Abstract
In this study, we identify and characterize a novel transmembrane adaptor protein, designated Lck-interacting membrane protein (LIME), as a binding partner of the Lck Src homology (SH)2 domain. LIME possesses a short extracellular domain, a transmembrane domain, and a cytoplasmic tail containing five tyrosine-based motifs. The protein is primarily expressed in hematopoietic cells and lung. Interestingly, LIME expression is up-regulated by TCR stimulation and sustained up to 24 h, suggesting that LIME acts throughout the early to late stages of T cell activation. LIME is localized to membrane rafts and distributed within the T cell–APC contact site. Upon TCR stimulation of Jurkat T cells, LIME associates with Lck as a tyrosine-phosphorylated protein. Experiments using Jurkat T cells expressing CD8–LIME chimera reveal that the protein associates with phosphatidylinositol 3-kinase, Grb2, Gads, and SHP2, and activates ERK1/2 and JNK but not p38. Moreover, overexpression of LIME in Jurkat T cells induces transcriptional activation of the IL-2 promoter. Our data collectively show that LIME is a raft-associated transmembrane adaptor protein linking TCR stimuli to downstream signaling pathways via associations with Lck.
Keywords: T cell receptor, signal transduction, lipid rafts, immunological synapse, T cell activation
Introduction
Engagement of the TCR triggers a signaling cascade involving protein tyrosine kinases (PTKs) (1). Membrane-proximal TCR signaling comprises redistribution and activation of the Src kinases, Lck and Fyn, phosphorylation of immunoreceptor tyrosine-based activation motifs (ITAM) within the CD3 complex, and subsequent recruitment of the tandem SH2 domain–containing tyrosine kinase, ZAP-70, to phosphorylated CD3-ζ ITAM. Activated ZAP-70 phosphorylates specific downstream substrates (2).
Recent studies on TCR-mediated signaling events have focused on the roles of glycolipid-enriched microdomains or detergent-insoluble rafts, which are characterized by enrichment with sphingolipids, cholesterol, and signaling proteins. The functional importance of membrane compartmentation in immune receptor signaling is confirmed by the report that disruption of rafts due to the depletion of membrane cholesterol results in attenuation of immune receptor–mediated calcium mobilization (3). A variety of signaling molecules are concentrated in rafts, including the Src family tyrosine kinase Lck. Upon TCR stimulation, ZAP-70 translocates from the cytoplasm to rafts and phosphorylates a transmembrane adaptor protein, linker for activation of T cells (LAT) (4). Phosphorylation of LAT provides a platform for the accumulation of critical signaling molecules, such as Grb2, Gads, phospholipase C (PLC)-γ, and phosphatidylinositol 3-kinase (PI3K) (5, 6, 7). Other members of the transmembrane adaptor protein group, including TCR-interacting molecule (TRIM) (8), SHP2-interacting transmembrane adaptor protein (SIT) (9), and phosphoprotein associated with glycosphingolipid-enriched microdomains (PAG)/Csk-binding protein (Cbp) (10, 11) additionally appear to play roles in T cells, although their exact functions remain to be established. Non–T cell activation linker (NTAL) (12) and linker for activation of B cells (LAB) (13) were reported recently as novel transmembrane adaptor proteins expressed in B cells but not T cells. These findings suggest that in immune receptor–mediated proximal signaling regulation at the level of transmembrane adaptors determines the outcome of downstream signaling events.
The various functions of Lck in T cells are implemented via both the catalytic activity of the kinase domain and protein–protein interactions through the regulatory SH2 and SH3 domains (14). In the resting state, the SH2 domain of Lck binds a COOH-terminal phosphotyrosine (pY505), resulting in a catalytically inactive conformation. Upon T cell activation, pY505 is dephosphorylated and subsequently the SH2 domain becomes free. The free SH2 domain is capable of interacting with tyrosine-based motifs on other signaling molecules, including Lad/RIBP (15, 16). The importance of the SH2 domain in T cell activation is further supported by the finding that a kinase-inactive Lck mutant is still able to activate signaling events, leading to IL-2 gene expression. In addition, deletion of SH2 results in abrogation of the transforming activity of Lck, confirming the importance of this domain in Lck function (17).
Accumulating studies report the possibility that specific subsets of signaling molecules are phosphorylated by individual PTK proteins involved in TCR signaling, namely Lck, Fyn, and ZAP-70. Downstream substrates of ZAP-70, such as SLP-76 and LAT, contain several phosphotyrosine sites and protein–protein interaction domains involved in the recruitment of other signaling molecules (18). Several reports suggest that Fyn-binding protein (FYB) and SKAP55 are specifically involved in Fyn signaling cascades in T cells. FYB associates with Fyn and serves as a binding partner for two Fyn-binding proteins, SKAP55 and SKAP55R (SKAP55-related protein), which are substrates for Fyn kinase in T cells (19, 20). However, the specific contribution of each PTK and its substrates to the downstream TCR signaling pathway remain to be determined.
Here, we describe the cDNA cloning and characterization of a 30-kD phosphoprotein designated Lck-interacting membrane protein (LIME) that interacts with the SH2 domain of Lck. LIME is a novel raft-associated transmembrane adaptor protein with five phosphotyrosine sites in the cytosolic domain, which is preferentially expressed in hematopoietic cells. Expression of this protein is up-regulated by TCR stimulation. We demonstrate that LIME is a substrate of Lck and a binding partner of the SH2 domain, both in vitro and in vivo. Importantly, LIME associates with signaling molecules such as PI3K, SHP2, Gads, and Grb2 and activates ERK and JNK and the IL-2 promoter. Based on these results, we propose that LIME is an important component of membrane-proximal TCR-mediated signaling.
Materials and Methods
Cloning of LIME cDNA.
All bait plasmids employed for the yeast two-hybrid system are described in earlier reports (15, 21, 22). LckSH2K (F505) encoding the SH2 and kinase domains (F505, the constitutively active form) was used as bait for yeast two-hybrid screening of a murine T cell lymphoma cDNA library (CLONTECH Laboratories, Inc.). Plasmids were transformed using the lithium acetate method (23). Isolation of positive clones and subsequent analyses were performed as described previously (15). Subsequently, to obtain cDNA encoding the whole open reading frame, a mouse lung cDNA library in λ-gt10 (CLONTECH Laboratories, Inc.) was screened using a 750-bp fragment of LIME cDNA isolated by yeast two-hybrid screening. After screening of 4 × 106 plaques, eight positive clones were selected, and the nucleotide sequences were determined.
Plasmids.
The expression plasmid encoding LIME, pLIME–FLAG, was constructed by PCR and subsequent insertion of the corresponding fragment into pcDNA3.1 (Invitrogen). The 3′ primer contained sequence encoding FLAG tag. The primers also had BamHI (5′ primer and 3′ primer) site extension to facilitate the subcloning. The PCR product was digested with BamHI and inserted into corresponding sites in pcDNA3.1(+) (CLONTECH Laboratories, Inc.). The expression plasmid encoding the dominant negative form of LIME (lacking aa 1–82 including transmembrane domain), pFLAG–LIMEΔN, was also constructed in the same way. For expression vector encoding LIME–green fluorescent protein (GFP), the open reading frame was inserted into pEGFP-N1 (CLONTECH Laboratories, Inc.). The point mutants of LIME were generated by substituting individual tyrosine for phenylalanine using a Quick mutagenesis kit (Stratagene). Expression plasmid encoding CD8–LIME chimera was constructed following the method described previously (24). Fragments corresponding to human CD8 α chain (aa 1–188) and LIME cytosolic region (aa 28–269) were generated by PCR. Primers encoding the 3′ sequences of the CD8 fragment and the 5′ sequences of the FLAG-tagged LIME fragment were designed to overlap, such that annealing of the two products yielded a hybrid template. From this template, the chimera was amplified using external primers containing XbaI sites. The CD8–LIME chimera was inserted into pcDNA3.1(+). pcDNA1–Lck (WT), pcDNA1–Lck (F505), pcDNA1–Lck (A273), and glutathione S-transferase (GST)–Lck SH2 constructs were described previously (15). The reporter plasmid pGL3/IL-2–luc contains the luciferase reporter gene downstream from the IL-2 promoter region including 548 bp 5′ of the transcription start site and was described previously (15).
Northern Blot Analysis.
Mouse and human multiple tissue Northern blots (CLONTECH Laboratories, Inc.) were probed with a [32P]dCTP-labeled PCR product encompassing the full open reading frame of LIME. Hybridization was performed at 42°C overnight in buffer containing 50% deionized formamide, 0.5% SDS, 5× SSC, 5× Denhardt's solution, and 0.1 μg/ml salmon sperm single-stranded DNA.
