Abstract
Regulation of transcription of the tryptophanase operon requires that translation of its leader peptide coding region, tnaC, be coupled with its transcription. We show in vitro that a transcription pause site exists at the end of the tnaC coding region and that translation of tnaC releases the paused transcription complex, coupling transcription with translation.
The enzyme tryptophanase catalyzes the degradation of l-tryptophan to indole, pyruvate, and ammonia (15). Bacterial species that have this enzyme can utilize tryptophan as a source of carbon, nitrogen, and energy (8). Indole, in addition to serving as a tryptophan precursor, appears to play other roles in bacterial populations. Recent evidence suggests that it may act as a volatile signal molecule during biofilm formation and quorum sensing (2, 18).
The tryptophanase (tna) operon of Escherichia coli contains two major structural genes, a promoter-proximal gene, tnaA, encoding tryptophanase, and a distal gene, tnaB, encoding a low-affinity tryptophan permease (1, 17). Preceding tnaA in the tna operon is a 319-nucleotide (nt) transcribed regulatory region that contains the coding region for a 24-residue leader peptide, TnaC. The 220-nt spacer sequence that separates the tnaC stop codon from the start codon of tnaA contains several transcription pause sites that can serve as regulated sites of Rho-dependent transcription termination (16, 17). The RNA sequence in the vicinity of the tnaC stop codon is rich in cytidylate residues (Fig. 1A). Such a C-rich sequence, termed a Rho utilization site (rut), is required for efficient Rho-dependent termination in the leader region of the tna operon (3, 5, 9, 12, 13).
FIG. 1.
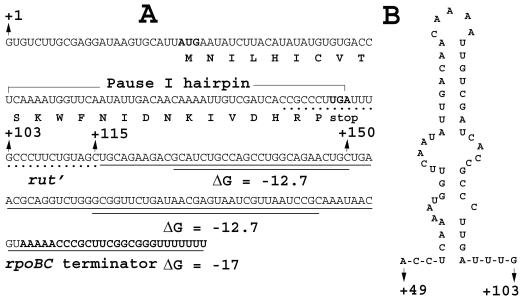
(A) Nucleotide sequence of a segment of the tna leader transcript (from nts +1 to +115) followed by the fused nucleotide sequence containing the rpoBC terminator. The nucleotide sequence at the distal end of the tnaC coding region that forms the pause I hairpin structure and a portion of the Rho factor utilization (rut′) site that is rich in C residues located in the vicinity of the tnaC stop codon are indicated. The rpoBC terminator sequence including a segment of the rpoC coding sequence is underlined. Note that there are predicted hairpin structures that might form in the segment of rpoBC coding sequence (underlined twice); the free energy of formation of each structure is shown. (B) Predicted RNA hairpin structure (pause I) formed by the end of the transcript of the tnaC coding region. The free energy of formation of this structure is −9.6 kcal/mol.
Transcription of the structural genes of the tna operon is regulated by a novel mechanism of transcription attenuation, tryptophan-induced transcription antitermination (7, 17). Synthesis of the tryptophan-containing leader peptide TnaC and the presence of high levels of free tryptophan are both required for induction (17). The combined action of the uncleaved nascent TnaC-peptidyl-tRNA, retained in the translating ribosome, and ribosome-bound tryptophan inhibits peptidyltransferase cleavage of the nascent TnaC-peptidyl-tRNA at the tnaC stop codon (4). Therefore, the uncleaved TnaC-peptidyl-tRNA remains associated with the translating ribosome. This ribosome, stalled at the tnaC stop codon, blocks Rho factor's access to its rut binding site on the tna leader transcript, thereby permitting the polymerase paused downstream to resume transcription before Rho can bind and cause transcription termination (5). Recent studies suggest that the tryptophan binding or induction site in the ribosome may be the site normally occupied by the aminoacyl moiety of a charged tRNA during translation (6).
