Primer extension 2 (P1, P2) | AJ556795 | sae1017, CY5 | TAAGATTAAGCAACATAATGCGATTTGTAG |
| Primer extension 3 (P2) | AJ556795 | sae1290, CY5 | CAATCTCTCCGAGTGGGACAACAATATC |
| Primer extension 5 (P2, P3) | AJ556795 | sae1772, CY5 | GTATCATGTTCTTGTGTTTTGGCAGTTA |
| Primer extension 6 (P3) | AJ556795 | sae1973, CY5 | TACGCATAGGGACTTCGTGACCATTTACAG |
GenBank accession number.
Numbers indicate nucleotide position in the corresponding reference sequence. U, upper primer; L, lower primer.
Cloning and sequencing.
A 3.5-kb fragment comprising the sae operon was amplified from strain ISP479C or strain Newman by using a high-fidelity polymerase (HF; Roche Diagnostics) and cloned into the PCR cloning vector pGEM-TEasy (Promega, Mannheim, Germany) or pCR2.1 (Invitrogen), respectively. The inserts were sequenced by ABI377 instrumentation using the DYEnamicET Terminator sequencing kit (Amersham Biosciences, Freiburg, Germany). Sequence analysis was performed with the HUSAR software package (Dkfz, Heidelberg, Germany).
Primer extension analysis.
In order to facilitate the identification of the appropriate promoters from which transcription is initiated, we mapped the 5′ ends of the different sae transcripts by primer extension as previously described (2) with some modifications. Briefly, we adapted the [γ-32P]ATP-based methodology to a fluorescence ALFexpressII sequencer (Amersham Biosciences), utilizing Cy5-fluorescein end-labeled oligonucleotides (Table 2) for reverse transcription and dye-primer sequencing. Sequencing reaction mixtures primed by oligonucleotides identical to those used for primer extension were applied on the gel in parallel lanes.
Promoter-fusion assay and measurement of promoter activity using xylE as a reporter gene.
The reporter plasmid pSN8388 (carrying the cap5 promoter fused to the promoterless reporter gene xylE in pCL4 [18]) was transduced from strain Newman into the isogenic sae mutant of strain Newman by using φ11. The presence of correct constructs was confirmed by restriction mapping of the isolated plasmid. After in vitro growth of the strains under different conditions, the bacterial cells were harvested and lysed, and XylE activity was determined as described previously (24). The promoter activity was expressed as XylE activity per milligram of protein.
Ligand affinity analysis for detection of FnBP.
Fibronectin-binding proteins were detected with biotinylated fibronectin as described previously (41). Briefly, cell wall extracts were prepared from an equivalent number of washed cells (1011 CFU/ml). After protoplast formation, cell wall proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Membranes were blocked in 5% dehydrated skim milk in phosphate-buffered saline and incubated first with biotinylated fibronectin and then with streptavidin-peroxidase complex (Pierce, Rockford, Ill.). Bound fibronectin was visualized with the ECL PlusTM Western blotting detection system according to the manufacturer's instructions (Amersham Biosciences). A high-molecular-weight prestained marker (Invitrogen) was used for determination of molecular size (in kilodaltons).
Coagulase assay.
Coagulase activity was measured by addition of 100 μl of serial twofold dilutions of culture supernatants to 200 μl of rabbit plasma. The titer was the reciprocal of the highest dilution of samples that caused clotting after a 4-h incubation at 37°C.
Endothelial cell binding and invasion assay.
Primary human umbilical vein endothelial cells (Labor Glatthaar, Reutlingen, Germany) were seeded in 0.2% gelatin-coated 24-well plates (BD Biosciences, Heidelberg, Germany) and grown in endothelial growth medium (Cell Systems, Katharinen, Germany) without antibiotics at 37°C in an atmosphere containing 5% CO2. Confluent monolayers of endothelial cells (∼105 cells per well) were washed three times with M199 (Sigma Chemical Co., Deisenhofen, Germany) and incubated for 20 min at 37°C with 3% bovine serum albumin (Sigma) in M199 to minimize background adherence. Endothelial cells were then incubated with bacteria from the mid-exponential-growth phase (5 × 106 CFU of S. aureus/well) at 37°C for 2 h. The supernatants containing the nonadherent bacteria were removed, and CFU were determined by plating aliquots on sheep blood agar plates. Cells were subsequently washed three times with M199 and treated with trypsin-EDTA and 0.25% Triton X-100 to lyse endothelial cells, after which the CFU of adherent and intracellular bacteria were counted. In order to determine the number of intracellular bacteria, extracellular bacteria were killed by adding lysostaphin (10 μg/ml) prior to cell lysis. Each assay was repeated at least once, and within each experiment all samples were processed at least in quadruplicate.
Detection of CP5.
