Abstract
We discovered a novel small heat shock protein (sHsp) named AgsA (aggregation-suppressing protein) in the thermally aggregated fraction from a Salmonella enterica serovar Typhimurium dnaK-null strain. The −10 and −35 regions upstream of the transcriptional start site of the agsA gene are characteristic of σ32- and σ72-dependent promoters. AgsA was strongly induced by high temperatures. The similarity between AgsA and the other two sHsps of Salmonella serovar Typhimurium, IbpA and IbpB, is rather low (around 30% amino acid sequence identity). Phylogenetic analysis suggested that AgsA arose from an ancient gene duplication or amplification at an early evolutionary stage of gram-negative bacteria. Here we show that overproduction of AgsA partially complements the ΔdnaK52 thermosensitive phenotype and reduces the amount of heat-aggregated proteins in both ΔdnaK52 and ΔrpoH mutants of Escherichia coli. These data suggest that AgsA is an effective chaperone capable of preventing aggregation of nonnative proteins and maintaining them in a state competent for refolding in Salmonella serovar Typhimurium at high temperatures.
The heat shock response is induced upon exposure of cells to a large variety of environmental stresses including heat shock and by pathophysiological and metabolic states (32, 33, 36). It consists of the compartment-specific induction of heat shock genes, which in gram-negative bacteria are under the common transcriptional control of σ32, the transcriptional activator of the heat shock regulon (3, 16, 35, 51). The heat shock proteins (Hsps) include the major cytosolic chaperones such as ClpB, the DnaK chaperone system, GroEL/ES, HtpG, IbpA, IbpB, and a number of proteases (4, 13, 16, 35, 42, 51). The ensemble of molecular chaperones and proteases constitutes a cellular system for de novo folding and quality control of proteins that relies on the ability of chaperones and proteases to refold or degrade misfolded proteins (4, 17). Hsp104 (ClpB) and the Hsp70 (DnaK) bichaperone system are known to disaggregate and refold the denatured proteins efficiently (14, 15, 31, 46).
Members of the small heat shock protein (sHsp) family are found in most organisms. The sHsps are characterized by (i) a molecular mass typically between 12 and 42 kDa, (ii) a conserved central domain generally referred to as the α-crystallin domain, (iii) the formation of large oligomeric complexes, and (iv) an ATP-independent chaperone activity (6, 20, 30, 47). A number of in vitro studies suggest that sHsps bind nonnative proteins, prevent their aggregation, and maintain them in a state competent for refolding by the ATP-dependent chaperones, the ClpB-DnaK bichaperone system, and GroEL/GroES (11, 20, 28, 47).
However, only a few reports on their intracellular functions have been published (1, 45). The Legionella pneumophila sHsp “gspA” mutant was shown to be more susceptible to in vitro stress stimuli (oxidative, heat, acid, and osmotic) (1). Kuczýnska-Wísnik et al. have shown that Escherichia coli IbpA and IbpB are able to prevent the aggregation of endogenous proteins induced by extreme heat shock (24). It has also been reported that the overproduction of sHsps makes eukaryotic and prokaryotic cells more resistant to stresses such as heat, hydrogen peroxide, and superoxide anion (21, 23, 27). However, the growth rates and survival of sHsp mutant strains from Saccharomyces cerevisiae and E. coli were not affected under a variety of stress conditions (21, 37).
So far, most chaperone studies for bacteria have been performed using the model strain E. coli. Salmonella enterica serovar Typhimurium is a facultatively intracellular pathogen that causes gastroenteritis in humans and systemic diseases similar to typhoid in mice. It is known that large numbers of general and specific chaperones as well as specific virulence factors are required to successfully colonize the host organism and to avoid clearance by the immune system. In this study, we discovered a novel sHsp named AgsA (aggregation-suppressing protein) in the fraction of thermally aggregated proteins from Salmonella serovar Typhimurium. Our data indicate that AgsA suppresses aggregation of nonnative proteins and denatured proteins and that it reduces the amount of heat-aggregated proteins in cells.
MATERIALS AND METHODS
Strains and culture conditions.
Bacterial strains are listed in Table 1. Luria broth (L broth) and SS medium were used with ampicillin (AMP; 50 μg/ml), chloramphenicol (CHL; 20 μg/ml), kanamycin (KAN; 20 μg/ml), and nalidixic acid (NAL; 25 μg/ml). To construct the strain carrying the agsA ibpAB triple mutation, bacteriophage P22 was propagated on CS2458 (ΔagsA::Km) and the resultant lysate was used for infection of CS2042 (ΔibpAB::Cm). The transductants were selected for KAN resistance.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α Z1 | F−endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA(Nalr) relA1 Δ(lacZYA-argF)U169 deoR [φ80dlac Δ(lacZ)M15] tetR lacIq Specr | 29 |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2 Tc::Mu(Kmr) λpir | 41 |
| S17-1λpir | RP4-2 Tet::Mu-Kan::Tn7 λpir | 41 |
| BB1553 | Same as MC4100 except for ΔdnaK52::Cm | 2 |
| CS5257 | BB1553 pBB528 (lacIq) pTKY605 (agsA) | This study |
| BB7224 | MC4100 ΔrpoH::Km suhX401 araD+ | 46 |
| CS5262 | BB7224 pDMI, 1(lacIq) pTKY605 (agsA) | This study |
| Salmonella serovar Typhimurium | ||
| χ3306 | SR-11 gyrA1816; virulent | Provided by R. Curtiss III |
| CS2042 | Same as χ3306 except for ΔibpAB::Cm | This study |
| CS2458 | Same as χ3306 except for ΔagsA::Km | This study |
| CS2021 | Same as χ3306 except for ΔdnaK::Cm | This study |
| CS2565 | Same as χ3306 except for ΔagsA::Km ibpAB::Cm | This study |
Cm, CHL resistance; Km, KAN resistance.
Construction of ΔibpAB, ΔagsA, and ΔdnaK mutants.