For tryptophan induction of the tna operon to occur, transcription and translation of the operon must be coordinated so that a translating ribosome may stall at the tnaC stop codon before RNA polymerase completes transcription of the leader region. This stalling would prevent Rho factor's access to the rut site in the leader transcript, thereby preventing transcription termination. The findings in previous studies (6) mentioned above do not explain how transcription and translation are coupled during transcription of the tna leader region. Of potential relevance, it was established in studies with the biosynthetic trp operon of E. coli that transcription pausing and ribosome release of the paused polymerase complex are the crucial events that establish that ribosome movement on the trp operon transcript and polymerase movement in the leader region of the operon proceed in unison (10). We reasoned that a similar mechanism might be operating in the tna operon; this would ensure that a ribosome would be engaged in translating tnaC before the transcribing RNA polymerase resumed transcription and synthesized the transcript segment that serves as the Rho factor binding site. Early studies using a purified transcription system did demonstrate the existence of several transcription pause sites in the tna leader region (16). One of these pause sites generated a major short pause RNA species, 103 nt in length (16). Nucleotides at positions 52 through 99 of this tna leader transcript were predicted to form a relatively stable hairpin structure (ΔG = −9.6 kcal/mol; Fig. 1B); this hairpin structure was proposed to be responsible for the transcription pause producing the 103-nt RNA species (16). Since the nucleotides forming this hairpin structure are located at the distal end of the tnaC coding region (Fig. 1A), if the paused RNA polymerase remained at this site until released by the ribosome translating tnaC, synchronization of transcription and translation would be achieved. The present study was performed to examine this possibility.
An S-30 system permitting transcription and translation of the tna operon and its regulation was employed in these analyses; the S-30 reaction conditions used have been described elsewhere (7). An appropriate tna operon promoter-leader region template was constructed. This DNA template was a self-ligated PCR fragment containing the region from positions −195 to +115 of the tna operon (relative to the tna transcription initiation site), followed by the attached rpoBC transcription terminator sequence (Fig. 1A) (7). Thus, in this template, the intact tna promoter was present, but the multiple transcription pause sites (7, 16) located between tnaC and tnaA coding sequences were removed and replaced by the rpoBC terminator and adjacent sequences. This template allowed us to focus on the role of the transcription pause that occurs at the end of tnaC; this presumed RNA hairpin structure is shown in Fig. 1B.
To monitor RNA synthesis in the S-30 extract, UTP was replaced by [α-33P]UTP. To enhance transcription pausing, neither CTP nor GTP was added to the S-30 system; low levels of CTP and GTP are present in the S-30 extract. We compared the rate of transcription under these conditions with the rate under the conditions we used before in reconstituting regulation of the tna operon expression in vitro (7). Previously 200 μM (each) CTP and GTP was present in the S-30 reaction mixture, and only a low concentration of labeled UTP was added. We observed no significant difference in the rate of transcription when we performed reactions with or without added CTP and GTP, suggesting that the low CTP and GTP concentrations in our S-30 extract are sufficient to allow transcription to proceed. Presumably, this is so because the labeled UTP is limiting in both reactions. Bicyclomycin, at 50 μg/ml, was generally added to each reaction mixture, as a specific inhibitor of transcription termination factor Rho (11). Since the rpoBC terminator is an intrinsic terminator, neither Rho factor nor bicyclomycin should have an effect on termination at the rpoBC site. Reaction mixtures were preincubated with bicyclomycin at 37°C for 10 min, and then 100 μg of rifampin (an inhibitor of transcription initiation) per ml and 2 μl of [α-33P]UTP (10 μCi/μl; Perkin-Elmer) were added. Time course samples were then taken to monitor transcription and transcription pausing. When used, the translation inhibitor chloramphenicol or puromycin was added together with the [α-33P]UTP.
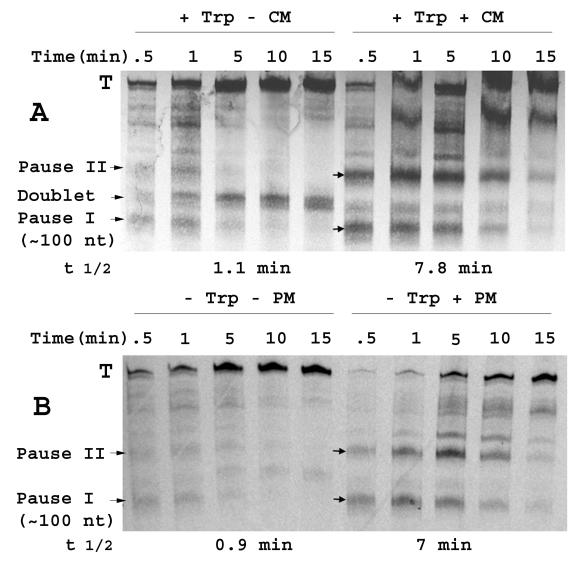
Using a modified single-round transcription approach (19) with our S-30 system, a short RNA species (Fig. 2A, pause I) was evident, particularly in the presence of a translation inhibitor (Fig. 2). This pause RNA species comigrated with a 100-nt RNA marker (Ambion, Austin, Tex.) run on the same urea-polyacrylamide gel (marker not shown). Also apparent was the ∼120-nt doublet RNA species produced only when inducing levels of tryptophan were present (7) (Fig. 2A, doublet). The RNA doublet is thought to be produced by nuclease attack on the mRNA, 3′ of the stalled ribosome, and partial degradation of the ribosome-bearing terminated transcripts (7). Consistent with this view, the intensity of the RNA doublet on the gel increased with time in this single-round transcription assay. In contrast, the intensity of the pause I RNA species decreased with time, as the RNA species terminating at the rpoBC terminator (Fig. 2A, T band) became more prominent. The half-life of pause I RNA detected in the reaction performed without added chloramphenicol was estimated as 1.1 min (Fig. 2A). Due to the presence of low levels of nucleoside triphosphates in our S-30 extract, we were unable to synchronize transcription initiation in our cell-free system. Thus, the modified single-round transcription approach used in our experiments differs from the single-round transcription procedure that allows synchronized transcription initiation (19). Thus, the apparent RNA pause half-life that can be estimated from our data is likely to be an overestimate of the true pause half-life.