Isolation of Murine Tissue and Primary T Cells.
Thymocytes, splenocytes, and lymphocytes were isolated from Balb/c mice. Resting CD4+ and CD8+ lymphoid T cells were isolated by positive selection using MACS.
Cells and Transfection.
Cell lines were obtained from American Type Culture Collection. Jurkat T (E6.1) and Raji B cell lines were maintained in RPMI1640 medium supplemented with 10% FBS. 293T cells were grown in DME containing 10% FBS, plated at a density of 2 × 106 cells per 100-mm dish, and transfected with 15 μg DNA using the calcium phosphate method (25). 48 h after transfection, cells were harvested and used for immunoprecipitation and Western blot analyses. Jurkat T cells were transfected by electroporation (BTX-co.). Cells (1.5 × 107) were combined with 20 μg DNA in an electroporation cuvette, mixed gently, pulsed once at 240 V for 25 ms, transferred into 10 ml complete medium, and incubated overnight at 37°C for further processing. Jurkat T cells stably expressing the CD8–LIME chimera were established by selection in the presence of 1 mg/ml G418. Stable cells were activated with 5 μg/ml anti-CD8 antibody in a total volume of 1 ml.
T Cell Activation.
Primary T cells were stimulated by cross-linking with the combination of anti-CD3 and anti-CD28 antibodies to provide both of TCR and costimulatory signals. Jurkat T cells were stimulated by cross-linking with anti-CD3 antibody.
Antibodies.
Polyclonal rabbit anti-Lck serum was generated by immunizing rabbits with GST-conjugated Lck (aa 66–224). Polyclonal anti-LIME antisera were generated against either GST fusion proteins or synthetic peptides. The first method involved production of polyclonal rabbit and mouse anti-LIME sera by immunizing with GST-fused LIME (aa 159–269). The second method involved immunization of rabbits with KLH-conjugated synthetic peptides. Out of the six different peptides tested, only one (GPLENVYESIKEMGL) (aa 255–269) effectively generated antiserum that was capable of detecting LIME on a Western blot. The other antibodies used in this study were obtained from commercial sources, including monoclonal anti-Lck, anti-Gad, and antiphosphotyrosine (4G10) (Upstate Biotechnology), anti-LAT, anti–ZAP-70, anti-PI3K P85, anti-Grb2, and anti-SHP2 (Santa Cruz Biotechnology, Inc.), anti-CD8, anti–mouse IgG, TRITC–anti–mouse IgG, anti-CD3, and anti-CD28 (BD Biosciences), FITC-CTx B subunit and antitalin (Sigma-Aldrich), anti–phospho-ERK1/2, anti–phospho-JNK, anti–phospho-p38, anti-ERK, and anti-JNK (Cell Signaling), and anti–Flag M2 (Eastman Kodak Co.) antibodies.
Immunoprecipitation and Western Blot Analysis.
For immunoprecipitation, cells were lysed in TNE buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1 mM Na3VO4, 5 mM NaF, 25 μg/ml aprotinin, 1 mM PMSF, 25 μg/ml leupeptin, and 1 mg/ml BSA) containing 1% Brij 35 or 1% Digitonin. After incubation for 1 h on ice, insoluble material was removed by centrifugation at 12,000 rpm for 20 min at 4°C. Subsequently, supernatant samples were precleared for 1 h with normal rabbit serum coupled to protein A agarose or mouse IgG coupled to agarose (Sigma-Aldrich) and subjected to immunoprecipitation with the appropriate antibodies and protein A agarose. Western blot analysis was performed using the ECL detection system. (Amersham Biosciences).
Purification of the Raft Fraction.
CD3-stimulated Jurkat cells (5 × 107) were lysed in 1 ml TNE buffer (25 mM Tris, 150 mM NaCl, 5 mM EDTA) containing 1% Triton. Lysates were homogenized with 10 strokes of a dounce homogenizer, mixed with 1 ml of 80% sucrose in TNE buffer, and transferred to a centrifuge tube. Samples were overlaid with 2 ml of 30% sucrose and 1 ml of 5% sucrose in TNE. After centrifugation for 16 h at 40,000 rpm in a Beckman Coulter SW41Ti rotor, 10 0.4-ml fractions were collected from the top of the gradient. Each fraction was precipitated with TCA and washed with 70% EtOH.
Immunofluorescense Staining and Confocal Laser Scan Microscopy.
Jurkat T cells were transfected by electroporation (BTX co.). After incubation for 20 h and washing with PBS, cells were fixed by immersion in 3.7% formaldehyde (Sigma-Aldrich) at room temperature for 15 min and subsequently in prechilled MeOH. Fixed cells were transferred to chamber slides and immediately air dried. Cells were incubated with anti-FLAG antibody (1:100 dilution) in 3% BSA for 30 min at 30°C. Subsequently, slides were exposed to TRITC-conjugated mouse secondary antibody and incubated for 30 min. After mounting, photographs were taken using a LSM510 Confocal Microscope (Carl Zeiss MicroImaging, Inc.). As a control, FITC–cholera toxin B subunits were added after fixation for GM1 staining.
T Cell–APC Conjugates Were Formed Using Raji B Cells as APC.
Jurkat T cells were transfected with expression plasmids encoding LIME–GFP or empty vector. Prior to conjugation, Raji B cells were incubated in RPMI1640 with 5 μg/ml SEE (Toxin Technology) at 37°C for 1 h. Jurkat T cells and Raji B cells were mixed at a 1:1 ratio and density of 106 cells/ml, immediately transferred to poly-l-lysine–coated slides, and incubated at 37°C for 15 min. Slides were fixed in 3.7% formaldehyde for 20 min at room temperature and processed as described above.
Metabolic Labeling of 293T Cells.
293T cells were transiently transfected with LIME expression plasmids in 6-well plates. 24 h after transfection, cells were labeled with 0.5 mCi 3H-palmitate in 1 ml DMEM containing 5% FCS and 5 mM sodium pyruvate for 4 h. Cell lysates were fractionated on SDS-PAGE gel, which was fixed, treated with Enlightening (Dupont) for 30 min, dried, and exposed to x-ray film for 4 wk.
In Vitro Binding.
FLAG–LIME was translated in vitro (Promega), phosphorylated using 40 units of purified Lck protein (Upstate Biotechnology Inc.) in kinase assay buffer (50 mM, Tris, pH 7.5, 1 mM DTT, 10 mM MnCl2, 50 mM NaCl, 0.1 mM ATP), and incubated with purified GST or GST–Lck SH2 proteins coupled to glutathione-sepharose beads. After washing, eluted proteins were separated by SDS-PAGE and subjected to Western analysis with anti-FLAG antibody.
Luciferase Assay.
Jurkat T cells (106) were cotransfected with appropriate combinations of IL-2–luc reporter construct and expression plasmids. After incubation for 24 h, cells were activated by further incubation for 14 h on a 35-mm dish coated with 5 μg/ml anti-CD3 antibody (UCHT1) and harvested. Luciferase activity was determined three times in duplicate for each experiment with a Berthold luminometer LB953. To control for the transfection efficiency, pCMV–β-gal plasmids were cotransfected as test plasmids and the luciferase activity was normalized by β-galactosidase activity (Promega).
Results
Identification of LIME as a Protein Binding to the SH2 Domain of Lck.
To isolate molecules that associate with the SH2 domain of Lck in a phosphorylation-dependent manner, a previously established tyrosine phosphorylation–dependent yeast two-hybrid screening system was used (15, 22). Using this system, we employed part of Lck encompassing the SH2 and kinase domains (SH2K) as bait. After screening 2 × 106 independent clones of a murine T cell lymphoma cDNA library, ∼70 positive clones were isolated. Here, we characterize a protein detected four times out of the 70 positive clones, designated LIME.
To further assess the binding specificity of LIME to Lck, we employed various control plasmids as described in Fig. 1 A. Although LIME interacted with Lck–SH2K (F505), the protein did not bind either SH2 or kinase domain independently, implying that association of LIME with Lck requires both SH2 and kinase activity. Moreover, LIME did not associate with either the SH2 mutant (SH2K [K154/F505]) that is incapable of binding the phosphotyrosine-based motif or a catalytically inactive mutant SH2K (A273/F505). These results collectively suggest that LIME specifically binds the Lck SH2 domain in a tyrosine phosphorylation–dependent manner.