To construct the ΔibpAB::Cm mutant CS2042, the DNA fragment between nucleotides (nt) −55 and 1089 in the ibpAB operon coding region with promoters was initially amplified by PCR and cloned into pT7Blue-2 (Novagen). The resultant plasmid, pTKY577, was cleaved at the BstXI and BglII sites (nt 226 and 831) in the cloned fragment, and the overhanging ends were filled in and ligated to the CHL resistance cassette, which was generated from BamHI-digested pNK2884 (50) and then filled. The resultant plasmid, pTKY580, was cleaved at the MluI and SalI sites in the vector. The generated ibpAB::Cm fragment was ligated to the MluI and SalI sites of pTKY513, which is a previously constructed transferable suicide vector (43). The resultant mutator plasmid, pTKY583, was introduced into strain SM10(λpir), which provides the π protein required for the replication of the suicide vector by transformation. The chromosomal ibpAB operon was replaced by the ibpAB::Cm construct by conjugative crosses as previously described (49). The mutant was selected for resistance to CHL and NAL. A double-crossover event resulting in the ΔibpAB::Cm mutant was assessed by its sensitivity to AMP. Disruption of the ibpAB operon was checked by immunoblotting of the heat-aggregated proteins with an anti-E. coli IbpB serum (data not shown).
The agsA mutant CS2458 was constructed in the same way as the ΔibpAB::Cm mutant. Synthetic oligonucleotides (5′-TTTCGTTAACCACTTAGAATTC) and (5′-GGCCAAGCTTATGATTTGTGTTCAATCGCC), which have an EcoRI or HindIII recognition sequence, were used as PCR primers. The amplified fragment containing the agsA coding region and its promoters was cloned into pHSG398 (44). The resultant plasmid was cleaved at the AhaII site (nt 256) in the cloned fragment and ligated to the KAN resistance cassette, which was generated from PstI-digested pUC4K (48) and then blunted. The resultant plasmid, pTKY584, was cleaved at the EcoRI and FspI sites in the vector, and the overhanging FspI end was filled in. The generated agsA fragment was ligated to the EcoRI and filled-in SalI sites of pTKY513. The resultant mutator plasmid, pTKY585, was introduced into strain S17-1(λpir). The chromosomal agsA gene was replaced by the agsA::Km construct by conjugative crosses. The mutant was selected for resistance to KAN and NAL. A double-crossover event resulting in the ΔagsA::Km mutant was assessed by its sensitivity to AMP. Disruption of the agsA gene was checked by PCR and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of the heat-aggregated proteins (data not shown).
To construct the dnaK mutant CS2021, the DNA fragment between nt 53 and 1000 in the dnaK coding region was initially amplified by PCR and cloned into pT7Blue-2. The resultant plasmid, pTKY505, was cleaved at the Eco47III site (nt 525) in the cloned fragment, filled in, and ligated to the CHL resistance cassette, which was generated from BamHI-digested pNK2884 and then filled in. The resultant plasmid, pTKY508, was cleaved at the SalI and SmaI sites in the vector. The generated ΔdnaK::Cm fragment was ligated to the SalI and SmaI sites of pCVD442 (10). The resultant mutator plasmid, pTKY507, was introduced into strain SM10(λpir). The chromosomal dnaK gene was replaced by the dnaK::Cm construct by conjugative crosses. The mutant was selected for resistance to CHL and NAL. A double-crossover event resulting in the ΔdnaK::Cm mutant was assessed by its sensitivity to AMP. Disruption of the dnaK gene was checked for a thermosensitive phenotype and by immunoblotting of the total proteins with an anti-DnaK serum (data not shown).
Construction of AgsA-overproducing plasmid.
To construct the AgsA-overproducing plasmid, the DNA fragment in the ribosome binding site and the agsA coding region was initially amplified by PCR. Synthetic oligonucleotides (5′-GGCCGAATTCAGGAGGTTAATGATGGCATCT) and (5′-GGCCAAGCTTATGATTTGTGTTCAATCGCC), which have an EcoRI or HindIII recognition sequence, were used as primers in a PCR. The amplified fragment was digested with EcoRI and HindIII and then cloned into pUHE21-2fdΔ12 (12). The resultant plasmid, pTKY605, allows the isopropyl-β-d-thiogalactopyranoside (IPTG)-controlled induction of AgsA.
Databases and protein sequence analysis.
The database of nucleotide sequences of completed and unfinished eukaryotic and prokaryotic genomes at the National Center for Biotechnology Information (NCBI; National Institutes of Health) was used. The nonredundant database was searched with the gapped BLAST program and PSI-BLAST as described on the website (http://www.ncbi.nlm.nih.gov/BLAST/). Sequences were aligned by homology searching of the maximum matching program with GENETYX-MAC 7.3. Multiple alignments were constructed with the CLUSTALW program. For the construction of a phylogenic tree, large inserts and ambiguously aligned regions were removed from the multiple alignments, and then commonly conserved domains were chosen (97 amino acid [aa] residues). Phylogenetic trees were constructed by using the PHYLIP package.
Isolation of aggregated proteins and gel electrophoresis.
Isolation of aggregated proteins was performed as described previously (46). Quantification of the amount of aggregated proteins was performed using the Bradford assay reagent (Bio-Rad, Hercules, Calif.) with bovine serum albumin as the standard. Gel electrophoresis was carried out according to the method of Laemmli (26) by using SDS-15% polyacrylamide gels and by staining with Coomassie brilliant blue.
Identification of protein.
To determine the N-terminal protein sequence, proteins were separated by two-dimensional gel electrophoresis and transferred onto a polyvinylidene difluoride membrane (Millipore). The proteins were stained with Coomassie brilliant blue, cut out, and analyzed with a Shimazu PPSD-21 protein sequencer.
Plating efficiency and spot test.
Cells were grown in L medium for 3 h at 30°C to mid-logarithmic phase. Aliquot of cultures were diluted from 10−1 to 10−5 in L medium. Aliquots (100 μl) were plated, and 5-μl aliquots were spotted onto L agar plates containing 250 μM IPTG. Plates were incubated at 30 or 37°C for 24 h, and colony numbers were determined afterwards.
Survival ratio of shsp mutants.