CP5 was detected by indirect immunofluorescence as described previously (32). Briefly, protein A was blocked by incubation with human immunoglobulin G, and CP5 antigen was detected by using monoclonal antibodies raised against CP5 (20) and secondary Cy3- or fluorescein isothiocyanate-labeled anti-mouse antibodies.
Nucleotide sequence accession numbers.
The sequence data obtained in this study have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers AJ556794 and AJ556795.
RESULTS
Identification of a sae Tn917 mutant.
In order to identify regulatory loci which act independently of agr, we screened a Tn917 library of the spontaneous agrC mutant ISP479Mu (ISP479Mu pLT4ts) for clones with differences in the hemolytic patterns observed on sheep blood agar plates after incubation in air alone versus air supplemented with 5% CO2. The parental strain ISP479Mu was nonhemolytic under regular air conditions but developed visible hemolytic activity after incubation in elevated CO2. One colony which remained nonhemolytic under elevated CO2 was selected for further characterization. Sequencing of the transposon insertion site revealed that Tn917 was inserted into saeS, encoding a previously described histidine kinase (11). In order to rule out the possibility that additional mutations contributed to the observed alterations of the phenotype, the mutation was transduced back into ISP479Mu as well as into other agr-negative (ALC355) and agr-positive (ISP479C, RN6390, Newman) strains. As in the previous characterization of a sae mutant strain (15), all the transductants showed a severe reduction in hemolysis on sheep blood agar plates (for agr-positive strains) or were nonhemolytic (in the agr-deficient background). None of the transductants led to visible clotting of rabbit plasma. Northern blot analysis also confirmed (12) that neither hla nor coa transcripts are detectable in sae mutant strains (data not shown). A single chromosomal integration of Tn917 could be verified for all derivatives of strain 8325-4 (AS1, AS2, and AS5) as shown by Southern hybridization of the genomic DNA digested with SmaI and separated by PFGE using a Tn917-specific probe (data not shown). According to the physical SmaI map designation of strain 8325 (29), the labeled DNA-band corresponded to fragment D. Derivatives of strain Newman showed a single insertion in a DNA fragment of a similar size.
Sequence analysis of the sae operon.
A 3.5-kb DNA fragment encompassing sae was amplified from strains ISP479C and Newman by using primers based on the published genomic sequence of strain N315 (22). The fragments were cloned and sequenced. The sequence derived from strain ISP479C (GenBank accession number AJ556795) was found to be identical to a homologous region of strain COL (available from The Institute for Genomic Research [TIGR] at http://www.tigr.org) but showed minor differences from those of N315, Mu50 (22), and MW2 (1). Sequence comparison between the amplified fragments of strains Newman (GenBank accession number AJ556794) and ISP479 (GenBank accession no. AJ556795) revealed two single-base-pair exchanges, one within saeR and the other within saeS. However, there were substantial differences from the already published saeRS sequence derived from strain Newman (GenBank accession number AF129010) (11). Reamplification and resequencing of all the regions in question yielded results identical to our initial sequencing of the sae fragment.
Sequence analysis revealed two additional ORFs (designated ORF3 and ORF4) upstream of saeR (Fig. 1A). ORF3 encodes a putative 157-amino-acid (157-aa) polypeptide with a molecular mass of 17.68 kDa and an isoelectric point of 9.76. Due to the presence of an N-terminal signal peptide sequence and four membrane-spanning stretches, the putative gene product of ORF3 is probably membrane associated. The sequence revealed no obvious homology to known proteins. A homologous ORF upstream of saeR, encoding only 60 aa, is annotated in the genomes of S. aureus strains N315 (NTSA0730), Mu50 (SAV0693) (22), and MW2 (MW0668) (1); its product is identical to the C terminus of the ORF3 product. The different annotations are due not to sequence discrepancies but to different interpretations of the sequence data. In each of the published genomes, the same internal ATG motif within ORF3 was interpreted as the translational start site. In contrast, sequence annotation by TIGR (http://www.tigr.org) supports our hypothesis that the putative start site is located further upstream, which is based on the results obtained with a trained GLIMMER program. ORF4 codes for a second putative protein, of 146 aa, with a molecular mass of 16.05 kDa and an isoelectric point of 9.87. There are strain-specific variations in the N-terminal part of the ORF4-deduced polypeptide from strains COL, ISP479C, and Newman versus those from strains N315 and MW2. Sequence analysis of the translated ORF4 sequence revealed neither a signal peptide nor hydrophobic stretches. There is no obvious homology to known proteins in the database.
