“ … the plague bacillus never dies or disappears for good; … it bides its time in bedrooms, cellars, trunks, and bookshelves; and perhaps the day would come when, for the bane and the enlightening of men, it would rouse up its rats again and send them forth to die in a happy city.” Albert Camus, The Plague.
Few other pathogens have caused as much devastation in human history as Yersinia pestis, the etiologic agent of the plague, or the Black Death, as the disease was aptly named in the Middle Ages. Y. pestis is generally transmitted through the bites of the rat flea Xenopsylla cheopis, while the two other related pathogenic Yersinia species, Y. pseudotuberculosis and Y. enterocolitica, are food-borne pathogens that cause various gastrointestinal syndromes (1). All three species of Yersinia are pathogenic for humans and rodents and, despite differences in the routes of entry into the host, all three infect lymphoid tissues and organs. The pathogenicity of Yersinia spp. largely results from the ability to resist innate host defense mechanisms such as phagocytosis and the induction of inflammatory responses by macrophages and neutrophils. Yersinia achieves this resistance by injecting into host cells effector proteins that can manipulate or inhibit normal immune responses.
Like many other gram-negative bacterial pathogens of animals and plants, Yersinia employs a specialized secretory apparatus called the type III secretion system (TTSS) to interact with host cells (1, 2). The TTSS is a multicomponent secretion machine that injects specialized proteins (TTSS effectors) into the cytosol of the host cell where they interact with a variety of host proteins and thus manipulate cellular behavior in ways that ultimately benefit the pathogen (3). The effector proteins (collectively referred to as Yops–Yersinia outer membrane proteins) and the proteins comprising the TTSS are all encoded on a 70-kb plasmid (2). The cellular functions of the Yops are currently under intense study and fall into two general categories: proteins facilitating the translocation of Yops into the host cells, and those actually secreted into the cytosol (1, 2). Notably, YopD, YopB, and LcrV (low calcium response protein V) appear to operate in the translocation of other Yops into the cytosol whereas YopE, YopH, YopJ (Yop P in Y. enterocolitica), YopM, YopO, and YopT function within the host cell (1, 2). The Yop proteins are Yersinia virulence factors that can interfere with phagocytosis, inhibit the antimicrobial oxidative burst, inhibit the production of inflammatory cytokines, such as TNF-α, and promote apoptosis in macrophages and neutrophils (1, 2). Like TTSS effectors of other bacterial pathogens, Yops function by mimicking activities of cellular proteins and either activate or inhibit cellular processes to promote the pathogen's survival and replication (4).
An article in this issue by Sing et al. (5) suggests a novel and unexpected function for the Yersinia virulence factor known as LcrV. LcrV was characterized some 50 yr ago as it is the major antigen of Yersinia (6, 7). In addition to its function in TTSS-mediated protein translocation, LcrV can also be secreted into the environment (6). Secreted LcrV has been previously shown to down-regulate host protective immune responses and this LcrV-mediated immune evasion is due to the ability of LcrV to induce IL-10 production by macrophages (8). Thus, pretreatment of wild-type peritoneal macrophages with recombinant LcrV (rLcrV) led to an inhibition of zymosan-induced TNF-α production. This effect was IL-10 dependent because it could be reversed by neutralizing antibodies specific to IL-10 (but not IL-4 or TGF-β: two other cytokines known to inhibit inflammatory responses of macrophages). Moreover, IL-10–deficient macrophages could still produce TNF-α after rLcrV pretreatment (8). Not surprisingly then, IL-10–deficient mice were found to be more resistant to Y. enterocolitica infection than wild-type mice (8). Following these findings, Sing et al. sought to identify cellular receptors responsible for LcrV-induced IL-10 production. Surprisingly, these receptors turned out to be CD14 and TLR2 (5).
The Toll-like receptors (TLRs) comprise a family of signaling receptors involved in innate immune recognition of microbes (9, 10). When TLRs engage their microbial ligands, they activate signaling pathways including nuclear factor (NF)-κB and mitogen-activated protein (MAP) kinases which leads to the production of proinflammatory cytokines such as TNF-α, IL-6, and IL-12 as well as to the induction of microbicidal mechanisms. In addition to activating effector cells of the innate immune system (such as macrophages and neutrophils), TLRs turn on many genes required for the initiation of pathogen-specific adaptive immune responses (9, 10). Indeed, mice deficient in individual TLRs or their signaling components are highly susceptible to a variety of microbial infections (10).