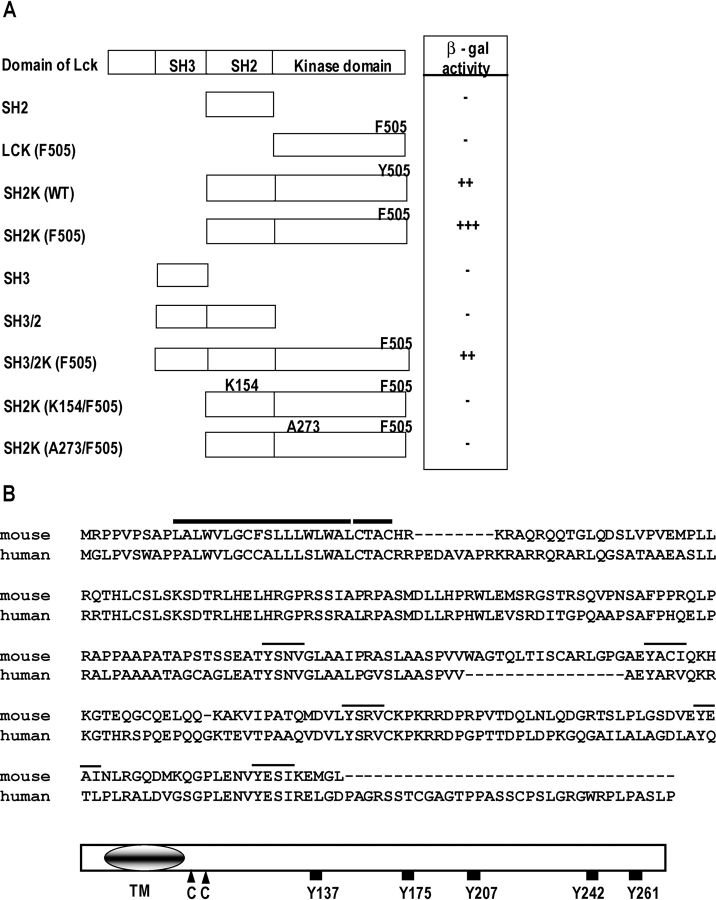
Figure 1.
cDNA cloning and the deduced amino acid sequence of LIME. (A) Binding specificity of LIME to Lck in a yeast two-hybrid system. Indicated regions of Lck were employed as bait and tested for binding to LIME. A part of LIME encompassing aa D52–270 was employed as prey. The binding affinity was scored as +++ (deep blue), ++ (intermediate blue), + (pale blue), and − (white) upon X-Gal (5-bromo-4-chloro-3-indoyl β-d-galactoside) staining. (B, top) The deduced amino acid sequences of mouse and human LIME and structural characteristics of the proteins. The transmembrane domain (TM) and CXXC motif are marked by bold lines. Putative tyrosine-based motifs are marked by thin lines on top of the sequence. Sequence data available from GenBank/EMBL/DDBJ under accession no. AF115339. (B, bottom) Schematic diagram of the predicted structure of murine LIME.
Structural Features of LIME.
The full-length open reading frame of LIME was subsequently obtained by screening a mouse lung cDNA library. The characterization of positive clones disclosed an open reading frame encoding 269 aa residues with a predicted molecular mass of 30 kD (Fig. 1 B).
The amino acid sequence of LIME is characteristic of a transmembrane adaptor protein. The open reading frame of murine LIME codes for a leaderless transmembrane protein with a short 9-aa extracellular region, an 18-aa hydrophobic membrane-spanning portion, and a 242-aa cytoplasmic tail. These structural features of LIME are similar to those of the recently identified immune cell-specific transmembrane adaptor proteins, LAT (4), TRIM (8), SIT (9), and Cbp/PAG (10, 11). LIME additionally contains a CXXC motif proximal to the transmembrane domain. In LAT, this sequence serves as a site for palmitoylation, which is essential for raft targeting of LAT (2). LIME contains five potential tyrosine phosphorylation sites in the cytosolic domain, which may play roles in forming signaling complexes with SH2 domain–containing molecules. These structural features indicate that LIME functions as a docking protein capable of recruiting various signaling molecules. Using the murine LIME sequence, we identified the human homologue from the NCBI database. Conceptual translation of human LIME cDNA showed 295 aa (Fig. 1 B). Moreover, alignment of the murine and human sequences showed extensive identity between the two proteins. In particular, the transmembrane domain, CXXC motif, and five potential phosphotyrosine motifs are conserved (Fig. 1 B).
Tissue Distribution of LIME.
On a Northern blot of mouse tissues, the LIME transcript was detected as a 1.5-kb band mainly restricted to the spleen and lung (Fig. 2 A). Among the human tissues, LIME mRNAs were observed in spleen and peripheral blood lymphocytes, although a much lower signal was detected in other tissues tested (Fig. 2 B). Therefore, it is likely that LIME is expressed predominantly in both hematopoietic cells and lung. LIME expression was additionally analyzed by Western blotting with anti-LIME antiserum. In vitro–translated LIME migrated as an ∼30-kD protein, concurrent with the size estimated from the deduced amino acid sequence (unpublished data). Western blot analysis of lymphoid tissues revealed that LIME was expressed at a comparable level in spleen and CD4+ and CD8+ T cells from LN but not in thymus, indicating that among T cell subsets LIME expression is restricted to peripheral T cells (Fig. 2 C). Various hematopoietic cell lines were additionally tested for LIME expression. Protein was detected in most of the hematopoietic cell lines tested, including K562 (erythroleukemia), Raw264.7 (macrophages), WEHI231, Raji (B cells), EL4 (thymus), and Jurkat (T cell) cell lines (Fig. 2 D). The results indicate that LIME is predominantly expressed in hematopoietic cells such as T, B, and macrophage.
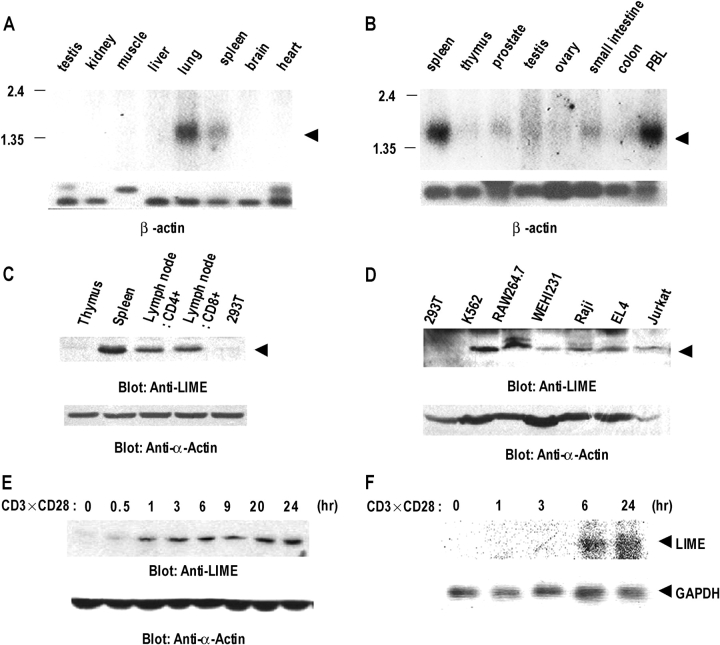
Figure 2.
Expression profiles of LIME. (A and B) Northern blot analyses on mouse (A) and human (B) tissues. Multiple Tissue Northern blots were performed using LIME cDNA as a probe. The positions of the LIME transcript are indicated by arrowheads. A β-actin cDNA probe was used as control (bottom). (C) Expression of LIME in murine lymphoid tissues. Western blot analyses were performed on cell lysates isolated from indicated tissues (2 × 106 cells/lane) using polyclonal mouse anti-LIME antibody. (D) Western blot analysis of LIME expression in various cell lines. Whole cell lysates (2 × 106 cells/lane) from indicated cell lines were immunoblotted with mouse anti-LIME antiserum. 293T, human embryonic kidney; K562, human erythroleukemic cells; Raw 264.7, macrophage; WEHI 231, murine B cell; Raji, human B cell; EL-4, murine thymoma; Jurkat, human T cell leukemia. The same membrane was immunoblotted with anti–α-actin antibody. (E) Western blot analysis of LIME expression upon T cell activation. Resting lymphoid T cells were stimulated in vitro with anti-CD3ɛ and anti-CD28 mAb for the indicated times. LIME expression was determined by Western blotting. (F) Northern blot analysis of LIME expression upon TCR activation. Resting lymphoid T cells were stimulated and processed as described in E. GAPDH cDNA probe was used as control (bottom).