FIG. 2.
(A) Single-round transcription analyses examining the effects of chloramphenicol on transcription pausing. The DNA template used to direct the S-30 system was a self-ligated PCR fragment containing the region from positions −195 to + 115 of the tna operon (relative to the tna transcription initiation site), followed by the rpoBC transcription terminator sequence. The corresponding mRNA sequence is shown in Fig. 1A. S-30 reaction mixtures (50-μl volume) with 1 mM tryptophan and 50 μg of bicyclomycin per ml but without CTP, GTP, and UTP were incubated at 37°C for 10 min. Then 20 μCi of [α-33P]UTP and 200 μg of rifampin per ml with (+) or without (−) 200 μg of chloramphenicol (CM) per ml were added together to the reaction mixture. Samples were taken at the indicated time points, the reactions were stopped by phenol extraction, and the samples were loaded on a urea-polyacrylamide RNA gel. Pauses that generate RNA species longer than 115 nt (pause II, etc.) correspond to hairpin structures that could form in the transcript produced from the incorporated rpoBC terminator (Fig. 1A); these pauses are not directly relevant to this study. The positions of the terminated (T) transcripts, RNA doublet, and transcript produced by pausing at the end of the tnaC coding region (pause I), are indicated. The half-life (t 1/2) of pause I was measured using a phosphorimager (7). (B) Effect of puromycin addition on transcription pausing. Single-round assays were conducted as described above for panel A in the absence of a high level of tryptophan. In addition, chloramphenicol was replaced by 200 μg of puromycin (PM) per ml, where indicated. The half-life (t 1/2) of pause I was measured using a phosphorimager (7).
Next we examined the effects of inhibiting tnaC translation on the stability of pause I RNA in the presence of inducing levels of added tryptophan. We found that when TnaC synthesis was inhibited by the addition of chloramphenicol, the initial level of pause I RNA detected was increased (Fig. 2A). The half-life of this pause I RNA increased significantly, from 1.1 min in the absence of chloramphenicol to 7.8 min in its presence (Fig. 2A). The RNA doublet was intense only when TnaC synthesis was allowed in the presence of inducing levels of tryptophan (Fig. 2A) (7). The role of inducing levels of l-tryptophan in stalling the ribosome translating TnaC has been established (6, 7).
Similar results were obtained with another translation inhibitor, puromycin, in comparable single-round transcription analyses performed in the absence of added tryptophan. Figure 2B shows the effect of puromycin addition on the stability of the pause I complex; its half-life increased from 0.9 min without puromycin to 7 min with puromycin. The RNA doublet band was not evident (Fig. 2B versus Fig. 2A), because tryptophan was not added to these reaction mixtures. We noticed a prominent ∼150-nt pause RNA species, pause II, migrating between pause I and the terminated transcripts, with either chloramphenicol or puromycin as an inhibitor (Fig. 2). This species exhibited a pattern of formation similar to that of pause I RNA in that its presence depended on the addition of a translation inhibitor. However, the intensity of pause I RNA was reduced with time in this single-round transcription assay, while by contrast, the intensity of this pause II RNA species initially increased with time before decaying. Apparently, RNA polymerase paused at site II ultimately elongates to terminate at the rpoBC terminator. We could not calculate the stability of the pause complex producing this RNA species, because RNA polymerases producing the pause I RNA species continued elongating to this pause site. Examination of the sequence of the rpoBC terminator fragment incorporated into our template revealed that two stem-loop hairpin structures could theoretically form after the pause I RNA structure (Fig. 1A). We believe these hairpin structures are responsible for the two major bands we see on the gel in addition to pause I RNA and the terminated RNA species (Fig. 2). It is possible that the pause at site I is responsible for the pause at site II. Perhaps the formation of pause I RNA prevents the formation of a third hairpin structure that would prevent the formation of pause II RNA. This third structure might contain a segment of the 3′ strand of pause I and a segment of the 5′ strand of pause II. Such a theoretical hairpin structure does in fact exist (not shown). Pause II RNA (∼150 nt) corresponds to one of the hairpin structures predicted for the rpoBC-associated sequences (Fig. 1A).