Cells were grown in L medium for 3 h at 30°C to mid-logarithmic phase. Aliquots (0.5 ml) of wild-type cultures and of each shsp mutant culture (0.5 ml) were mixed. An aliquot (100 μl) of the mixed culture was diluted in L medium without heat treatment. The rest of the culture (900 μl) was shifted to 70°C for 1 min and then diluted in L medium. The diluted cultures were plated onto an L agar plate, and total cell numbers were counted after incubation for 24 h at 30°C. Among them, the numbers of viable bacteria of strains carrying the ΔagsA::Km and ΔibpAB::Cm mutations were determined by selecting KAN- and CHL-resistant bacteria, respectively. The survival ratios were calculated as the ratio of the number of surviving shsp mutant cells to the number of surviving wild-type cells (shsp/WT) in the heat-treated sample divided by the ratio (shsp/WT) for the non-heat-treated sample.
RESULTS
Identification of a new sHsp, AgsA.
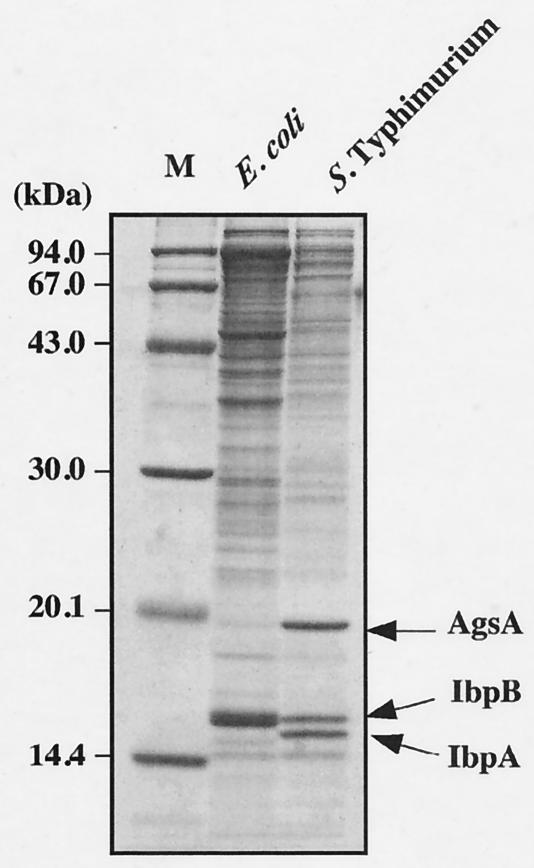
Our aim was to analyze the physiological role of sHsps in Salmonella serovar Typhimurium. Since sHsps have been known to coaggregate with heat-denatured proteins in an E. coli ΔdnaK52 strain (31, 46), we isolated and analyzed heat-aggregated proteins in a dnaK-null mutant of Salmonella serovar Typhimurium by SDS-PAGE (Fig. 1). We used a previously developed protocol for cell lysis and aggregate purification, which greatly increased the sensitivity of detection of aggregated protein species and minimized contamination of membrane proteins (46). We found that three major proteins (14.5, 15.0, and 18.0 kDa) were accumulated in the aggregated fraction (Fig. 1). It was generally believed that most of the members of the Enterobacteriaceae have two sHsps, IbpA and IbpB. The 14.5- and 15.0-kDa proteins were identified by immunoblotting using a specific antiserum against E. coli IbpB (31) as the IbpA and IbpB homologues of Salmonella serovar Typhimurium, respectively (data not shown). The 18.0-kDa protein was identified to the product of an uncharacterized open reading frame (ORF) of the Salmonella serovar Typhimurium genome by determination of its N-terminal amino acid sequence as shown in Fig. 2. The translated amino acid sequence of this protein shows weak but significant similarity to the sHsps from the eukaryotes and to the structurally related α-crystallin lens proteins (Fig. 3A). Thus, the 18.0-kDa protein is a member of the sHsp protein family. We named this protein AgsA (aggregation-suppressing protein) on the basis of its function as described below.
FIG. 1.
Detection of aggregated proteins in dnaK-null mutants. dnaK-null mutant cells were grown to mid-exponential growth at 30°C and incubated further at 42°C for 1 h. The aggregated proteins were isolated and analyzed by SDS-15% PAGE as described in Materials and Methods. M, molecular weight marker; E. coli, BB1553 (ΔdnaK52); Salmonella serovar Typhimurium, CS2021 (dnaK::Cm).
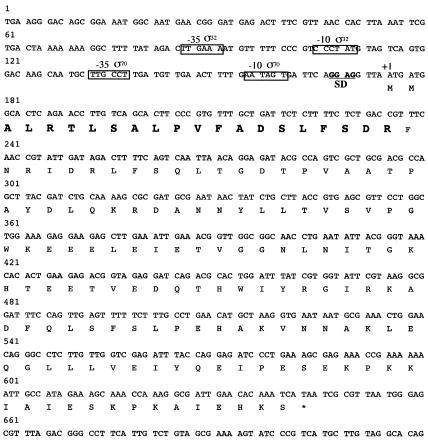
FIG. 2.
Nucleotide sequence of the Salmonella serovar Typhimurium agsA gene. The nucleotide sequence of the agsA gene was taken from the completed Salmonella serovar Typhimurium LT2 genomic sequence at the National Center for Biotechnology Information (National Institutes of Health) by BLAST search. Position +1 marks the start of the AgsA ORF, which is translated. Bold letters show the N-terminal amino acid sequence that was determined by N-terminal sequencing. The underlined region represents the sequence of the ribosome binding site (SD). The putative promoter regions (−35 to −10) are boxed. σ32, putative σ32 promoter; σ70, putative σ70 promoter.
FIG. 3.
(A) Alignment of the most highly conserved carboxy-terminal amino acid sequence of AgsA and various sHsps. Positions where four proteins have identical amino acids are boldfaced. Two or more identical amino acids in one position are indicated by plain text. Gaps are indicated by dashes. (B) Phylogenetic tree of the sHsps. Phylogenetic trees were constructed by using the PHYLIP package as described in Materials and Methods. Proteins (with accession numbers in parentheses) are as follows: mouse, Hsp27 (A53423); rat, Hsp27 (JN0924); dog, Hsp27 (JC4244); human, Hsp27 (HHHU27); chicken, Hsp25 (A39644); bullfrog, αB-crystallin (S54824); African clawed frog, Hsp30C (JN0274); alfalfa, Hsp18 (S16248); soybean, Hsp17.5 (T07629); common tobacco, Hsp18p (T03958); rice, Hsp17 (T04171); wheat, Hsp17 (HHWT17); Arabidopsis thaliana, Hsp22 (S71188); common sunflower, Hsp17.9 (S46310); Methanobacterium themoautotrophicum, Hsp17 (F69214); tomato, Hap17.6 (T07602); tomato (chloroplast), Hsp21 (T06324); common tobacco (chloroplast), Hsp26a (T02018); maize (plastid), Hsp26 (T03379); Thermotoga maritima, Hsp17 (T46658); Stigmatella aurantaca, Hsp SP21 (A49942); Saccharomyces cerevisiae, Hsp26 (P15992); Schizosaccharomyces pombe, Hsp16 (T40376); Pseudomonas aeruginosa IbpA (D83256); Escherichia coli IbpB (G65170); Legionella pneumophila, GspA (S49042); Vibrio cholerae, Hsp16 (D82373); Escherichia coli IbpA (A45245).