Having determined that LIME is expressed in T cells, we were interested in establishing whether expression is regulated upon TCR stimulation. Resting T cells were isolated from mouse LNs, stimulated by CD3 and CD28 cross-linking and analyzed for the levels of LIME protein and mRNA. Interestingly, LIME protein was up-regulated upon TCR stimulation, reaching the maximum level within 1 h and was sustained up to 24 h (Fig. 2 E). In addition, LIME mRNA was also up-regulated but with a slower kinetics. The level of LIME mRNA reached a peak level at 6 h and was sustained up to 24 h (Fig. 2 F). The observed difference in kinetics of LIME expression suggests that LIME expression is regulated at both protein and mRNA levels. The inducible expression pattern also suggests that the protein operates throughout the early to late stages of T cell activation.
LIME Localizes To Membrane Rafts and Immunological Synapses.
To ascertain the function of LIME, we determined the localization pattern of the protein. Jurkat T cells were either left unstimulated or stimulated with anti-CD3 antibody and stained with a combination of rabbit anti-LIME antiserum and Texas red–conjugated anti–rabbit IgG. As a control for membrane raft fraction, the FITC–cholera toxin B subunit was employed for detecting GM1, an established marker of raft. Before TCR stimulation, LIME was evenly distributed throughout the plasma membrane. The protein translocated to the capping site after TCR stimulation (Fig. 3 A). Regardless of the absence or presence of TCR stimulation, the staining pattern of LIME overlapped exactly with that of the cholera toxin B subunit, suggesting that the protein specifically localizes to membrane raft.
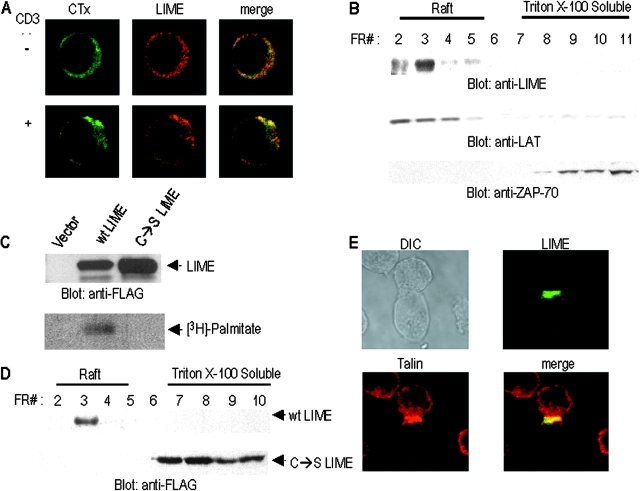
Figure 3.
LIME localizes to membrane raft and is distributed within the immunological synapse upon contact with APC. (A) LIME cocaps with GM1. Jurkat T cells were either left unstimulated (−, top) or stimulated (+, bottom) with anti-CD3 (UCHT1). Subsequently, cells were stained with FITC-labeled cholera toxin B subunit (CTx) or anti-LIME antibody and Texas red–conjugated anti–rabbit IgG. (B) LIME localizes to raft. Jurkat T cells were stimulated with anti-CD3 for 10 min and solubilized in 1% Triton X-100. Lysates were subjected to sucrose gradient ultracentrifugation, and fractions (numbered 2–11 from top to bottom) were analyzed by immunoblotting with anti-LIME, anti-LAT, or anti-ZAP70 antibodies. LAT was employed as an established control marker of raft fraction. (C) LIME is palmitoylated. 293T cells were transfected with the expression plasmids encoding FALG-tagged WT LIME or mutant LIME in which Cys28 and Cys31 were mutated to serine (C→S LIME) and metabolically labeled with 3H-palmitate for 4 h. LIME proteins were immunoprecipitated with anti-FLAG antibody, and the immunoprecipitates were analyzed by immunoblot (top) and autoradiography (bottom). (D) Palmitoylation of membrane-proximal cysteines is required for localization of LIME to membrane raft. Cell lysates were prepared as described in C and subjected to sucrose gradient centrifugation. The fractions were analyzed by immunoblotting with anti-FLAG antibody. (E) Localization of LIME to the APC–T cell contact site. Jurkat T cells transfected with expression plasmids encoding LIME–GFP were mixed at a ratio of 1:1 with SEE-pulsed Raji B cells. Cells were fixed and examined by confocal microscopy. Recruitment of LIME to T cells was characterized by a bright band at the cell interface.
To biochemically confirm raft localization of LIME, we fractionated Jurkat T cell lysates by sucrose gradient ultracentrifugation and analyzed fractions by Western blotting with anti-LIME antiserum. The protein was mainly detected in the raft fraction, even in the absence of TCR stimulation (Fig. 3 B). A control experiment with LAT as a marker for raft fractions and ZAP-70 for nonraft fractions confirmed that fractionation was performed properly. We conclude that LIME is a constitutive component of raft.
Next, the presence of C28XXC31 motif in the membrane-proximal region of LIME led us to test whether LIME is palmitoylated on this motif and if the palmitoylation is responsible for localization of LIME in membrane raft. Cys 28 and Cys 31 of LIME were mutated to serine (C→S LIME). Subsequently, WT or C→S LIME was transiently expressed in 293T cells in the presence of 3H-palmitate and subjected to immunoprecipitation. The immunoprecipitates were analyzed by immunoblotting (Fig. 3 C, top) or autoradiography (Fig. 3 C, bottom). WT LIME but not C→S LIME was labeled indicating that LIME is palmitoylated on membrane-proximal cysteines. Subsequently, the localization of WT and C→S LIME was studied by sucrose gradient fractionation. As shown in Fig. 3 D, C→S LIME localized in nonraft fraction, indicating that the palmitoylation of LIME is required for its localization to membrane raft.
Having determined the localization of LIME to membrane raft, we were interested in trafficking of the protein upon T cell–APC conjugation. Jurkat T cells were transfected with constructs expressing GFP–LIME and allowed to conjugate with SEE-pulsed Raji B cells as APC. Using confocal microscopy, we observed that LIME localized to the T cell–APC contact site with a pattern overlapping that of talin, a synapse marker (Fig. 3 E). In view of the results in the preceeding paragraphs, we conclude that LIME is a constitutive component of membrane raft that is recruited to the immunological synapse upon conjugation of T cells with APC. The results additionally suggest that the adaptor protein is involved in T cell activation signaling.
LIME Associates with Lck in T Cells.
We next investigated the association between LIME and Lck in Jurkat T cells upon TCR stimulation. Jurkat T cell lysates, either left unstimulated or stimulated by CD3 cross-linking, were subjected to immunoprecipitation with anti-Lck mAb. Normal mouse IgG was used as a negative control for immunoprecipitation. Analysis of immunoprecipitates by Western blotting with anti-LIME antiserum revealed LIME only in anti-Lck immunoprecipitates of CD3-stimulated cells (Fig. 4 A, top). Subsequent reprobing with antiphosphotyrosine antibody showed that the protein coprecipitating with Lck was phosphorylated (Fig. 4 A, middle). Our results confirm that LIME associates with Lck in the phosphorylated form, and interactions are induced by TCR stimulation.
Figure 4.