In other experiments, we tested an analogous DNA template in which the tnaC start codon, ATG, was replaced by a stop codon, TAG. This template gave a pause I RNA complex with a half-life of 6.5 min in the single-round transcription assay. Moreover, addition of chloramphenicol or puromycin to the reaction mixture did not further increase the stability of the pause I complex (data not shown). Thus, eliminating translation of tnaC by start codon replacement also increased pause I RNA half-life, but here addition of chloramphenicol or puromycin had no effect.
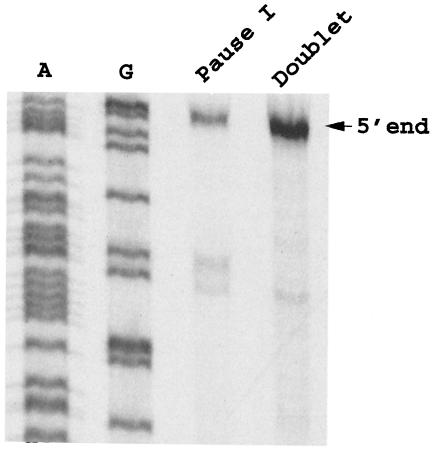
The 5′ end of the ∼100-nt RNA species of the pause I complex was determined by primer extension (14). Pause I RNA and the doublet RNA species from the gels shown in Fig. 2A were recovered, and their 5′ ends were determined. The oligonucleotide used in the primer extension reaction mixtures was complementary to nts +84 to +99 of the tna transcript. The 5′-end nucleotide of the pause 1 RNA species was the same as that of the doublet. The 5′ end corresponds to nucleotides at the tna transcription initiation site (Fig. 3). Therefore, we believe that the 3′ end of the pause I RNA is approximately nt 103. The 3′ end of what we believe is the same pause RNA species was determined previously using a purified transcription system (16).
FIG. 3.
Primer extension analysis designed to determine the 5′ ends of pause I RNA and the doublet RNA. Pause I RNA and the doublet RNA species from the gels shown in Fig. 2A were cut out and eluted, and their 5′ ends were determined by a primer extension approach. The oligonucleotide used in the primer extension reactions is complementary to nts +84 to +99 of the tna transcript. Dideoxy sequencing ladders were obtained with the same primer used for primer extension analysis. Lanes A and G are the Sequenase reaction lanes for dideoxyadenine and dideoxyguanidine, respectively.
On the basis of previous in vivo and in vitro studies, the following model is proposed to explain the events involved in transcription termination and antitermination regulation of the tna operon of E. coli. Tryptophan induction (antitermination) is believed to be achieved by the inhibition of Rho factor-dependent transcription termination at one or more transcription pause sites in the leader region of the operon. Tryptophan is thought to act by binding to the translating ribosome bearing the nascent TnaC-peptidyl-tRNA and inhibiting release factor-mediated cleavage of this peptidyl-tRNA. Inhibition of release factor cleavage would inhibit ribosome release, and the stalled ribosome would therefore block the ability of the Rho factor to bind to its rut attachment site adjacent to the tnaC stop codon. Inhibition of Rho action would result in antitermination. A key assumption in this model is that translation of the tnaC coding region must be closely coupled to transcription of the leader region so that the translating ribosome can block Rho binding.
In this report, we demonstrate that RNA polymerase does pause at the end of the tnaC coding region, allowing time for a ribosome to bind to the transcript and begin translation. We also show that the translating ribosome decreases the lifetime of this transcription pause complex. Pause release thus allows the paused RNA polymerase to resume transcription. This coupling of translation with transcription would therefore permit the translating ribosome to reach the tnaC stop codon and determine whether the Rho factor could gain access to the transcript. Preventing completion of translation of tnaC significantly prolonged the half-life of the critical transcription pause that occurs at the end of the tnaC coding region. We recognize that the artificial conditions we used for transcription and translation in the in vitro S-30 system are substantially different from the conditions that exist in vivo. However, the finding that the ribosome synthesizing TnaC can release the critical transcription pause complex in vitro strongly supports a role for the translating ribosome in achieving coupling of transcription and translation in the tna leader region. Thus, transcription pausing, ribosome-mediated pause release, and tryptophan-induced ribosome stalling are all essential events in the mechanism of regulation of tna operon expression. This coupling resembles the coupling of transcription with translation found to be essential in studies on attenuation regulation of the trp biosynthetic operon of E. coli (10). Transcription pausing in a transcribed leader region has also been proposed to be involved in attenuation regulation of the trp operon of Bacillus subtilis, although in this example transcription is coupled with the binding of a regulatory protein (20).