The TLRs belong to a class of pattern recognition receptors (PRRs) because they recognize conserved molecular signatures (or patterns) common to entire classes of microorganisms. These molecular signatures or pathogen associated molecular patterns (PAMPs) are essential for microbial survival and are therefore highly conserved. It is this evolutionary conservation of PAMPs that makes them perfect targets for innate immune recognition which must rely on a limited number of germline-encoded receptors (11). The PAMPs recognized by TLRs are representative of bacterial, fungal, and viral pathogens including the TLR4 ligand, LPS (product of gram-negative bacteria), the TLR2 ligands, peptidoglycan and zymosan (gram positive bacteria and yeast cell wall products, respectively), the TLR9 ligand, unmethylated CpG DNA (presumably of bacterial and viral origin), the TLR3 ligand, double-stranded RNA (viral product), and the TLR5 ligand, flagellin (bacterial protein; references 12–20). Some TLRs require accessory proteins for ligand recognition. For example, CD14 functions as a ligand binding accessory receptor protein that aids TLR2 and TLR4 in recognition of peptidoglycan and LPS, respectively (9, 10).
It is important to note that unlike virulence factors, microbial products recognized by TLRs are not unique to pathogens and evolved to perform physiological functions unrelated to host-pathogen interactions. Indeed, most, if not all, PAMPs existed long before TLRs or the host. Conversely, virulence factors evolved as a result of adaptation to the host; they manipulate cellular proteins in order to increase the adaptive fitness of the pathogen in the unique environment provided by the host (Table I). Therefore, interaction of LcrV with TLR2 should not be mistaken for pattern recognition. Rather, it is an example of a virulence factor that appears to exploit the pattern recognition system to the advantage of the pathogen. The details of the mechanism of this manipulation, however, remain unclear.
Table I.
Comparison of PAMPs and Virulence Factors
| PAMPs | Virulence Factors |
|---|---|
| Invariant products of conserved metabolic pathways |
Differ between different pathogens |
| Evolved to perform essential physiological functions of microorganisms |
Evolved as a result of adaptation to the host |
| Not unique to pathogens. Produced by all microorganisms of a given class |
Unique to pathogens |
| Targets of innate immune recognition |
Not recognized directly by PRRs. Some virulence factors may interact with PRRs to manipulate the host innate immune response |
Sing et al. found that secretion of IL-10 in response to rLcrV was abrogated in CD14 and TLR2-deficient macrophages. Further, rLcrV was shown to activate an NF-κB reporter gene in CD14 and TLR2-transfected cells (5). As suggested by previous studies (8), the LcrV homologue from P. aeroginosa (PcrV) had no TLR2-inducing activity. Remarkably, the TLR2 stimulating region of LcrV maps to a short 19 amino acid long NH2-terminal sequence that is divergent between LcrV and PcrV (8). Finally, the implication that LcrV triggers IL-10 secretion through CD14/TLR2 to downmodulate the immune response was further substantiated by the fact that TLR2-deficient mice were actually more resistant to Y. enterocolitica infection than wild-type animals (5).
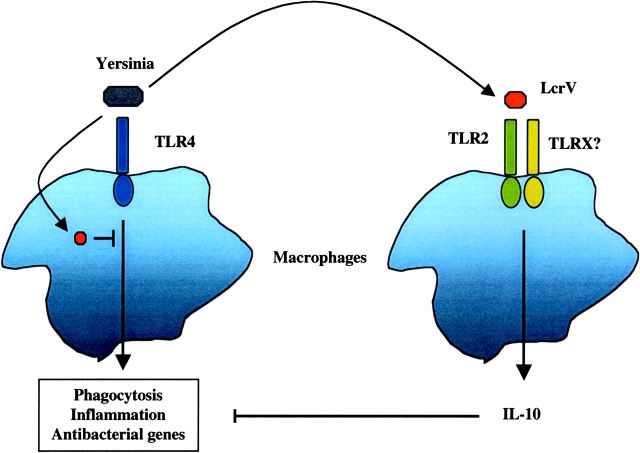
This provocative work provides one of the first examples of a microbial strategy of immune evasion based on manipulation of the TLR system. However, there are several limitations to this study that make interpretation of some of the results difficult and these issues will need to be resolved in the future. First, the immunosuppressive effect of LcrV was tested with regard to TNF-α production induced by zymosan. As Yersinia are gram-negative bacteria, a more biologically relevant stimulus would have been LPS. Nevertheless, the main source of confusion is the fact that when triggered by its microbial ligands, TLR2, like other TLRs, induces the production of pro-inflammatory cytokines, such as TNF-α, IL-6, and IL-12 (19, 21). It remains to be determined whether LcrV induces these cytokines in addition to IL-10. If LcrV promotes IL-10 secretion without inducing a proinflammatory response, this would imply that LcrV initiates a different signaling pathway than that triggered by other TLR2 ligands. Although it is formally possible, it seems highly unlikely that that could be the case. TLR2 functions as a heterodimer with either TLR1 or TLR6 (22–25), and possibly with some other, as yet uncharacterized TLRs. Different ligands engage distinct TLR2 heterodimers: tripalmitoylated lipoproteins, for example, engage a TLR2/TLR1 complex, whereas peptidoglycan and dipalmitoylated lipoproteins, such as the ones found in mycoplasma, are detected via a TLR2/TLR6 complex (22–25). Conceivably, LcrV could engage TLR2 with another TLR, and this complex could induce the IL-10 gene, rather than the genes encoding proinflammatory cytokines (Fig. 1) . This possibility could easily be addressed by a careful comparison of IL-10 and TNF-α induction by LcrV versus peptidoglycan or lipoproteins. Unfortunately, without such comparison it is difficult to conclude whether there is something unique in the way LcrV activates TLR2.