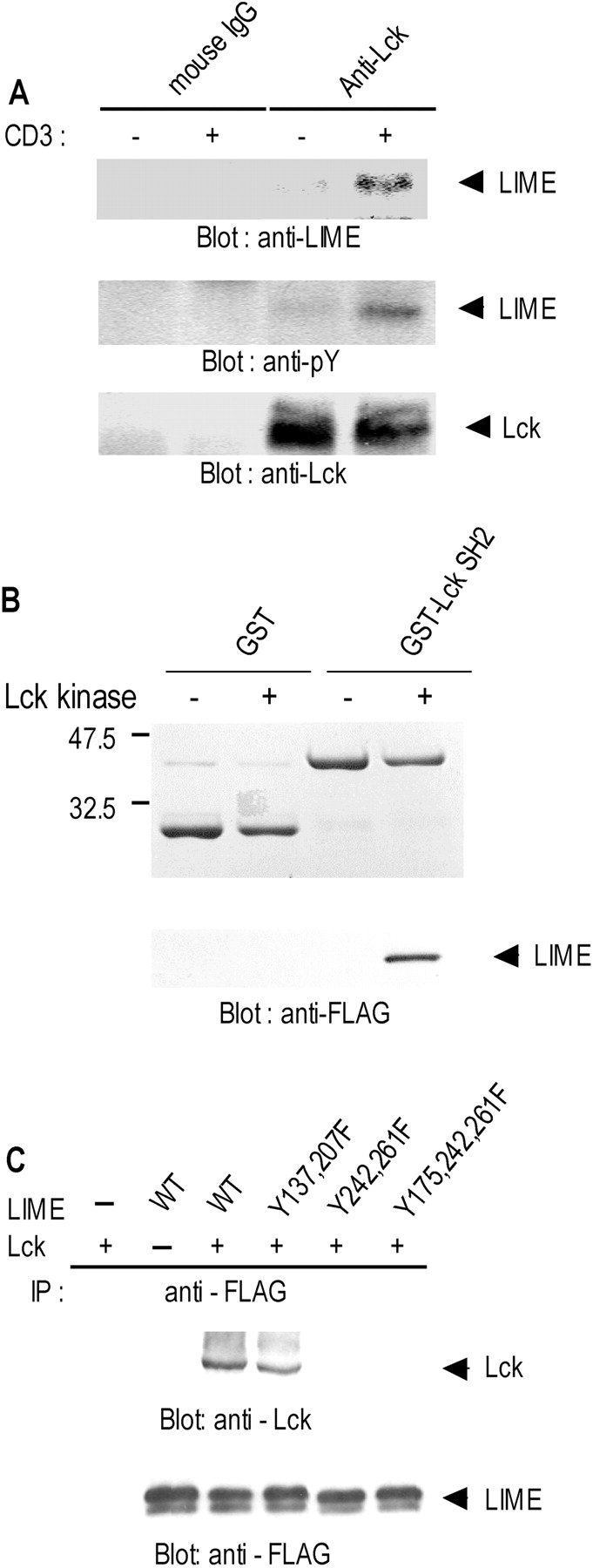
Binding of LIME to Lck. (A) In vivo association between LIME and Lck in T cells. Jurkat T cells were either left unstimulated or stimulated for 5 min by CD3 cross-linking. Digitonin lysates (1%) were subjected to precipitation with anti-Lck mAb or mouse IgG. Precipitates were analyzed by immunoblotting with anti-LIME and anti-phosphotyrosine mAbs. The amount of immunoprecipitated Lck is shown in the bottom panel. (B) LIME binds the Lck SH2 domain. Purified GST and GST–Lck SH2 fusion proteins were separated by SDS-PAGE and subjected to Coomassie staining (top). Subsequently, in vitro–translated FLAG–LIME was subjected to in vitro phosphorylation by purified Lck kinase and incubated with GST or GST–Lck SH2–coupled beads. Eluted proteins were separated by SDS-PAGE and immunoblotted with anti-FLAG antibody (bottom). (C) Mapping of sites in LIME responsible for binding to Lck. Tyrosine mutants of LIME were coexpressed with Lck in 293T cells by transient transfection. LIME mutants were immunoprecipitated with anti-FLAG mAb and analyzed by Western blotting with anti-Lck polyclonal antiserum (top). To control the amounts of precipitated LIME derivatives, the same membrane was blotted with anti-FLAG mAb after stripping (bottom).
Association of LIME with Lck Requires Tyrosine Phosphorylation.
To address whether phosphorylation of LIME by Lck is required for binding, we investigated associations between LIME and the Lck SH2 domain in vitro (Fig. 4 B). FLAG-tagged LIME was expressed by in vitro translation and subjected to phosphorylation in vitro by purified Lck kinase. The reaction mixture was subsequently incubated with GST or GST–Lck SH2 fusion proteins coupled to glutathione-sepharose beads, and bound proteins were analyzed by Western blotting with anti-FLAG antibody. LIME was detected in GST–Lck SH2 precipitates only in the presence of Lck kinase (Fig. 4 B). These results indicate that LIME functions as a direct binding partner of Lck SH2 and is a substrate of Lck kinase.
To identify the LIME motifs responsible for binding to Lck, we coexpressed tyrosine mutants of LIME with Lck F505 in 293T cells by transient transfection and examined binding abilities. Initially, the five tyrosines in the cytosolic domain of LIME were individually substituted using site-directed mutagenesis and examined for binding to Lck. All of the single tyrosine mutants coimmunoprecipitated with Lck F505, suggesting that multiple tyrosines are involved in binding (unpublished data). Accordingly, we generated double or triple tyrosine mutants and examined binding. As shown in Fig. 4 C, double LIME mutants (Y242F and Y261F) failed to bind Lck. We conclude that the specific amino acids of LIME that bind Lck SH2 are Y242 (YEAI) and Y261(YESI).
LIME Activates ERK and JNK.
To determine the signaling pathways regulated by LIME, we established Jurkat T cell lines stably expressing chimera of the CD8 extracellular domain and the cytoplasmic domain of LIME (Fig. 5 A). Since Jurkat T cells are devoid of CD8 surface expression, CD8 cross-linking is expected to induce LIME-specific signals. Initially, chimera expression levels were quantitated by Western blotting using anti-FLAG mAb (Fig. 5 B, top), and CD8–LIME chimeric molecules were detected at the expected mol. wt. of 49 kD. Anti-Grb2 mAb was used as a control (Fig. 5 B, bottom). One of the most commonly used criteria to assess early activation events is MAPK phosphorylation (26). We assessed whether LIME-specific signals led to MAPK activation, using a cell line expressing CD8–LIME chimera (Fig. 5 C). After cross-linking with CD8 antibody, ERK1/2, JNK, and p38 activation were analyzed by Western blotting with antibodies specific for the phosphorylated forms. Phosphorylation of ERK1/2 and JNK, but not p38, was detected (Fig. 5 C). As controls, the same blots were analyzed by Western blot with anti-ERK1/2 and anti-JNK antibodies. Our results demonstrate that LIME is involved in the activation of ERK and JNK pathways in T cells.
Figure 5.
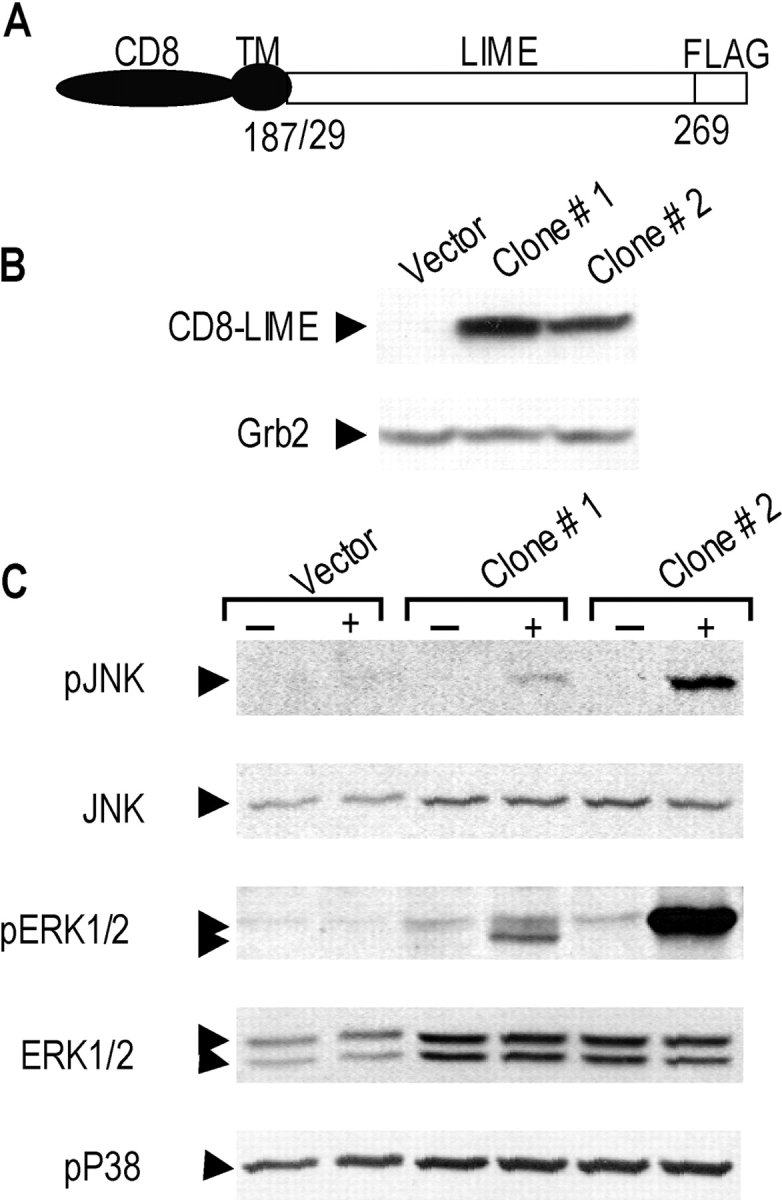
LIME mediates activation of ERK and JNK. (A) Structure of CD8–LIME chimera used in this study. The COOH termini of chimera were tagged with FLAG. (B) Jurkat T cell lines stably expressing CD8–LIME were established, and expression levels of chimeric proteins in two independent clones (clone #1, #2) were determined by Western blotting using anti-FLAG mAb. The blot was stripped and reincubated with anti-Grb2 mAb as a control. (C) LIME activates ERK1/2 and JNK but not p38. Jurkat T cells stably expressing the CD8–LIME chimera were left unstimulated or stimulated by CD8 cross-linking. Phosphorylation of MAP kinases was determined by blotting with antibodies specific for phosphorylated JNK, ERK1/2, and p38. The same blot was stripped and reprobed with anti-ERK1/2 or JNK.