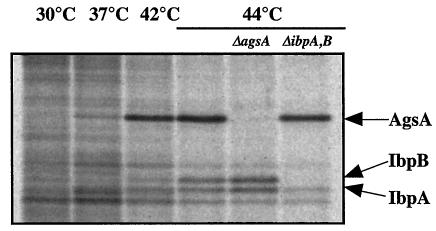
Analysis of the area upstream of agsA revealed that the transcription is possibly regulated by two promoters (Fig. 2). One is similar to the consensus sequence of a promoter that is recognized by the σ70 housekeeping transcription factor, while another promoter seems to be recognized by the σ32 heat shock transcription factor (8). In contrast to ibpA and ibpB, which form an operon together (7), it is likely that the agsA gene is transcribed in a monocistronic mRNA that could be translated to a polypeptide of 156 aa (including the N-terminal Met residue) with a molecular mass of 17.7 kDa. The result of N-terminal sequencing and determination of the predicted amino acid sequence suggested that AgsA contains neither Met nor Cys. This was confirmed by the result in which pulse-labeling of proteins with [35S]Met-Cys failed to detect the stimulated synthesis of an sHsp corresponding to the molecular weight of AgsA by heat shock (data not shown). On the other hand, pulse-labeling of proteins with a 14C-labeled amino acid mixture revealed a single sHsp corresponding to AgsA at high temperature. We observed a weak induction of AgsA at 37°C and strong induction at 42 and 44°C, stronger than that of IbpAB (Fig. 4). We also confirmed that there was no induction of AgsA in a ΔagsA strain, even at a high temperature (44°C) (Fig. 4).
FIG. 4.
Heat induction of AgsA. Cells of strains χ3306 (wild type), CS2042 (ΔibpAB), and CS2458 (ΔagsA) were grown for 4 h at 30°C in M9 glucose minimum medium. Aliquots of the cultures were pulse-labeled for 2 min with an l-U-14C-labeled amino acid mixture (Amersham Pharmacia Biotech) at 15 min after the shift from 30°C to the indicated temperature. Equal amounts of total proteins were analyzed by SDS-PAGE.
Homology alignment and phylogenetic analysis.
It is known that the carboxy-terminal domain called the α-crystallin domain displays the highest degree of conservation within the sHsp family (9). Homology alignment with eukaryotic sHsps revealed that AgsA has the highest similarity to soybean sHsps over the carboxy-terminal region of the protein (42% identical) (Fig. 3A). A BLAST search by comparison with the prokaryote genome database at the NCBI uncovered an ORF encoding a Klebsiella pneumoniae homologous protein with high similarity throughout the entire region (70% amino acid sequence identity). Another gene coding for a homologous protein (42% amino acid sequence identity) was found in the genome of Buchnera sp. APS, which is an endocellular bacterial symbiont harbored by pea aphids. The similarities between AgsA and IbpA or IbpB of Salmonella serovar Typhimurium are rather low (sequence identity to AgsA, 32% for IbpA [E values, 8e−17] and 31% for IbpB [E values, 5e−17]). In contrast to other members of the Enterobacteriaceae, including E. coli, that encode IbpA and IbpB homologues but not AgsA in their genomes, K. pneumoniae has three different sHsp homologues corresponding to AgsA, IbpA, and IbpB. Interestingly, Buchnera sp. APS has only one sHsp, which shows the highest homology to AgsA. A phylogenetic analysis suggested that AgsA arose from an ancient gene duplication or amplification at an early evolutionary stage of gram-negative bacteria that was followed by sequence divergence (Fig. 3B).
Role of sHsps in bacterial thermotolerance.
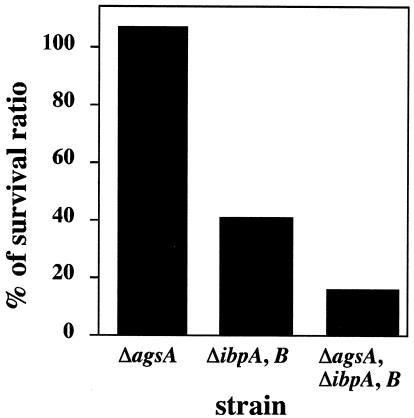
It is believed that sHsps constitute the system for protecting against irreversible aggregation of cellular proteins and assist in protein refolding by the ClpB-DnaK bichaperone system and GroESL after heat shock of E. coli (4, 11, 47). However, it has also been reported that no substantial differences were observed in the thermoresistance and tolerance phenotypes between ibpAB mutants and their parental strain (21). We examined the temperature sensitivity for growth (at 44°C), protein aggregation (at 44°C), and thermotolerance (at 52°C) phenotype of Salmonella serovar Typhimurium agsA and ibpAB mutant cells. The shsp mutant did not show any temperature-sensitive phenotype, accumulation of aggregated proteins, or defect in thermotolerance at the indicated temperatures (data not shown). It is well known that the ClpB-DnaK bichaperone system has a function crucial for the acquisition of heat resistance, thermotolerance, and protection of cellular proteins. It seems that the most defects of agsA and ibpAB mutations are sequestered by the function of the DnaK-ClpB bichaperone system. Therefore, we used the more intense condition to assess the role of the sHsps in the thermotolerance of Salmonella serovar Typhimurium. We determined the survival rates after transient exposure to a lethal temperature (70°C) (Fig. 5). When wild-type cells were shifted from 30 to 70°C, cell viability rapidly decreased, yielding about 0.01% viable cells after 60 s and no viable cells after 90 s of incubation. To avoid experimental error, we mixed the same numbers of wild-type and mutant cells and then exposed the cultures to 70°C for 1 min. The survival ratio was determined as described in Materials and Methods, and the relative ratio is shown in Fig. 5. In the agsA mutant strain, no decrease in viability was observed compared to the wild-type strain (relative survival rate, 113%). The ibpAB mutant showed a significantly lower viability (42%). The addition of the agsA::Km mutation to ibpAB mutants resulted in a further decrease in viability (15%). These results suggest that both the agsA and ibpAB genes play important roles in the survival of Salmonella serovar Typhimurium at lethal temperatures.