Figure 1.
Yersinia employ Yop effector proteins to inhibit the antimicrobial functions of macrophages. The Toll-like receptor family of proteins recognizes PAMPs expressed by bacteria and initiates a signaling cascade leading to activation of macrophages. Yersinia injects effector proteins (Yops) via the type III secretion system into host macrophages. Injected Yops inhibit phagocytosis, inflammation, and the synthesis of antibacterial proteins. One of the Yops, LcrV, is secreted and can act at a distance. LcrV utilizes TLR2 to induce the production of the antiinflammatory cytokine, IL-10, further dampening the innate immune response.
Another salient issue that these experiments would address is whether the immunosuppressive activity of LcrV is related to the phenomenon of LPS tolerance. When macrophages are stimulated with LPS, they become refractory to a second LPS stimulation. This unresponsiveness to secondary LPS stimulation (known as “LPS tolerance”) is maximal at around 24 h and subsides at longer time points. Although first described for LPS, this tolerance applies to other TLR ligands as well (26, 27). The mechanisms of tolerance are not completely understood and appear to be distinct for different TLRs (26, 27). Two features of LcrV activity suggest that LcrV-induced inhibition of TNF-α production is not due to tolerance induction via TLR2. First, the time sufficient for blocking zymosan-induced TNF-α production (3 h) is much less than what is required for the induction of maximal unresponsiveness due to a tolerance mechanism (24 h). Second, tolerance to TLR2 and TLR4 ligands is known to be IL-10 independent (27), whereas LcrV effects appear to be completely IL-10 dependent. A direct comparison of the effect of LcrV with the effects of other TLR ligands on macrophage responsiveness would be necessary to determine whether LcrV engages a signaling pathway distinct from other TLR ligands.
Many, if not all, TLR ligands can induce IL-10 secretion by macrophages as a part of a negative feedback control that limits the inflammatory response. This IL-10 induction, however, is typically delayed compared with the induction of proinflammatory cytokines. Therefore, the kinetics of IL-10 production induced by LcrV and by other TLR2 (as well as TLR4) ligands would also be very informative.
These questions notwithstanding, this study describes a very exciting finding of a novel mechanism of immune evasion that contributes to the virulence of one of the deadliest pathogens known to humankind. Without a doubt, many more examples of microbial interference with TLRs and their signal transduction pathways will be discovered, and these will help both to understand the pathogen's virulence mechanisms and to elucidate the complexity of innate immune recognition.