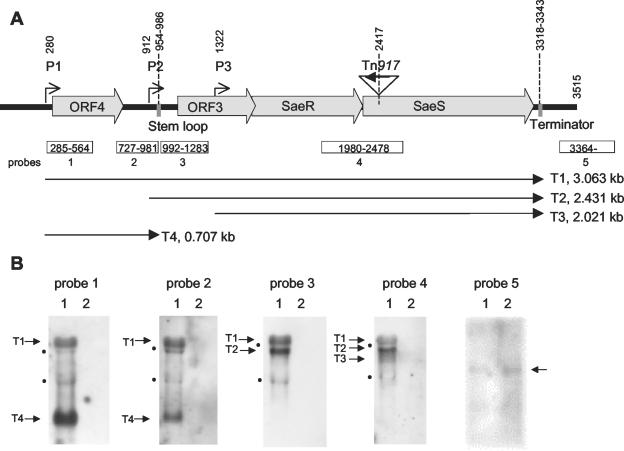
FIG. 1.
(A) Scheme of the sae operon based on the sequence of pCWSAE7 (GenBank accession number AJ556795). Transcriptional start points (P1, P2, and P3) were determined by primer extension assays. (B) Northern blot analysis of RNA of strain Newman (lanes 1) and its isogenic sae mutant (lanes 2) grown to the post-exponential-growth phase. The blots were hybridized using different DIG-labeled PCR fragments within the sae operon (probes 1 to 4 [Table 2]) and downstream of the sae operon (probe 5 [Table 2]). The sizes of the labeled transcripts were determined by comparison with a molecular weight marker run in parallel. The positions of the rRNA are marked by black dots.
Transcriptional analysis of the sae operon.
Northern blot analysis with different probes up- and downstream of saeRS showed the presence of three transcripts (T1, T2, and T3) probably initiating at different promoters but all terminating at the same stem-loop sequence downstream of saeS (Fig. 1). Interestingly, by using a probe specific for ORF4 (probes 1 and 2), one additional transcript (T4; 0.7 kb) could be detected (Fig. 1B). This transcript probably terminates at a stem-loop sequence upstream of ORF3, since probes downstream of the predicted stem-loop sequence (probes 3 and 4) did not detect T4. Bands of the size of the rRNA molecules are detectable by using different probes. We assume that this is an artifact rather than being due to additional specific transcripts, since these rRNA signals are usually seen when larger transcripts (e.g., fnbA [see Fig. 4A]) are analyzed using nonradioactive probes.
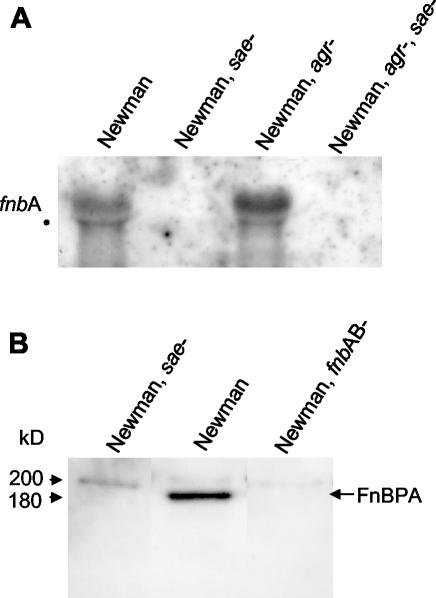
FIG. 4.
(A) Northern blot analysis of equal amounts (2 μg) of total RNA isolated from strains grown to the mid-exponential phase. Hybridization was performed using a DIG-labeled PCR fragment specific for fnbA. The position of the 23S rRNA is marked by a black dot. (B) Detection of FnBP by ligand blotting using biotinylated fibronectin. Cell wall proteins were isolated from bacteria grown to mid-exponential phase and separated on an SDS-5% polyacrylamide gel.
When RNA from the sae mutant strain was analyzed (Fig. 1B, lanes 2) no sae-specific mRNA was detectable with probes 1 to 4. The sae-downstream probe 5 reacted with a transcript present both in the wild type and in the sae mutant strain, indicating that the marked transcript does not belong to the sae operon.
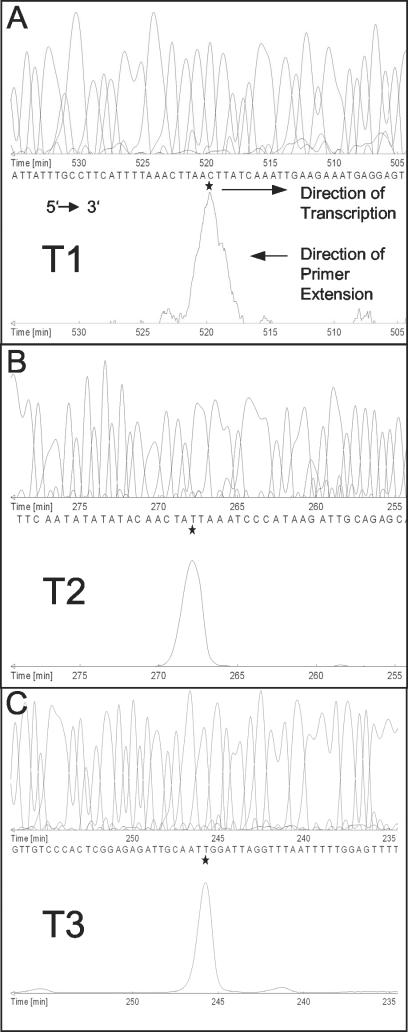
To determine the transcriptional start points more precisely, primer extension experiments were performed using RNA from strain Newman grown to the post-exponential-growth phase. Each start point (P1, P2, and P3) was verified with at least two different primers. The P1 and P2 transcriptional start points could be clearly mapped to positions 280 and 912, respectively. The P3 start point was mapped to position 1322 (Fig. 2). However, the results for the starting point of T3 were more ambiguous, because the primers used to map P3 generated additional signals (data not shown). This is consistent with the results of sae-specific Northern blot analysis of strain Newman, in which T3 appeared to be less distinct than T1 and T2 (see Fig. 1B, 3A, and 3B). However, in other S. aureus strains, a clearer and stronger signal corresponding to T3 is detectable (see Fig. 3B).