Association of LIME with Grb2, Gads, PI3K, and SHP2 in T Cells.
Since LIME contains five tyrosines in the cytoplasmic tail, we hypothesized that SH2-containing signaling molecules could be recruited by the protein. Using a Jurkat T cell line stably expressing CD8–LIME chimera, we analyzed the binding partners of LIME. Immunoprecipitation of CD8–LIME with anti-FLAG antibody and subsequent Western blotting with anti-pY antibody showed that several tyrosine phosphorylated proteins with approximate mol. wt. of 85, 76, 70, 63, 40 kD coimmunoprecipitated with LIME (Fig. 6 A). Based on the molecular weights of phosphoproteins coimmunoprecipitating with LIME, we analyzed precipitates with a panel of antibodies specific for SH2 domain–containing signaling molecules. As shown in Fig. 6 B, the p85 regulatory subunits of PI3K, SHP2, Gads, and Grb2 were specifically detected in CD8–LIME immunoprecipitates. The data imply that LIME is present in signaling complexes containing various SH2 domain–containing molecules.
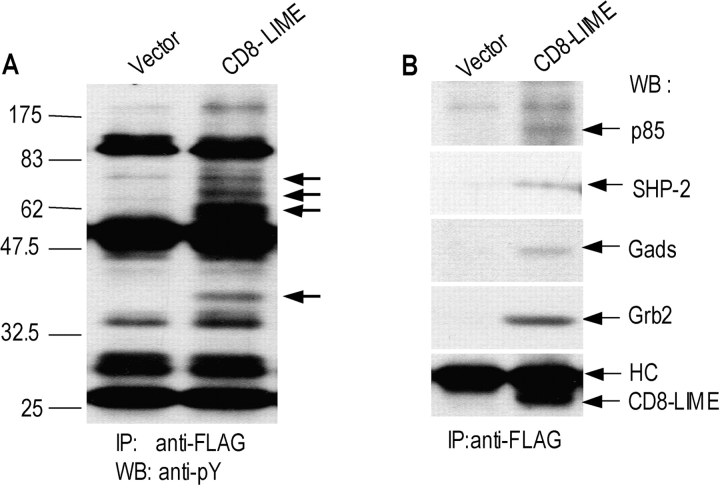
Figure 6.
LIME recruits various SH2 domain–containing signaling proteins. (A) Jurkat T cells expressing CD8–LIME chimera were analyzed by immunoprecipitation with anti-FLAG antibody and subsequent Western blotting with anti-pY 4G10 mAb. Tyrosine-phosphorylated bands detected only in immunoprecipitates of CD8–LIME chimera–expressing cells are marked by arrows. (B) The same blot was stripped and reprobed with anti-PI3K p85 subunit, anti-SHP2, anti-Gads, anti-Grb2, and anti-FLAG antibodies. HC, IgG heavy chain.
LIME Induces TCR-mediated IL-2 Gene Expression.
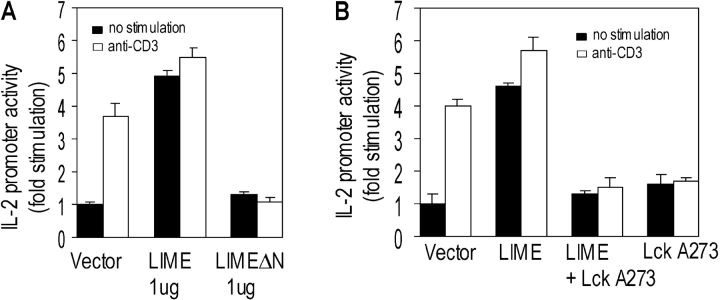
The data presented in the previous paragraphs show that LIME is involved in TCR-mediated signaling. To further assess the functional roles of the protein, we examined the effects of overexpression of WT or mutant LIME (LIMEΔN [lacking aa 1–82]) on IL-2 promoter–driven gene expression in Jurkat T cells. Since LIMEΔN lacks the transmembrane domain but retains potential binding sites for signaling molecules, it presumably acts as a dominant-negative form by sequestering LIME-associated proteins. The same approach was successfully employed for another transmembrane adaptor protein, LAT (27). Overexpression of LIME in the absence of CD3 stimulation led to sixfold induction of IL-2 promoter activity, indicating that the protein is involved in T cell signaling events leading to IL-2 gene expression (Fig. 7 A). Moreover, the transient overexpression of LIMEΔN, a dominant-negative form, resulted in reduced CD3-stimulated reporter activity by ∼75% (Fig. 7 A), indicating a requirement of LIME in TCR-mediated IL-2 gene expression.
Figure 7.
LIME mediates TCR cross-linking–induced IL-2 promoter activation. (A and B) Jurkat T cells were cotransfected with 0.75 μg each of IL-2–luc reporter and empty vector or the indicated amounts of plasmids. Cells were either left unstimulated or stimulated with anti-CD3 antibody and subsequently assayed for luciferase activity. Results are depicted as fold stimulation compared with activity in unstimulated cells transfected with empty vector. Each experiment was performed three times in duplicates and normalized by β-galactosidase activity from cotransfected pCMV–β-gal.
To further determine the functional significance of interaction between LIME and Lck in T cell activation, we examined the effects of Lck A273 mutant overexpression on LIME-mediated IL-2 promoter activation in Jurkat T cells. Lck A273 lacks kinase activity and was established previously as a catalytically inactive form of Lck. Overexpression of Lck A273 completely inhibited LIME-mediated IL-2 promoter activity (Fig. 7 B), indicating involvement of Lck in LIME-mediated signaling leading to IL-2 gene activation. The results collectively suggest that LIME function is Lck dependent and the protein plays an essential role in transmitting TCR-mediated T cell activation signals to IL-2 gene expression via association with Lck.
Discussion
One of the most widely accepted paradigms in TCR signal transduction is that TCR proximal signaling is initiated by the activated Src-type kinases, Lck and Fyn, leading to ITAM phosphorylation on the TCR subunit (2). Phosphorylated ITAM promotes recruitment and subsequent stimulation of ZAP-70 kinase. Adaptor proteins, LAT and SLP-76, which are established ZAP-70 substrates, recruit several other proteins involved in downstream signaling pathways (4). In contrast, although the importance of Lck in T cell activation is well established, its downstream targets and effectors remain to be determined. Here, we report the identification and characterization of a novel Lck substrate, LIME, which may potentially bridge the gap between Lck-mediated membrane-proximal events and downstream signaling pathways in T cell activation.
To date, four different transmembrane adaptor proteins, specifically, LAT (6), TRIM (8), SIT (9), and Cbp/PAG (10,11) have been reported in T cells. These proteins display similar structures but different functions. In particular, during T cell activation the function of LAT is well defined (5). Although LIME and LAT share common characteristics in terms of localization to the immunological synapse, binding proteins, activation of MAPK, and IL-2 promoter, several differences are observed between the two proteins. First, whereas LAT is a substrate of ZAP-70, LIME is identified as a substrate and a binding partner of Lck. With regard to this issue, we examined the possibility of LIME as a substrate for Syk family kinases by coexpressing LIME and Syk in 293T cells. Our results show that LIME is not phosphorylated by Syk (unpublished data). Therefore, it appears that LIME is a substrate of Lck kinase but not ZAP-70. Second, tissue expression patterns of LIME and LAT are distinct. In addition to T cells where LAT is expressed, LIME is detected in most of hematopoietic cells tested. Third, LIME expression is up-regulated upon TCR stimulation (Fig. 2 E), indicating that the protein may have a sustained role at a later stage of T cell activation or in effector T cells. These characteristics suggest that LIME plays unique roles related to Lck signaling and a sustained role throughout the later stages of T cell activation.