FIG. 5.
sHsps are necessary for improvement of survival at 70°C. Cells of strain χ3306 (wild type) and mutants CS2458 (ΔagsA), CS2042 (ΔibpAB), and CS2565 (ΔagsA and ΔibpAB) were grown to mid-exponential growth at 30°C. The same numbers of wild-type and shsp mutant cells were mixed and then sifted at 70°C for 1 min. The numbers of viable shsp mutant cells were determined by using a drug resistance marker (Km for ΔagsA; Cm for ΔibpAB). Survival ratios were calculated as described in Materials and Methods. Each value is the average from two different experiments.
The heat sensitivity of a ΔdnaK52 mutant is partially suppressed by the overproduction of AgsA.
To examine the function of AgsA, we constructed the agsA recombinant plasmid pTKY605, which contains an IPTG-regulatable PA1lacO1 promoter. For this experiment, we chose an E. coli ΔdnaK52 mutant (BB1553) which expresses DnaJ at low levels and carries a spontaneous suppressor mutation in rpoH, which partially inactivates σ32 (2). Cells of this strain grow with approximately wild type rates at 30°C but remain temperature sensitive for growth above 37°C. Since E. coli has no agsA gene on the chromosome, we decided to monitor the chaperone function of AgsA.
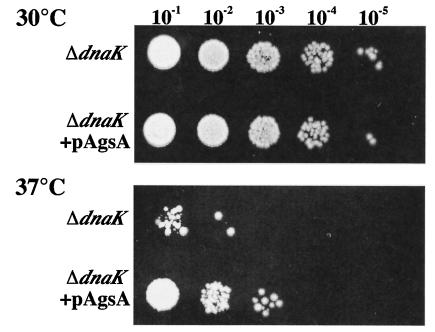
To assess the role of AgsA in the state of thermoresistance, we determined the plating efficiency at a sublethal temperature (37°C) for ΔdnaK52 cells. When ΔdnaK52 cells were incubated at 37°C, the growth of the cells was inhibited, and only about 0.02% of the cells could form colonies. Interestingly, the plating efficiency of a ΔdnaK52 derivative carrying an AgsA-overproducing plasmid at 37°C was increased to 1.7%, suggesting that AgsA could partially complement the thermosensitive phenotype. This phenotype was also confirmed by using a spot test as shown in Fig. 6. These results suggest that AgsA partially compensates DnaK function and assists the growth of ΔdnaK52 cells under sublethal conditions. We observed no significant differences in colony formation between induced and noninduced conditions (data not shown). This could be due to the accumulation of AgsA in cells carrying pTKY605 without an inducer, as indicated in Fig. 7A.
FIG. 6.
AgsA partially suppresses the ΔdnaK52 phenotype. ΔdnaK52 (strain BB1553) and ΔdnaK52 + pAgsA (strain CS5257) cells were grown in L medium at 30°C for 3 h. Various dilutions (10−1 to 10−5) were spotted onto L agar plates supplemented with 250 μM IPTG and were incubated at 30 or 37°C for 24 h.
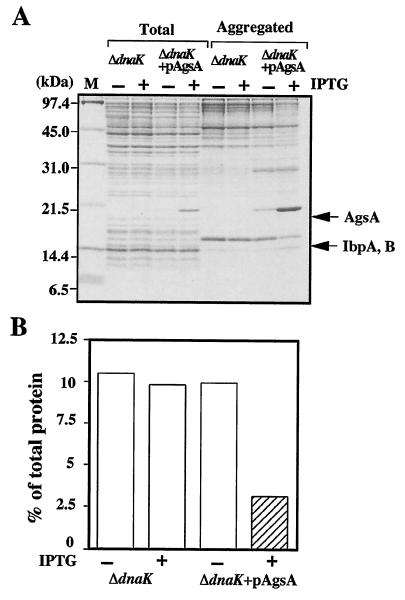
FIG. 7.
AgsA prevents heat-induced aggregation of proteins in the ΔdnaK52 mutant at 42°C. (A) ΔdnaK52 (strain BB1553) and ΔdnaK52 + pAgsA (strain CS5257) cells were grown in L medium that was either left unsupplemented (−) or supplemented with (+) 1 mM IPTG at 30°C for 2 h. Cells were cultured further at 42°C for 1 h. Aggregated proteins were isolated as described in Materials and Methods. Equal amounts of proteins (20 μg) were analyzed by SDS-15% PAGE. (B) The amount of aggregated protein was quantified by Bradford assay and calculated in relation to total protein content (set at 100%). Total, total cell lysate; Aggregated, aggregated proteins; M, molecular weight marker. Each value is the average from at least three different experiments.
Suppression of protein aggregation by AgsA in ΔdnaK52 and ΔrpoH mutants.
The Hsp70 (DnaK) chaperone system is known to refold the denatured proteins efficiently, and the ΔdnaK52 mutation causes a large accumulation of aggregation protein at nonpermissive temperatures (46). This phenotype of the ΔdnaK52 mutant allowed us to analyze the chaperone function of AgsA. Thus, we examined the ability of AgsA protein to prevent protein aggregation in E. coli ΔdnaK52 mutant cells at 42°C (Fig. 7A). Overproduction of AgsA did not perturb cell growth (data not shown). When AgsA was overproduced in ΔdnaK52 cells by IPTG, a large amount of AgsA was produced (around 5% of total proteins) and copurified in the aggregated fraction (around 10% of aggregated proteins). Under nonoverproduced conditions, cells already had significant amounts of AgsA protein (around 0.5% of total proteins), which copurified in the aggregated fraction (around 1% of aggregated proteins). We then examined protein aggregation in ΔdnaK52 cells with and without overproduction of AgpA. Without IPTG, about 10% of total proteins were aggregated in ΔdnaK52 cells at 42°C (Fig. 7B). Interestingly, overproduction of AgsA decreased the amount of aggregated protein to 3% of total proteins at 42°C, suggesting that AgsA is able to prevent protein aggregation. Since the ΔdnaK52 cells have only a small amount of aggregated proteins, less than 1% of total protein at 30°C, we failed to monitor the effect of AgsA overproduction reproducibly.