FIG. 2.
Mapping of the 5′ ends of three sae transcripts by fluorescence-based primer extension analysis. The precise base mapping was done by comparing the migration of the extended product with a parallel sequencing reaction primed by an identical Cy5-labeled oligonucleotide. The sequence and product traces are given in complement reverse order, thus corresponding to the 5′-to-3′ orientation. The initiation start nucleotide of the mRNA is marked by a star. The sae mRNAs of 3 (A), 2.4 (B), and 2 (C) kb are labeled T1, T2, and T3, respectively.
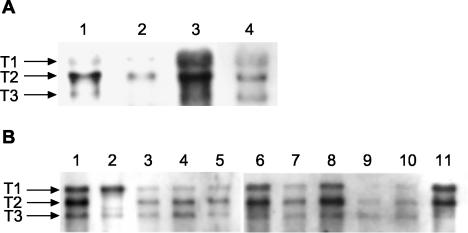
FIG. 3.
Northern blot analysis using a DIG-labeled PCR fragment specific for saeRS (probe 4 [Table 2]). (A) RNA isolated from strains Newman (lanes 1 and 3) and ISP479C (lanes 2 and 4) grown to the mid-exponential-growth phase (lanes 1 and 2) or the post-exponential growth phase (lanes 3 and 4). (B) RNAs of strain Newman (lane 11) and different clinical isolates (lane 1, GN81; lane 2, GN435I; lane 3, W674; lane 4, W678; lane 5, W681; lane 6, W713; lane 7, CFN724; lane 8, CFN726; lane 9, W684; lane 10, W710) grown to the post-exponential-growth phase. Lanes 1 and 2, nasal isolates from healthy controls; lanes 3 to 6, 9, and 10, wound isolates; lanes 7 and 8, isolates from patients with cystic fibrosis.
Differential transcription of sae during the growth phase and in clinical isolates.
Northern blot analysis with a saeRS-specific probe revealed that the three transcripts T1 to T3 were differentially expressed during the growth phase, with maximal expression in the post-exponential-growth phase (Fig. 3A). The overall expression of the sae transcripts was higher in strain Newman than in strain ISP479C. In order to examine whether sae is equally expressed in different clinical isolates, we analyzed sae transcripts in 40 epidemiologically independent clinical isolates after subculturing the strains to the post-exponential-growth phase. The three sae mRNAs were detectable in all the isolates. However, the total amounts of the transcripts were different for different strains. Furthermore, there was a strain-specific pattern with respect to the relative intensity of the three transcripts. The sae-related mRNA patterns of 10 clinical isolates and strain Newman are shown in Fig. 3B. Interestingly, in one isolate (Fig. 3B, lane 2) the size of the T2 transcript was obviously diminished.
Phenotypic characterization of sae mutants with respect to FnBPA and cell invasion.
In order to further characterize the sae regulon, we analyzed the impact of sae on the transcription of fnbA. No fnbA-specific transcript was detectable in strains ISP479C and RN6390 by Northern blot analysis. When RNA from strain Newman was used, maximum transcription of fnbA was seen in bacteria from the exponential-growth phase. fnbA transcription is inhibited by agr, as shown by increased fnbA mRNA levels in the agr mutant strain (Fig. 4A) (33, 41). In contrast, fnbA mRNA was not detectable in the sae mutant or the agr sae double mutant of strain Newman (Fig. 4A). Thus, sae seems to be necessary for fnbA activation independently of agr. In the sae mutant strain, no FnBPA was detectable by ligand affinity blotting using cell wall extracts from bacteria in the exponential-growth phase (Fig. 4B). Besides FnBPA, one additional faint band with an apparent molecular mass of 200 kDa was visible in all preparations. This additional putative FnBP is also present in the fnbAB mutant and is not regulated by sae.