Several data in this report show that LIME acts as a signaling molecule in the early stages of T cell activation. Although LIME is specifically up-regulated upon TCR stimulation, a considerable amount of protein is detected in resting T cells (Fig. 2 C). Furthermore, 5 min after TCR stimulation LIME associates with Lck and localizes to the capping site, raft fractions, and immunological synapse (Fig. 3). It is plausible that on localization to synapse LIME provides a docking site upon which signaling complexes containing Grb2, Gads, PI3K, and SHP2 are assembled. Once recruited to the LIME-based signaling complex, Grb2 and p85 subunits of PI3K may activate the Ras-MAPK and PI3K pathways, respectively, whereas Gads may act as an adaptor for the recruitment of SLP-76 and PLC-γ as shown previously for LAT (6). In this regard, we additionally tested PLC-γ binding to LIME using coimmunoprecipitation studies and detected weak binding (unpublished data), possibly reflecting an indirect mode of recruitment of PLC-γ though Gads and SLP-76.
The finding that LIME expression is up-regulated upon TCR stimulation and sustained for at least 24 h suggests that in addition to its role in early T cell activation the protein may have specific functions at later stages of T cell activation or in effector T cells. Although there are many different possibilities for the function of LIME at later stages of T cell activation, one possible scenario is that LIME acts in the stabilization of APC-T cell conjugate. Previously, it has been shown that sustained TCR engagement up to 20 h (28) or at least 2 h (29) in length is required for activation of naive T cells. Furthermore, it has been shown that APC–T cell conjugation is dependent on Lck but not ZAP-70 (30–32), suggesting a unique role of Lck in conjugate stabilization. Therefore, as a binding partner of Lck LIME may be involved in stabilization of the APC–T cell conjugate. In relation to this possibility, Lck has been shown to play a role in the intermediate to late stages of T cell activation (33). The investigators additionally demonstrated that intracellular pools of Lck associated with recycling endosomes translocating to the immunological synapse, and inhibition of Lck kinase activity within up to 24 h of conjugation leads to a severe reduction in T cell activation (33). Therefore, although supporting data are insufficient at this stage, we hypothesize that LIME acts as a mediator of Lck signaling in the stabilization of APC–T cell conjugates and thereby contributes to the full activation of T cells. Alternatively, LIME may play an undefined role in events occurring at later stages of T cell activation.
Structurally, LIME is similar to other transmembrane adaptor proteins, such as LAT (4), Cbp/PAG (10,11), NTAL (12), and LAB (13). All of these proteins are characterized by the presence of a short extracellular domain, a transmembrane domain, and a long cytoplasmic tail containing multiple phosphotyrosine sites. In the membrane-proximal region of the cytoplasmic side, LIME contains a CXXC motif, which is also present in LAT, Cbp/PAG, NTAL, and LAB. In LAT, the motif was shown to serve as a palmitoylation site essential for targeting to raft (34). The results in this paper show that LIME is palmitoylated on the CXXC motif and the palmitoylation is essential for localization to raft (Fig. 3, C and D). In the cytoplasmic domain, murine LIME contains two overlapping, proline-rich SH3-binding motifs (RQLPRAPPAAP), which correspond to the type I SH3 binding motif, RxxPxxP (35, 36). However, this sequence is not conserved in human LIME, and it remains to be established whether it binds SH3. As expected from the presence of multiple tyrosines in the cytoplasmic domain, LIME associates with SH2-containing signaling molecules, including PI3K, Gad, Grb2, and SHP2 (Fig. 6 B). In accordance with previous binding specificity studies between various SH2-containing proteins and phosphotyrosine sites (37, 38), pY242EAI and pY261ESI motifs of LIME closely resemble the Lck SH2 domain binding motif, pYEEI, and are actually mapped as binding sites for Lck SH2 (Fig. 4 C). Additionally, pY137SNV is a potential binding site for Grb2 and Gads (defined as pYxN) (39). Another noteworthy point is the presence of an immunoreceptor tyrosine-based inhibition motif (ITIM)3 with a consensus sequence of (I/L/V)xYxx(L/V) (40). The immunoreceptor tyrosine-based inhibition motif in LIME, VLY207SRV, significantly corresponds to the consensus sequence and may serve as a binding site for SHP2.
In addition to its role in T cell activation, LIME may function as an adaptor in other cell types. Expression analyses revealed LIME protein in most of hematopoietic cells tested such as T, B, macrophage, and erythroleukemic cell lines. In this regard, LIME differs from LAT, NTAL, and LAB; LAT is restricted to T and mast cells, whereas NTAL and LAB are not expressed in T cells. Thus, it is possible that LIME acts as a docking protein in response to BCR stimulation. In addition, the detection of a LIME transcript in lung tissue suggests that the protein may have a specific role in lung tissue.
In summary, we have identified a novel transmembrane adaptor protein, LIME, as a binding partner of Lck upon TCR stimulation. The protein is expressed in T cells, and upon TCR stimulation expression is up-regulated, suggesting that LIME functions in the early to late stages of T cell activation. Further studies with knockout mice are proposed for characterizing the function of LIME in detail.
Acknowledgments
We thank members of the laboratory for materials and helpful discussions, and Dr. Jaesang Kim for critical reading of the manuscript.
This work was supported by funds for the National Research Laboratory and by the Korea Science Foundation through the Center for Cell Signaling Research at Ewha Woman's University.
Abbreviations used in this paper: Cbp, Csk-binding protein; GFP, green fluorescent protein; GST, glutathione S-transferase; ITAM, immunoreceptor tyrosine-based activation motif; LAB, linker for activation of B cells; LAT, linker for activation of T cells; LIME, Lck-interacting membrane protein; NTAL, non–T cell activation linker; PAG, phosphoprotein associated with glycosphingolipid-enriched microdomains; PI3K, phosphatidylinositol 3-kinase; PLC, phospholipase C; PTK, protein tyrosine kinases; pY, phosphotyrosine; SH, Src homology; SIT, SHP2-interacting transmembrane adaptor protein; TRIM, TCR-interacting molecule.
E.M. Hur and M. Son contributed equally to this work.