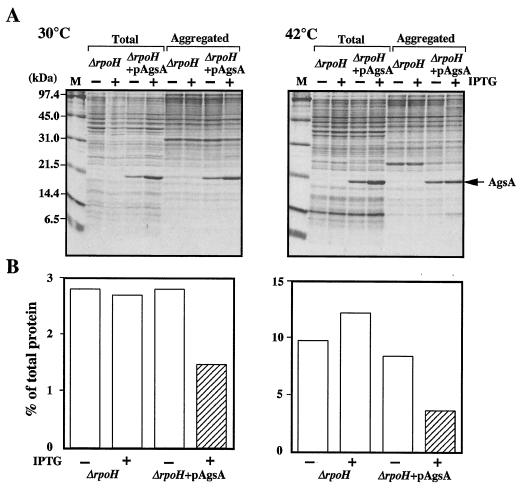
We also examined function of AgsA by overproduction in a ΔrpoH strain (BB7224). This mutant lacks σ32, is therefore largely devoid of all major cytosolic chaperones except for GroEL/GroES, and has lower levels of proteases (25). The ΔrpoH strain carries an insertion element in the promoter region of the groES groEL operon, which drives constitutive expression of this operon and results in fourfold-increased GroEL/GroES levels compared with those in the wild type. This increase in these levels allows ΔrpoH cells to grow, albeit slowly, at temperatures up to 40°C (25). After growth at 30°C, the levels of heat shock proteins relative to those in the wild type were below 1% for DnaK, 20% for HtpG, 10% for IbpB, 20% for ClpB, and about 50% for Lon and HslVU (46). Lack of these chaperones and proteases results in a large accumulation of aggregation protein, that is, about 3 and 10% of total proteins were aggregated at 30 and 42°C, respectively (Fig. 8). It seems that this phenotype allows us to perform fine analysis of the AgsA chaperone function without effects of other chaperones and proteases. When AgsA was overproduced in ΔrpoH cells to the same extent as in ΔdnaK52 cells (5% of total proteins) at 30 and 42°C, large amounts of AgsA were copurified in the aggregated fraction (corresponding to 10% of aggregated proteins) at both temperatures (Fig. 8). In the ΔrpoH cells overproducing AgsA, the amount of aggregated proteins decreased to 42% compared to that in noninduced cells at 42°C (Fig. 8), suggesting the ability of AgsA to suppress protein aggregation in ΔrpoH cells. Interestingly, overproduced AgsA also suppressed protein aggregation at a lower temperature, that is, the amount of aggregated protein decreased to 52% compared to that in noninduced cells at 30°C (Fig. 8). Taken together, our data clearly indicate that the AgsA is capable of protecting cellular proteins against aggregation.
FIG. 8.
AgsA prevents aggregation of proteins in a ΔrpoH mutant. (A) ΔrpoH (strain BB7224) and ΔrpoH + pAgsA (strain CS5262) cells were grown in L medium that was either left unsupplemented (−) or supplemented with (+) 1 mM IPTG at 30°C for 2 h. Cells were cultured further at 30 or 42°C for 1 h. Aggregated proteins were isolated as described in Materials and Methods. Equal amounts of proteins (20 μg) were analyzed by SDS-15% PAGE. (B) The amount of aggregated protein was quantified by Bradford assay and calculated in relation to total protein content (set at 100%). Each value is the average from at least three different experiments.
DISCUSSION
In this study, we discovered a novel sHsp (AgsA) in the aggregated proteins accumulated in Salmonella serovar Typhimurium ΔdnaK cells expressed at high temperatures (Fig. 1). sHsps are classified into two classes, class A and B (34). Class A contains only bacterial proteins, and the similarity is not restricted to the α-crystallin domain but extends into the flanking amino- and carboxy-terminal regions. Class B proteins are much more divergent in length and sequence. They include prokaryotic as well as eukaryotic members from a wide variety of organisms. Homology alignment of AgsA with Salmonella serovar Typhimurium IbpA and IbpB shows weak similarity (32% sequence identity to AgsA for IbpA, and 31% for IbpB), though the homology extends into the flanking regions (data not shown). These findings suggest that AgsA belongs to class A, and it seems to have arisen from the same phylogenetic origin by duplication or amplification at an early evolutionary stage of Enterobacteria (Fig. 3B). Homologues of AgsA were found in the genomes of the K. pneumoniae and in a Buchnera sp. The mutualism between Buchnera and its host is so obligatory that neither organism in the association can reproduce independently. This symbiotic bacterium lost many genes, including essential genes, and depends completely on its host (40). The genome size of the Buchnera sp. is only one-seventh the size of the E. coli genome. Nevertheless, an AgsA homologue is still maintained in Buchnera. These results suggest that the AgsA homologue may have an unknown important function for the symbiosis of Buchnera.
We showed that overproduction of AgsA partially suppressed the ΔdnaK52 thermosensitive phenotype at 37°C. However, AgsA overproduction could not rescue the lethality of the ΔdnaK52 mutant at 42°C and did not result in a significant increase in viability at 50°C (data not shown). These data suggest that AgsA cannot assist in refolding for strongly damaged proteins but can protect the partially damaged proteins and assist their spontaneous refolding. However, AgsA could prevent aggregation of strongly damaged proteins at high temperatures (Fig. 7 and 8). Rajaraman et al. reported that human α-crystallin interacts with unfolded proteins to reactivate them at an early stage of denaturation and to protect them from aggregation at a later stage (38). The function of AgsA may be analogous to that of human α-crystallin. To know the function of sHsps in Salmonella serovar Typhimurium, we examined the effect of a lethal temperature (70°C) on the viability of the wild type and agsA, ibpAB, and agsA ibpAB triple mutants (Fig. 5). Compared to the wild-type strain, an agsA-null mutant exhibited no loss of viability at lethal temperatures. In contrast, an ibpAB mutant showed significantly lower viability at high temperatures. The agsA ibpAB triple mutant exhibited a stronger defect than the ibpAB mutant. These data show that the major contribution to thermotolerance comes from IbpAB, and this advantage may account for the high conservation of IbpAB in Enterobacteriaceae.