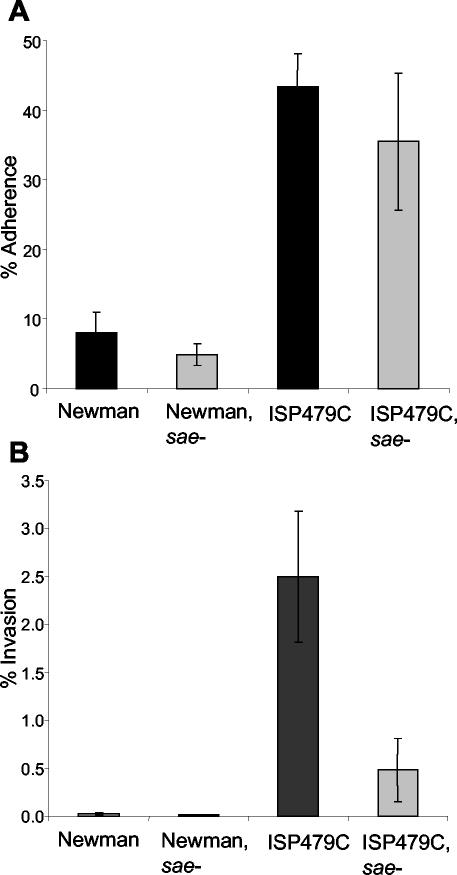
Since FnBPs were shown to be essential for the uptake of S. aureus by endothelial cells (30), we examined the interaction of the sae mutant strains with endothelial cells. There was no significant difference between the adherence of sae mutants to endothelial cells and that of their respective parental strains (Fig. 5A). However, the sae mutant of strain ISP479C was significantly less invasive than the parent strain (Fig. 5B) (P < 0.0001 by a two-tailed t test). Only 0.025% of strain Newman bacteria and 0.007% of bacteria of the sae mutant of strain Newman were incorporated by endothelial cells after 2 h of infection.
FIG. 5.
(A) Adherence of S. aureus to primary human endothelial cells. Strains were grown to the mid-exponential-growth phase. Cell monolayers (∼105 cells/well) were incubated with 5 × 106 bacteria/well for 2 h at 37°C. The percentage of adherence was expressed as 100 × (CFU of adherent bacteria/number of inoculated bacteria). (B) Internalization of S. aureus by endothelial cells. After 2 h of incubation, extracellular bacteria were removed by lysostaphin treatment. The percentage of invasion was expressed as 100 × (CFU of internalized bacteria/number of inoculated bacteria). Each data point represents the mean of four determinations. Error bars, standard deviations.
Phenotypic characterization of sae mutants with respect to CP5.
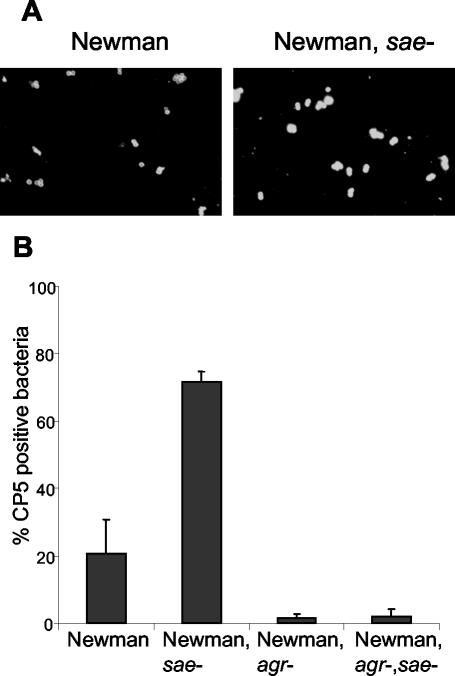
Since strain 8325 and its derivatives are CP5 negative (39), the impact of sae on the expression of CP5 was evaluated in the CP5-positive strain Newman and its isogenic mutants. The sae mutant expressed more capsular antigen than strain Newman, as shown by indirect immunofluorescence (Fig. 6A). Following standard liquid culture, 20% of strain Newman bacteria were shown to be CP5 positive, whereas in the sae mutant 78% of bacteria showed detectable CP5 expression (Fig. 6B) (P < 0.001 by a two-tailed t test). Examination of the agr single mutant and the agr sae double mutant showed that significantly fewer bacteria were encapsulated in the latter than in the parental strain or the sae single mutant. In order to analyze whether sae inhibits CP5 expression on the transcriptional level, cap5-promoter activity was determined by using a xylE-reporter fusion that was introduced into strain Newman and its sae mutant. cap5 promoter activity was significantly higher in the sae-negative background than in the sae-positive background of the parental strain (Fig. 7A).
FIG. 6.