References
- 1.Cornelis, G.R. 2000. Molecular and cell biology aspects of plague. Proc. Natl. Acad. Sci. USA. 97:8778–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornelis, G.R., A. Boland, A.P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M.P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galan, J.E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 284:1322–1328. [DOI] [PubMed] [Google Scholar]
- 4.Staskawicz, B.J., M.B. Mudgett, J.L. Dangl, and J.E. Galan. 2001. Common and contrasting themes of plant and animal diseases. Science. 292:2285–2289. [DOI] [PubMed] [Google Scholar]
- 5.Sing, A., D. Rost, N. Tvardovskaia, A. Roggenkamp, A. Wiedemann, C.J. Kirschning, M. Aepfelbacher, and J. Heesemann. 2002. Yersinia V-antigen exploits toll-like receptor-2 and CD14 for interleukin 10–mediated immunosuppression. J. Exp. Med. 196:1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fields, K.A., M.L. Nilles, C. Cowan, and S.C. Straley. 1999. Virulence role of V antigen of Yersinia pestis at the bacterial surface. Infect. Immun. 67:5395–5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, V.T., C. Tam, and O. Schneewind. 2000. LcrV, a substrate for Yersinia enterocolitica type III secretion, is required for toxin targeting into the cytosol of HeLa cells. J. Biol. Chem. 275:36869–36875. [DOI] [PubMed] [Google Scholar]
- 8.Sing, A., A. Roggenkamp, A.M. Geiger, and J. Heesemann. 2002. Yersinia enterocolitica evasion of the host innate immune response by V antigen-induced IL-10 production of macrophages is abrogated in IL-10-deficient mice. J. Immunol. 168:1315–1321. [DOI] [PubMed] [Google Scholar]
- 9.Medzhitov, R. 2001. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1:135–145. [DOI] [PubMed] [Google Scholar]
- 10.Akira, S., K. Takeda, and T. Kaisho. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675–680. [DOI] [PubMed] [Google Scholar]
- 11.Janeway, C.A., Jr. 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54:1–13. [DOI] [PubMed] [Google Scholar]
- 12.Poltorak, A., X. He, I. Smirnova, M.Y. Liu, C.V. Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, et al. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 282:2085–2088. [DOI] [PubMed] [Google Scholar]
- 13.Qureshi, S.T., L. Lariviere, G. Leveque, S. Clermont, K.J. Moore, P. Gros, and D. Malo. 1999. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4). J. Exp. Med. 189:615–625 [published erratum at 189:1518]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature. 408:740–745. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi, F., K.D. Smith, A. Ozinsky, T.R. Hawn, E.C. Yi, D.R. Goodlett, J.K. Eng, S. Akira, D.M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 410:1099–1103. [DOI] [PubMed] [Google Scholar]
- 16.Alexopoulou, L., A.C. Holt, R. Medzhitov, and R.A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 413:732–738. [DOI] [PubMed] [Google Scholar]
- 17.Hoshino, K., O. Takeuchi, T. Kawai, H. Sanjo, T. Ogawa, Y. Takeda, K. Takeda, and S. Akira. 1999. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 162:3749–3752. [PubMed] [Google Scholar]
- 18.Schwandner, R., R. Dziarski, H. Wesche, M. Rothe, and C.J. Kirschning. 1999. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J. Biol. Chem. 274:17406–17409. [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi, O., K. Hoshino, T. Kawai, H. Sanjo, H. Takada, T. Ogawa, K. Takeda, and S. Akira. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 11:443–451. [DOI] [PubMed] [Google Scholar]
- 20.Underhill, D., A. Ozinsky, A. Hajjar, A. Stevens, C. Wilson, M. Bassetti, and A. Aderem. 1999. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 401:811–815. [DOI] [PubMed] [Google Scholar]
- 21.Takeuchi, O., A. Kaufmann, K. Grote, T. Kawai, K. Hoshino, M. Morr, P.F. Muhlradt, and S. Akira. 2000. Cutting edge: preferentially the R-stereoisomer of the mycoplasmal lipopeptide macrophage-activating lipopeptide-2 activates immune cells through a toll-like receptor 2- and MyD88-dependent signaling pathway. J. Immunol. 164:554–557. [DOI] [PubMed] [Google Scholar]
- 22.Ozinsky, A., D.M. Underhill, J.D. Fontenot, A.M. Hajjar, K.D. Smith, C.B. Wilson, L. Schroeder, and A. Aderem. 2000. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc. Natl. Acad. Sci. USA. 97:13766–13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeuchi, O., T. Kawai, P.F. Muhlradt, M. Morr, J.D. Radolf, A. Zychlinsky, K. Takeda, and S. Akira. 2001. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int. Immunol. 13:933–940. [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi, O., S. Sato, T. Horiuchi, K. Hoshino, K. Takeda, Z. Dong, R.L. Modlin, and S. Akira. 2002. Cutting edge: role of toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 169:10–14. [DOI] [PubMed] [Google Scholar]
- 25.Alexopoulou, L., V. Thomas, M. Schnare, Y. Lobet, J. Anguita, R.T. Schoen, R. Medzhitov, E. Fikrig, and R.A. Flavell. 2002. Hyporesponsiveness to vaccination with Borrelia burgdorferi OspA in humans and in TLR1- and TLR2-deficient mice. Nat. Med. 8:878–884. [DOI] [PubMed] [Google Scholar]
- 26.Sato, S., O. Takeuchi, T. Fujita, H. Tomizawa, K. Takeda, and S. Akira. 2002. A variety of microbial components induce tolerance to lipopolysaccharide by differentially affecting MyD88-dependent and -independent pathways. Int. Immunol. 14:783–791. [DOI] [PubMed] [Google Scholar]
- 27.Sato, S., F. Nomura, T. Kawai, O. Takeuchi, P.F. Muhlradt, K. Takeda, and S. Akira. 2000. Synergy and cross-tolerance between toll-like receptor (TLR) 2- and TLR4-mediated signaling pathways. J. Immunol. 165:7096–7101. [DOI] [PubMed] [Google Scholar]