It has been reported that the overproduction of E. coli IbpA and IbpB may possibly reduce the amount of aggregated proteins by long exposure (0.5 to 4 h) at an extremely high temperature, 50°C (24). It is also indicated that some sHsps form a stable multimer conformation at low temperatures and cannot associate with denatured proteins (19, 22, 39). Here we showed that overproduction of AgsA could reduce the amount of aggregated proteins even at a permissive temperature (30°C) in ΔrpoH cells. This data suggest that AgsA allows the forming of some active conformation even at low temperatures. Since Salmonella serovar Typhimurium synthesized only a small amount of AgsA at a low temperature (Fig. 4), it is impossible to discuss some important physiological function of AgsA in this condition at present. ΔrpoH cells have only small amounts of heat shock chaperones and proteases except for GroEL/GroES. It is well known that the protein aggregates can be suppressed by the DnaK chaperone system and the GroEL/GroES chaperone (but with lesser effects and to lesser extents than DnaK) and subsequently refolded into the native state. No other chaperones were reported to provide effective prevention of endogenous protein aggregation in the cells. We showed that AgsA overproduction could suppress protein aggregation in ΔrpoH cells. Since ΔrpoH and ΔdnaK52 mutants have the GroEL/GroES chaperone in cells, we cannot exclude the possibility that AgsA may be functional only with GroEL/GroES. However, it is known that GroEL/GroES has limiting folding capacity, and their substrates are believed to be <60 kDa (5, 18). Our data clearly showed that AgsA could protect against aggregation of proteins larger than 60 kDa (Fig. 8). For aggregation prevention, AgsA seems to be functional without the help of another chaperone such as the DnaK-ClpB bichaperone system, GroEL/GroES, HtpG, IbpA, or IbpB.
Acknowledgments
We thank M. Mayer for critical reading of this paper and helpful suggestions. We also express our gratitude to B. Bukau and A. Mogk for providing the anti-IbpB antibody, strain BB1553, and plasmid pDMI,1.
This work was supported by grants-in-aid for scientific research (13470058 to T.Y. and 13771371 to T.T.) from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government.
REFERENCES
- 1.Abu Kwaik, Y., L. Y. Gao, O. S. Harb, and B. J. Stone. 1997. Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol. Microbiol. 24:629-642. [DOI] [PubMed] [Google Scholar]
- 2.Bukau, B., and G. Walker. 1990. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 9:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukau, B. 1993. Regulation of the E. coli heat shock response. Mol. Microbiol. 9:671-680. [DOI] [PubMed] [Google Scholar]
- 4.Bukau, B., F. X. Schmid, and J. Buchner. 1999. Assisted protein folding, p. 3-10. In B. Bukau (ed.), Molecular chaperones and folding catalysts—regulation, cellular function and mechanism. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 5.Bukau, B., E. Deuerling, C. Pfund, and E. A. Craig. 2000. Getting newly synthesized proteins into shape. Cell 101:119-122. [DOI] [PubMed] [Google Scholar]
- 6.Caspers, G. J., J. A. Leunissen, and W. W. de Jong. 1995. The expanding small heat-shock protein family, and structure predictions of the conserved “alpha-crystallin domain.” J. Mol. Evol. 40:238-248. [DOI] [PubMed] [Google Scholar]
- 7.Chuang, S. E., V. Burland, G. Plunkett III, D. L. Daniels, and F. R. Blattner. 1993. Sequence analysis of four new heat-shock genes constituting the hslTS/ibpAB and hslVU operons in Escherichia coli. Gene 134:1-6. [DOI] [PubMed] [Google Scholar]
- 8.Cowing, D. W., J. C. Bardwell, E. A. Craig, C. Woolford, R. W. Hendrix, and C. A. Gross. 1985. Consensus sequence for Escherichia coli heat shock gene promoters. Proc. Natl. Acad. Sci. USA 82:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Jong, W. W., J. A. Leunissen, and C. E. Voorter. 1993. Evolution of the alpha-crystallin/small heat-shock protein family. Mol. Biol. Evol. 10:103-126. [DOI] [PubMed] [Google Scholar]
- 10.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrnsperger, M., S. Graber, M. Gaestel, and J. Buchner. 1997. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 16:221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamer, J., H. Bujard, and B. Bukau. 1992. Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor σ32. Cell 29:833-842. [DOI] [PubMed] [Google Scholar]
- 13.Georgopoulos, C., K. Liberek, M. Zylicz, and D. Ang. 1994. Properties of the heat shock proteins of Escherichia coli and the autoregulation of the heat shock response, p. 209-250. In R. I. Morimoto, A. Tissières, and C. Georgopoulos (ed.), Biology of heat shock proteins and molecular chaperones. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 14.Glover, J. R., and S. Lindquist. 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94:73-82. [DOI] [PubMed] [Google Scholar]
- 15.Goloubinoff, P., A. Mogk, A. P. Zvi, T. Tomoyasu, and B. Bukau. 1999. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. USA 96:13732-13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross, C. A. 1996. Function and regulation of the heat shock proteins, p. 1382-1399. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol 1. ASM Press, Washington, D.C.