(A) CP5 expression determined by indirect immunofluorescence of strain Newman and its isogenic sae mutant grown to stationary phase at 37°C in CYPG. Bacteria were marked with CP5-specific monoclonal antibodies and stained with Cy3-conjugated anti-mouse antibodies. (B) Percentages of CP5-positive bacteria. Three microscopic fields with about 50 bacteria each were evaluated, and the percentage of CP5-positive bacteria was determined by comparing the number of fluorescent bacteria with the total number of bacteria stained with DAPI (4′,6′-diamidino-2-phenylindole).
FIG. 7.
(A) cap5 promoter activity in strain Newman and its sae mutant containing the xylE fusion construct after growth in air alone and in air with 5% CO2. Promoter activity was expressed as XylE activity per milligram of total protein. (B) Percentages of CP5-positive bacteria of strain Newman and its isogenic sae mutant after growth in air alone and in air with 5% CO2. Three microscopic fields with 50 bacteria each were evaluated, and the percentage of CP5-positive bacteria was determined by comparing the number of fluorescent bacteria with the total number of bacteria stained with DAPI (4,′6′-diamidino-2-phenylindole).
As CP5 has been shown to be down-regulated by elevated CO2 concentrations (19), we tested the impact of CO2 on the sae-dependent up-regulation of CP5 by introducing a cap5 promoter-reporter gene fusion plasmid. Elevated CO2 concentrations resulted in down-regulation of the cap5 promoter in strain Newman as well as in the sae mutant (Fig. 7A). Accordingly, when the expression of CP5 was investigated by indirect immunofluorescence, an inhibitory effect of CO2 could be observed in both strains (Fig. 7B).
DISCUSSION
Here we characterized the sae operon, an independent global regulator for virulence gene expression in S. aureus. Sequence analysis revealed two additional ORFs upstream of the two-component system saeRS. Both ORFs are predicted to code for putative proteins with yet unknown functions. The predicted protein encoded by ORF3 is probably membrane associated, and that encoded by ORF4 is probably cytosolic. Transciptional analysis leads to the assumption that both ORFs are functionally linked to the saeRS two-component regulatory system. ORF3 is cotranscribed with saeRS in the major transcripts T1 and T2. ORF4 is cotranscribed with ORF3 and saeRS (T1) but is also contained in the monocistronic T4 transcript. All transcripts, including the monocistronic message T4, are absent in the sae mutant strain, although the transposon insertion site was shown to be localized further downstream within saeS. Thus, SaeRS is probably necessary for transcriptional initiation from P1. Transcriptional initiation from multiple promoters of regulatory genes has also been described for sarA (2) and sigB (10). This indicates that multiple control elements are essential for the fine tuning of regulatory pathways. Recently, transcriptional analysis of the sae operon has been published (13) Two promoters upstream of saeRS were mapped, one of which corresponds to the P2 promoter described here. In contrast, the second promoter is not identical to any of the promoters characterized by us. The discrepancy may be due to strain variation. The promoter activity of P2 was confirmed by promoter fusion experiments using a 1.15-kb upstream fragment excluding P1 (13). Thus, the possibility that the proposed T2 transcript is a degradation product derived from T1 can be ruled out.
Although saeRS clearly constitutes a classic two-component regulatory system, two additional ORFs are probably involved in its function, resulting in a four-component system. Two-component regulators as part of an operon with additional genes can be seen in quorum-sensing systems, such as agr (21). Recently, the regulatory operon rgf was shown to modulate genes involved in adhesion and virulence in Streptococcus agalactiae (37). rgf shows the same gene organization as the sae operon: four ORFs which are cotranscribed and predicted to encode one unknown protein, and one putative peptide with a signal sequence followed by a typical response regulator and a histidine kinase. The authors speculate that rgf may function as a quorum-sensing system in S. agalactiae. However, so far there is no evidence that S. aureus is equipped with additional quorum-sensing systems besides agr.
We further characterized the effects of sae on target genes and show that in addition to hla and coa (12), sae also activates fnbA. Previously it had been shown that sarA exerts a very similar effect on fnbA (41) and coa expression (unpublished data). Both sae and sarA are required for fnbA and coa transcription independently of agr, whereas agr leads to the inhibition of both genes (33, 40). Since no direct interaction between sae and sarA could be detected (data not shown), one may assume that sae interacts directly with the target genes. However, a comparison of the upstream sequences of fnbA and coa revealed no obvious common motifs. Additional factors may be needed to explain the observed regulatory pattern of fnbA and coa expression. This is further emphasized by the observation that both genes are strictly repressed during the late growth cycle, which cannot be explained by any of the regulatory loci studied so far.