Acknowledgments
We thank Deirdre Fahy for helpful comments on the manuscript.
This work was supported in part by a grant from the National Science Foundation (MCB-0093023) (to C.Y.).
REFERENCES
- 1.Deeley, M. C., and C. Yanofsky. 1981. Nucleotide sequence of the structural gene for tryptophanase of Escherichia coli K-12. J. Bacteriol. 147:787-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Martino, P., A. Merieau, R. Phillips, N. Orange, and C. Hulen. 2002. Isolation of an Escherichia coli strain mutant unable to form biofilm on polystyrene and to adhere to human pneumocyte cells: involvement of tryptophanase. Can. J. Microbiol. 48:132-137. [DOI] [PubMed] [Google Scholar]
- 3.Gollnick, P., and C. Yanofsky. 1990. tRNATRP translation of leader peptide codon 12 and other factors that regulate expression of the tryptophanase operon. J. Bacteriol. 172:3100-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong, F., K. Ito, Y. Nakamura, and C. Yanofsky. 2001. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNAPro. Proc. Natl. Acad. Sci. USA 98:8997-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong, F., and C. Yanofsky. 2002. Analysis of tryptophanase operon expression in vitro: accumulation of TnaC-peptidyl-tRNA in a release factor 2 depleted S-30 extract prevents Rho-factor action, simulating induction. J. Biol. Chem. 277:17095-17100. [DOI] [PubMed] [Google Scholar]
- 6.Gong, F., and C. Yanofsky. 2002. Instruction of translating ribosome by nascent peptide. Science 297:1864-1867. [DOI] [PubMed] [Google Scholar]
- 7.Gong, F., and C. Yanofsky. 2001. Reproducing tna operon regulation in vitro in an S-30 system. Tryptophan induction inhibits cleavage of TnaC peptidyl-tRNA. J. Biol. Chem. 276:1974-1983. [DOI] [PubMed] [Google Scholar]
- 8.Hopkins, F. G., and S. W. Cole. 1903. A contribution to the chemistry of proteid. Part II. The constitution of tryptophane, and the action of bacteria upon it. J. Physiol. (London) 29:451-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konan, K. V., and C. Yanofsky. 2000. Rho-dependent transcription termination in the tna operon of Escherichia coli: roles of the boxA sequence and the rut site. J. Bacteriol. 182:3981-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landick, R., J. Carey, and C. Yanofsky. 1985. Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc. Natl. Acad. Sci. USA 82:4663-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magyar, A., X. Zhang, F. Abdi, H. Kohn, and W. R. Widger. 1999. Identifying the bicyclomycin binding domain through biochemical analysis of antibiotic-resistant Rho proteins. J. Biol. Chem. 274:7316-7324. [DOI] [PubMed] [Google Scholar]
- 12.Platt, T. 1994. Rho and RNA: models for recognition and response. Mol. Microbiol. 11:983-990. [DOI] [PubMed] [Google Scholar]
- 13.Richardson, J. P. 2002. Rho-dependent termination and ATPases in transcript termination. Biochim. Biophys. Acta 1577:251-260. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Snell, E. E. 1975. Tryptophanase: structure, catalytic activities, and mechanism of action. Adv. Enzymol. 42:389-446. [DOI] [PubMed] [Google Scholar]
- 16.Stewart, V., R. Landick, and C. Yanofsky. 1986. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J. Bacteriol. 166:217-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart, V., and C. Yanofsky. 1985. Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coli K-12. J. Bacteriol. 164:731-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, D., X. Ding, and P. N. Rather. 2001. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183:4210-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winkler, M. E., and C. Yanofsky. 1981. Pausing of RNA polymerase during in vitro transcription of the tryptophan operon leader region. Biochemistry 20:3738-3744. [DOI] [PubMed] [Google Scholar]
- 20.Yakhnin, A. V., and P. Babitzke. 2002. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism in vitro. Proc. Natl. Acad. Sci. USA 99:11067-11072. [DOI] [PMC free article] [PubMed] [Google Scholar]