- 17.Hartl, F. U. 1996. Molecular chaperones in cellular protein folding. Nature 381:571-580. [DOI] [PubMed] [Google Scholar]
- 18.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 19.Haslbeck, M., S. Walke, T. Stromer, M. Ehrnsperger, H. E. White, S. Chen, H. R. Saibil, and J. Buchner. 1999. Hsp26: a temperature-regulated chaperone. EMBO J. 18:6744-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakob, U., M. Gaestel. K. Engel, and J. Buchner. 1993. Small heat shock proteins are molecular chaperones. J. Biol. Chem. 268:1517-1520. [PubMed] [Google Scholar]
- 21.Kitagawa, M., Y. Matsumura, and T. Tsuchido. 2000. Small heat shock proteins, IbpA and IbpB, are involved in resistances to heat and superoxide stresses in Escherichia coli. FEMS Microbiol. Lett. 184:165-171. [DOI] [PubMed] [Google Scholar]
- 22.Kitagawa. M., M. Miyakawa, Y. Matsumura, and T. Tsuchido. 2002. Escherichia coli small heat shock proteins, IbpA and IbpB, protect enzymes from inactivation by heat and oxidants. Eur. J. Biochem. 269:2907-2917. [DOI] [PubMed] [Google Scholar]
- 23.Knauf, U., H. Bielka, and M. Gaestel. 1992. Over-expression of the small heat-shock protein, hsp25, inhibits growth of Ehrlich ascites tumor cells. FEBS Lett. 309:297-302. [DOI] [PubMed] [Google Scholar]
- 24.Kuczýnska-Wísnik. D., S. Kedzierska, E. Matuszewska, P. Lund, A. Taylor, B. Lipinska, and E. Laskowska. 2002. The Escherichia coli small heat-shock proteins IbpA and IbpB prevent the aggregation of endogenous proteins denatured in vivo during extreme heat shock. Microbiology 148:1757-1765. [DOI] [PubMed] [Google Scholar]
- 25.Kusukawa, N., and T. Yura. 1988. Heat shock protein GroE of Escherichia coli: key protective roles against thermal stress. Genes Dev. 2:874-882. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacterophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Landry, J., P. Chretien, H. Lambert, E. Hickey, and L. A. Weber. 1989. Heat shock resistance conferred by expression of the human HSP27 gene in rodent cells. J. Cell Biol. 109:7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, G. J., A. M. Roseman, H. R. Saibil, and E. Vierling. 1997. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 16:659-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merck, K. B., P. J. Groenen, C. E. Voorter, W. A. de Haard-Hoekman, J. Horwitz, H. Bloemendal, and W. W. de Jong. 1993. Structural and functional similarities of bovine alpha-crystallin and mouse small heat-shock protein. A family of chaperones. J. Biol. Chem. 268:1046-1052. [PubMed] [Google Scholar]
- 31.Mogk, A., T. Tomoyasu, P. Goloubinoff, S. Rudiger, D. Roder, H. Langen, and B. Bukau. 1999. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 18:6934-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morimoto, R. I. 1993. Cell in stress: transcriptional activation of heat shock genes. Science 269:1409-1410. [DOI] [PubMed] [Google Scholar]
- 33.Morimoto, R. I., P. E. Kroeger, and J. J. Cotto. 1996. The transcriptional regulation of heat shock genes: a plethora of heat shock factors and regulatory conditions, p. 139-163. In U. Feige, R. I. Morimoto, I. Yahara, and B. S. Polla (ed.), Stress-inducible cellular responses. Birkhäuser-Verlag, Basel, Switzerland. [DOI] [PubMed]
- 34.Münchbach, M., A. Nocker, and F. Narberhaus. 1999. Multiple small heat shock proteins in Rhizobia. J. Bacteriol. 181:83-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neidhardt, F. C., and R. A. VanBogelen. 1987. Heat shock response, p. 1334-1345. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 2. ASM Press, Washington, D.C.
- 36.Nover, L. 1991. Inducers of HSP synthesis: heat shock and chemical sensors, p. 5-41. In L. Nover (ed.), Heat shock response. CRC Press, Boca Raton, Fla.
- 37.Petko, L., and S. Lindquist. 1986. Hsp26 is not required for growth at high temperatures, nor for thermotolerance, spore development, or germination. Cell 45:885-894. [DOI] [PubMed] [Google Scholar]
- 38.Rajaraman, K., B. Raman, T. Ramakrishna, and C. M. Rao. 2001. Interaction of human recombinant αA- and αB-crystallins with early and late unfolding intermediates of citrate synthase on its thermal denaturation. FEBS Lett. 497:118-123. [DOI] [PubMed] [Google Scholar]
- 39.Shearstone, J. R., and F. Baneyx. 1999. Biochemical characterization of the small heat shock protein IbpB from Escherichia coli. J. Biol. Chem. 274:9937-9945. [DOI] [PubMed] [Google Scholar]
- 40.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 41.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:787-796. [Google Scholar]
- 42.Squires, C. L., S. Pedersen, B. M. Ross, and C. Squires. 1991. ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 173:4254-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takaya, A., T. Tomoyasu, A. Tokumitsu, M. Morioka, and T. Yamamoto. 2002. The ATP-dependent Lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J. Bacteriol. 184:224-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeshita, S., M. Sato, M. Toba, W. Masahashi, and T. Hashimoto-Gotoh. 1987. High-copy-number and low-copy-number plasmid vectors for lacZ α-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63-74. [DOI] [PubMed] [Google Scholar]
- 45.Thomas, J. G., and F. Baneyx. 1998. Roles of the Escherichia coli small heat shock proteins IbpA and IbpB in thermal stress management: comparison with ClpA, ClpB, and HtpG in vivo. J. Bacteriol. 180:5165-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomoyasu, T., A. Mogk, H. Langen, P. Goloubinoff, and B. Bukau. 2001. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40:397-413. [DOI] [PubMed] [Google Scholar]
- 47.Veinger, L., S. Diamant, J. Buchner, and P. Goloubinoff. 1998. The small heat-shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J. Biol. Chem. 269:11032-11037. [DOI] [PubMed] [Google Scholar]
- 48.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto, T., T. Hanawa, S. Ogata, and S. Kamiya. 1996. Identification and characterization of the Yersinia enterocolitica gsrA gene, which protectively responds to intracellular stress induced by macrophage phagocytosis and to extracellular environmental stress. Infect. Immun. 64:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto, T., H. Sashinami, A. Takaya, T. Tomoyasu, H. Matsui, Y. Kikuchi, T. Hanawa, S. Kamiya, and A. Nakane. 2001. Disruption of the genes for ClpXP protease in Salmonella enterica serovar Typhimurium results in persistent infection in mice, and development of persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect. Immun. 69:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yura, T., and K. Nakahigashi. 1999. Regulation of the heat-shock response. Curr. Opin. Microbiol. 2:153-158 [DOI] [PubMed] [Google Scholar]