We could demonstrate that sae is necessary for fnbA transcription and that accordingly the sae mutant did not express FnBPA and was internalized significantly less by endothelial cells. It has been shown convincingly by others that FnBPs are essential and sufficient for bacterial uptake by endothelial cells (30, 35). However, there are striking differences between strains. Whereas in strain Newman more fnbA-specific transcripts are detectable than in strain ISP479C, only the latter is effectively taken up by endothelial cells. It remains to be determined why strain Newman is not internalized by these cells, even though it expresses FnBPA. One may speculate that strain-specific differences in the sequence, the surface localization of the protein, or proteolytic processing account for these discrepancies. It is also evident that the adhesion of S. aureus to endothelial cells is mediated by additional factors not affected by sae, since there was no difference in adhesion between sae mutant strains and the parental strains.
Since bacterial adhesion to endothelial cells is inhibited by CP5 (32), we also examined the effect of sae on CP5 expression in strain Newman. sae leads to the repression of cap transcription, as shown by promoter fusion assays. Accordingly, sae mutant bacteria express significantly more CP5. In contrast, agr was shown to activate the cap operon (6, 23, 32). Thus, agr and sae counteract the target gene expression not only of coa and fnbA but of cap as well. However, this is most probably not due to cross-inhibition of sae and agr. This is based on the observation that in sae mutant bacteria, the expression of the effector molecule RNAIII of the agr locus is not altered from that in the parental strain (12) (data not shown). Additionally, sae and agr exert similar and additive effects on hla transcription, which is activated by both regulators.
Since Hla (28) and CP5 (19) are affected in opposite manners by sae as well as by elevated CO2, we analyzed whether sae might function as a CO2 sensor. However, the CO2 responses with respect to CP5 expression were similar in the sae mutant strain and the parental strain. Thus, the appropriate signal for sae activation still remains to be determined. Since hla activation during device-related infection was shown to be dependent on sae (17), we assume that sae contributes to the regulatory adaptation of S. aureus during infection.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (Wo573/2-2).
We are grateful to Wencke Gabel for excellent technical assistance. We thank Jean-Michel Fournier for kindly donating the monoclonal anti-capsular antibody and Chia Lee for providing the cap promoter vector pSN8388.
REFERENCES
- 1.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, M. G., J. H. Heinrichs, and A. L. Cheung. 1996. The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178:4563-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung, A. L., and G. Zhang. 2002. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 7:d1825-d1842. [DOI] [PubMed] [Google Scholar]
- 5.Chien, Y.-T., and A. L. Cheung. 1998. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J. Biol. Chem. 273:2645-2652. [DOI] [PubMed] [Google Scholar]
- 6.Dassy, B., T. Hogan, T. J. Foster, and J. M. Fournier. 1993. Involvement of the accessory gene regulator (agr) in expression of type 5 capsular polysaccharide. J. Gen. Microbiol. 139:1301-1306. [DOI] [PubMed] [Google Scholar]
- 7.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase: mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 8.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 9.Garvis, S., J. M. Mei, J. Ruiz-Albert, and D. W. Holden. 2002. Staphylococcus aureus svrA: a gene required for virulence and expression of the agr locus. Microbiology 148:3235-3243. [DOI] [PubMed] [Google Scholar]
- 10.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of σB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558-566. [DOI] [PubMed] [Google Scholar]
- 11.Giraudo, A. T., A. Calzolari, A. A. Cataldi, C. Bogni, and R. Nagel. 1999. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 177:15-22. [DOI] [PubMed] [Google Scholar]
- 12.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53-58. [DOI] [PubMed] [Google Scholar]
- 13.Giraudo, A. T., C. Mansilla, C. Ana, C. Raspanti, and R. Nagel. 2003. Studies on the expression of regulatory locus sae in Staphylococcus aureus. Curr. Microbiol. 46:246-250. [DOI] [PubMed] [Google Scholar]
- 14.Giraudo, A. T., H. Rampone, A. Calzolari, and R. Nagel. 1996. Phenotypic characterization and virulence of a sae− agr− mutant of Staphylococcus aureus. Can. J. Microbiol. 42:120-123. [DOI] [PubMed] [Google Scholar]
- 15.Giraudo, A. T., C. G. Raspanti, A. Calzolari, and R. Nagel. 1994. Characterization of a Tn551 mutant of Staphylococcus aureus defective in the production of several exoproteins. Can. J. Microbiol. 40:677-681. [DOI] [PubMed] [Google Scholar]
- 16.Goerke, C., S. Campana, M. G. Bayer, G. Döring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA, and sae of Staphylococcus aureus on the induction of α-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40:1439-1448. [DOI] [PubMed] [Google Scholar]
- 18.Herbert, S., S. W. Newell, C. Lee, K. P. Wieland, B. Dassy, J. M. Fournier, C. Wolz, and G. Doring. 2001. Regulation of Staphylococcus aureus type 5 and type 8 capsular polysaccharides by CO2. J. Bacteriol. 183:4609-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbert, S., D. Worlitzsch, B. Dassy, A. Boutonnier, J. M. Fournier, G. Bellon, A. Dalhoff, and G. Döring. 1997. Regulation of Staphylococcus aureus capsular polysaccharide type 5: CO2 inhibition in vitro and in vivo. J. Infect. Dis. 176:431-438. [DOI] [PubMed] [Google Scholar]
- 20.Hoeger, P. H., W. Lenz, A. Boutonnier, and J. M. Fournier. 1992. Staphylococcal skin colonization in children with atopic dermatitis: prevalence, persistence, and transmission of toxigenic and nontoxigenic strains. J. Infect. Dis. 165:1064-1068. [DOI] [PubMed] [Google Scholar]
- 21.Kornblum, J., B. N. Kreiswirth, S. J. Projan, H. F. Ross, and R. P. Novick. 1990. agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, p. 373-402. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH, New York, N.Y.