References
- 1.Wange, R.L., and L.E. Samelson. 1996. Complex complexes: signaling at the TCR. Immunity. 5:197–205. [DOI] [PubMed] [Google Scholar]
- 2.Weiss, A., and D.R. Littman. 1994. Signal transduction by lymphocyte antigen receptors. Cell. 76:263–274. [DOI] [PubMed] [Google Scholar]
- 3.Cherukuri, A., M. Dykstra, and S.K. Pierce. 2001. Floating the raft hypothesis: lipid rafts play a role in immune cell activation. Immunity. 14(6):657–660. [DOI] [PubMed] [Google Scholar]
- 4.Zhang, W., J. Sloan-Lancaster, J. Kitchen, R.P. Trible, and L.E. Samelson. 1998. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 92:83–92. [DOI] [PubMed] [Google Scholar]
- 5.Wange R.L. 2000. LAT, the linker for activation of T cells: a bridge between T cell-specific and general signaling pathways. Sci. STKE. 2000:RE1. [DOI] [PubMed]
- 6.Zhang, W., R.P. Trible, M. Zhu, S.K. Liu, C.J. McGlade, and L.E. Samelson. 2000. Association of Grb2, Gads, and phospholipase C-gamma 1 with phosphorylated LAT tyrosine residues. Effect of LAT tyrosine mutations on T cell antigen receptor-mediated signaling. J. Biol. Chem. 275:23355–23361. [DOI] [PubMed] [Google Scholar]
- 7.Zhang, W., and L.E. Samelson. 2000. The role of membrane-associated adaptors in T cell receptor signalling. Semin. Immunol. 12:35–41. [DOI] [PubMed] [Google Scholar]
- 8.Bruyns, E., A. Marie-Cardine, H. Kirchgessner, K. Sagolla, A. Shevchenko, M. Mann, F. Autschbach, A. Bensussan, S. Meuer, and B. Schraven. 1998. T cell receptor (TCR) interacting molecule (TRIM), a novel disulfide-linked dimer associated with the TCR–CD3-zeta complex, recruits intracellular signaling proteins to the plasma membrane. J. Exp. Med. 188:561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marie-Cardine, A., H. Kirchgessner, E. Bruyns, A. Shevchenko, M. Mann, F. Autschbach, S. Ratnofsky, S. Meuer, and B. Schraven. 1999. SHP2-interacting transmembrane adaptor protein (SIT), a novel disulfide-linked dimer regulating human T cell activation. J. Exp. Med. 189:1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawabuchi, M., Y. Satomi, T. Takao, Y. Shimonishi, S. Nada, K. Nagai, A. Tarakhovsky, and M. Okada. 2000. Transmembrane phosphoprotein Cbp regulates the activities of Src-family tyrosine kinases. Nature. 404:999–1003. [DOI] [PubMed] [Google Scholar]
- 11.Brdicka, T., D. Pavlistova, A. Leo, E. Bruyns, V. Korinek, P. Angelisova, J. Scherer, A. Shevchenko, I. Hilgert, and J. Cerny. 2000. Phosphoprotein associated with glycosphingolipid-enriched microdomains (PAG), a novel ubiquitously expressed transmembrane adaptor protein, binds the protein tyrosine kinase csk and is involved in regulation of T cell activation. J. Exp. Med. 191:1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brdicka, T., M. Imrich, P. Angelisova, N. Brdickova, O. Horvath, J. Spicka, I. Hilgert, P. Luskova, P. Draber, P. Novak, et al. 2002. Non–T cell activation linker (NTAL): a transmembrane adaptor protein involved in immunoreceptor signaling. J. Exp. Med. 196:1617–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janssen, E., M. Zhu, W. Zhang, S. Koonpaew, and W. Zhang. 2003. LAB: a new membrane-associated adaptor molecule in B cell activation. Nat. Immunol. 4:117–123. [DOI] [PubMed] [Google Scholar]
- 14.Xu, H., and D.R. Littman. 1993. A kinase-independent function of Lck in potentiating antigen-specific T cell activation. Cell. 74:633–643. [DOI] [PubMed] [Google Scholar]
- 15.Choi, Y.B., C.K. Kim, and Y. Yun. 1999. Lad, an adapter protein interacting with the SH2 domain of p56lck, is required for T cell activation. J. Immunol. 163:5242–5249. [PubMed] [Google Scholar]
- 16.Rajagopal, K., C.L. Sommers, D.C. Decker, E.O. Mitchell, U. Korthauer, A.I. Sperling, C.A. Kozak, P.E. Love, and J.A. Bluestone. 1999. RIBP, a novel Rlk/Txk- and itk-binding adaptor protein that regulates T cell activation. J. Exp. Med. 190:1657–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reynolds, P.J., T.R. Hurley, and B.M. Sefton. 1992. Functional analysis of the SH2 and SH3 domains of the lck tyrosine protein kinase. Oncogene. 7:1949–1955. [PubMed] [Google Scholar]
- 18.Drevot, P., C. Langlet, X.J. Guo, A.M. Bernard, O. Colard, J.P. Chauvin, R. Lasserre, and H.T. He. 2002. TCR signal initiation machinery is pre-assembled and activated in a subset of membrane rafts. EMBO J. 21:1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, J., H. Kang, M. Raab, A.J. da Silva, S.K. Kraeft, and C.E. Rudd. 1998. FYB (FYN binding protein) serves as a binding partner for lymphoid protein and FYN kinase substrate SKAP55 and a SKAP55-related protein in T cells. Proc. Natl. Acad. Sci. USA. 95:8779–8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marie-Cardine, A., E. Bruyns, C. Eckerskorn, H. Kirchgessner, S.C. Meuer, and B. Schraven. 1997. Molecular cloning of SKAP55, a novel protein that associates with the protein tyrosine kinase p59fyn in human T-lymphocytes. J. Biol. Chem. 272:16077–16080. [DOI] [PubMed] [Google Scholar]
- 21.Park, D., Y.B. Choi, M.K. Han, U.H. Kim, J. Shin, and Y. Yun. 2001. Adaptor protein Lad relays PDGF signal to Grb2 in lung cells: a tissue-specific PDGF signal transduction. Biochem. Biophys. Res. Commun. 284:275–281. [DOI] [PubMed] [Google Scholar]
- 22.Park, D., and Y. Yun. 2001. Tyrosine phosphorylation-dependent yeast two-hybrid system for the identification of the SH2 domain-binding proteins. Mol. Cells. 12:244–249. [PubMed] [Google Scholar]
- 23.Gietz, D., A. St Jean, R.A. Woods, and R.H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis, L.A., C.D. Chung, J. Chen, J.R. Parnes, M. Moran, V.P. Patel, and M.C. Miceli. 1997. The Lck SH2 phosphotyrosine binding site is critical for efficient TCR-induced processive tyrosine phosphorylation of the ζ-chain and IL-2 production. J. Immunol. 159:2292–2300. [PubMed] [Google Scholar]
- 25.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugawara, T., T. Moriguchi, E. Nishida, and Y. Takahama. 1998. Differential roles of ERK and p38 MAP kinase pathways in positive and negative selection of T lymphocytes. Immunity. 9:565–574. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, W., B.J. Irvin, R.P. Trible, R.T. Abraham, and L.E. Samelson. 1999. Functional analysis of LAT in TCR-mediated signaling pathways using a LAT-deficient Jurkat cell line. Int. Immunol. 11:943–950. [DOI] [PubMed] [Google Scholar]
- 28.Iezzi, G., K. Karjalainen, and A. Lanzavecchia. 1998. The duration of antigenic stimulation determines the fate of naive and effector T cells. Immunity. 8:89–95. [DOI] [PubMed] [Google Scholar]
- 29.Lee, K.H., A.D. Holdorf, M.L. Dustin, A.C. Chan, P.M. Allen, and A.S. Shaw. 2002. T cell receptor signaling precedes immunological synapse formation. Science. 295:1539–1542. [DOI] [PubMed] [Google Scholar]
- 30.Morgan, M.M., C.M. Labno, G.A. Van Seventer, M.F. Denny, D.B. Straus, and J.K. Burkhardt. 2001. Superantigen-induced T cell:B cell conjugation is mediated by LFA-1 and requires signaling through Lck, but not ZAP-70. J. Immunol. 167(10):5708–5718. [DOI] [PubMed] [Google Scholar]
- 31.Wong, J., D. Straus, and A.C. Chan. 1998. Genetic evidence of a role for Lck in T-cell receptor function independent or downstream of ZAP-70/Syk protein tyrosine kinases. Mol. Cell. Biol. 18:2855–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanchard, N., V. Di Bartolo, and C. Hivroz. 2002. In the immune synapse, ZAP-70 controls T cell polarization and recruitment of signaling proteins but not formation of the synaptic pattern. Immunity. 17:389–399. [DOI] [PubMed] [Google Scholar]
- 33.Ehrlich, L.I., P.J. Ebert, M.F. Krummel, A. Weiss, and M.M. Davis. 2002. Dynamics of p56lck translocation to the T cell immunological synapse following agonist and antagonist stimulation. Immunity. 17:809–822. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, W., R.P. Trible, and L.E. Samelson. 1998. LAT palmitoylation: its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation. Immunity. 9:239–246. [DOI] [PubMed] [Google Scholar]
- 35.Yu, H., J.K. Chen, S. Feng, D.C. Dalgarno, A.W. Brauer, and S.L. Schreiber. 1994. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 76:933–945. [DOI] [PubMed] [Google Scholar]
- 36.Collette, Y., and D. Olive. 1997. Non-receptor protein tyrosine kinases as immune targets of viruses. Immunol. Today. 18:393–400. [DOI] [PubMed] [Google Scholar]
- 37.Songyang, Z., S.E. Shoelson, M. Chaudhuri, G. Gish, T. Pawson, W.G. Haser, F. King, T. Roberts, S. Ratnofsky, R.J. Lechleider, et al. 1993. SH2 domains recognize specific phosphopeptide sequences. Cell. 72:767–778. [DOI] [PubMed] [Google Scholar]
- 38.Songyang, Z., and L.C. Cantley. 1995. Recognition and specificity in protein tyrosine kinase-mediated signaling. Trends Biochem. Sci. 20:470–475. [DOI] [PubMed] [Google Scholar]
- 39.Liu, S.K., N. Fang, G.A. Koretzky, and C.J. McGlade. 1999. The hematopoietic-specific adaptor protein Gads functions in T-cell signaling via interactions with the SLP-76 and LAT adaptors. Curr. Biol. 9:67–75. [DOI] [PubMed] [Google Scholar]
- 40.Vely, F., S. Olivero, L. Olcese, A. Moretta, J.E. Damen, L. Liu, G. Krystal, J.C. Cambier, M. Daeron, and E. Vivier. 1997. Differential association of phosphatases with hematopoietic co-receptors bearing immunoreceptor tyrosine-based inhibition motifs. Eur. J. Immunol. 27:1994–2000. [DOI] [PubMed] [Google Scholar]