- 22.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 23.Luong, T., S. Sau, M. Gomez, J. C. Lee, and C. Y. Lee. 2002. Regulation of Staphylococcus aureus capsular polysaccharide expression by agr and sarA. Infect. Immun. 70:444-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manna, A. C., M. G. Bayer, and A. L. Cheung. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 180:3828-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182:3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 27.Novick, R. P. 2000. Pathogenicity factors and their regulation, p. 392-407. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 28.Ohlsen, K., K. P. Koller, and J. Hacker. 1997. Analysis of expression of the alpha-toxin gene (hla) of Staphylococcus aureus by using a chromosomally encoded hla::lacZ gene fusion. Infect. Immun. 65:3606-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pattee, P. A., H. C. Lee, and J. P. Bannantine. 1990. Genetic and physical mapping of the chromosome of Staphylococcus aureus, p. 41-58. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH, New York, N.Y.
- 30.Peacock, S. J., T. J. Foster, B. J. Cameron, and A. R. Berendt. 1999. Bacterial fibronectin-binding proteins and endothelial cell surface fibronectin mediate adherence of Staphylococcus aureus to resting human endothelial cells. Microbiology 145:3477-3486. [DOI] [PubMed] [Google Scholar]
- 31.Peng, H. L., R. P. Novick, B. N. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pöhlmann-Dietze, P., M. Ulrich, K. B. Kiser, G. Döring, J. C. Lee, J. M. Fournier, K. Botzenhart, and C. Wolz. 2000. Adherence of Staphylococcus aureus to endothelial cells: influence of capsular polysaccharide, global regulator agr, and bacterial growth phase. Infect. Immun. 68:4865-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saravia-Otten, P., H.-P. Müller, and S. Arvidson. 1997. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J. Bacteriol. 179:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlichting, C., C. Branger, J. M. Fournier, W. Witte, A. Boutonnier, C. Wolz, P. Goullet, and G. Döring. 1993. Typing of Staphylococcus aureus by pulsed-field gel electrophoresis, zymotyping, capsular typing, and phage typing: resolution of clonal relationships. J. Clin. Microbiol. 31:227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha, B., P. Francois, Y. A. Que, M. Hussain, C. Heilmann, P. Moreillon, D. Lew, K. H. Krause, G. Peters, and M. Herrmann. 2000. Heterologously expressed Staphylococcus aureus fibronectin-binding proteins are sufficient for invasion of host cells. Infect. Immun. 68:6871-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smeltzer, M. S., M. E. Hart, and J. J. Iandolo. 1993. Phenotypic characterization of xpr, a global regulator of extracellular virulence factors in Staphylococcus aureus. Infect. Immun. 61:919-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spellerberg, B., E. Rozdzinski, S. Martin, J. Weber-Heynemann, and R. Lutticken. 2002. rgf encodes a novel two-component signal transduction system of Streptococcus agalactiae. Infect. Immun. 70:2434-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wann, E. R., B. Dassy, J. M. Fournier, and T. J. Foster. 1999. Genetic analysis of the cap5 locus of Staphylococcus aureus. FEMS Microbiol. Lett. 170:97-103. [DOI] [PubMed] [Google Scholar]
- 40.Wolz, C., D. McDevitt, T. J. Foster, and A. L. Cheung. 1996. Influence of agr on fibrinogen binding in Staphylococcus aureus Newman. Infect. Immun. 64:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolz, C., P. Pöhlmann-Dietze, A. Steinhuber, Y.-T. Chien, A. C. Manna, W. J. van Wamel, and A. L. Cheung. 2000. agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36:230-243. [DOI] [PubMed] [Google Scholar]
- 42.Yarwood, J. M., J. K. McCormick, M. L. Paustian, V. Kapur, and P. M. Schlievert. 2002. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J. Bacteriol. 184:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Youngman, P. 1987. Plasmid vectors for recovering and exploiting Tn917 transpositions in Bacillus and other gram-positive bacteria, p. 79-103. In K. G. Hardy (ed.), Plasmids: a practical approach. IRL Press Limited, Oxford, United Kingdom.