Abstract
The host range of φCPAR39 is limited to four Chlamydophila species: C. abortus, C. caviae, C. pecorum, and C. pneumoniae. Chp3 (a newly discovered bacteriophage isolated from C. pecorum) shares three of these hosts (C. abortus, C. caviae, and C. pecorum) but can additionally infect Chlamydophila felis. The ability to support replication was directly correlated with the binding properties of the respective bacteriophages with their host species. Binding studies also show that φCPAR39 and Chp3 use different host receptors to infect the same host cells: cell binding is sensitive to proteinase K treatment, confirming that the chlamydiaphage receptors are proteinaceous in nature.
Chlamydiae have an obligate intracellular developmental cycle that alternates between the infectious elementary body and the replicative form, the reticulate body (RB). Five bacteriophages have been isolated from the chlamydiae. The first chlamydial bacteriophage to be characterized (13) was found by thin-section transmission electron microscopy (EM) of avian Chlamydophila psittaci RBs (14). Although samples containing Chp1 were later lost, a second bacteriophage (φCPG1) (7) was found to infect Chlamydophila caviae (8) and a third bacteriophage, Chp2, was isolated from Chlamydophila abortus (10). The Chlamydophila pneumoniae strain AR39 genome-sequencing project revealed the presence of a double-stranded DNA extrachromosomal element (11), which was subsequently shown to be the replicative form of bacteriophage φCPAR39 (4). Recently Chp3 was isolated from Chlamydophila pecorum (6) and was found to share 97.1% nucleotide sequence identity with Chp2. Chlamydiaphages φCPAR39 and φCPG1 also show 97.1% identity, whereas the two groups (Chp2-Chp3 and φCPAR39-φCPG1) share 93.3% identity. All the chlamydiaphages share similar features; they are small icosahedral T = 1 particles containing circular, single-stranded DNA genomes, and molecular characterization showed that they belong to the virus family Microviridae (9-11, 17).
Recently we described the host range of Chp2 and showed that Chp2 was unable to infect C. caviae or C. pneumoniae. It was therefore of interest to investigate the host range of the very closely related Chp3 and the slightly more diverse bacteriophages φCPAR39 and φCPG1, which were isolated from host chlamydiae resistant to Chp2. In our work, monolayers of BGMK (Buffalo green monkey kidney) or HEp2 cells in 25-cm2 flasks were infected by centrifugation at 1,000 × g for 1 h in medium containing cycloheximide (1 μg/ml) and gentamicin (25 μg/ml) with C. pecorum bearing Chp3 or C. pneumoniae (11) bearing φCPAR39. At 72 h postinfection the culture medium was replaced with a small volume of phosphate-buffered saline (PBS), and the flasks were frozen at −70°C. One hundred flasks of phage-infected chlamydiae were prepared, stored frozen, and then processed as a single batch. The flasks were frozen and thawed three times to lyse the chlamydial RBs and release phage. Any remaining monolayer that had not detached after this procedure was scraped off. The suspension was centrifuged at 2,000 × g for 15 min to sediment the cell debris. The supernatant was passed through a 0.45-μm-pore-size filter followed by a 0.22-μm-pore-size filter. The filtrate was centrifuged at 100,000 × g in a Beckman SW28 rotor for 3 h, and the resultant pellet was washed with PBS and centrifuged at 80,000 × g for 40 min. The pellet was finally suspended in PBS, vortexed with glass beads, and stored at −70°C as a partially purified bacteriophage preparation. A standard inoculum of Chp3 or φCPAR39 shown to be capable of infecting >99% of C. pecorum or C. pneumoniae IOL 207 by inclusion staining was mixed with each of a range of chlamydial species for 30 min at room temperature. The mixture was diluted in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, cycloheximide (1 μg/ml), and gentamicin (25 μg/ml), and BGMK cells cultured on coverslips were infected by centrifugation at 1,000 × g for 1 h. A set of coverslips were prepared in parallel with unchallenged chlamydiae. When inclusions were clearly visible (from 48 to 72 h postinfection), monolayers were fixed in ice-cold methanol. The susceptibility of chlamydiae to infection with φCPAR39 or Chp3 was monitored by fluorescence microscopy by using a phage-specific monoclonal antibody (MAb 55) as previously described (4). The tropism of φCPAR39 and Chp3 was tested by experimental infection of all chlamydial species and is summarized in Table 1. φCPAR39 produces a characteristic inclusion staining pattern when infecting C. pneumoniae, showing the formation of enlarged RBs (4). This phenomenon was observed in two independent isolates of C. pneumoniae when infected with φCPAR39. By contrast, infection of C. caviae, C. pecorum, and C. abortus by φCPAR39 gives whole inclusion staining (data not shown), indicative of complete RB lysis, and C. caviae was susceptible to Chp3 infection. In the case of C. caviae infection with Chp3, lysis occurred rapidly. This led us to recheck the effects of Chp2 on C. caviae, which we had previously scored as negative. Repeated infections of C. caviae with Chp2 showed that it is highly susceptible to Chp2 infection. We speculate that previous results were originally scored negative for infection by Chp2 because the life cycle of Chp2 in C. caviae is so rapid that it lyses RBs early in the developmental cycle before inclusions fully develop, making their detection difficult.
TABLE 1.
Host range and binding properties of φCPAR39 and Chp3a
| Genus and species | Strain | Source or reference | Chlamydial host species | φCPAR39
|
Chp3
|
||
|---|---|---|---|---|---|---|---|
| Sensitivity | Binding | Sensitivity | Binding | ||||
| Chlamydophila abortus | A22 | 16 | Ovine | S | B | S | B |
| Chlamydophila psittaci | Cal 10 | 5 | Avian | − | No | − | No |
| Chlamydophila pecorum | E58 | VR-628 | Bovine | S | B | S | B |
| Chlamydophila felis | FP | 2 | Feline | − | No | S | B |
| Chlamydophila pneumoniae | CWL 029 | VR-1310 | Human | S | B | − | No |
| TW 183 | VR-2282 | Human | S | ND | − | ND | |
| Chlamydophila caviae | GPIC | VR-813 | Human | S | B | S | B |
| − | − | ||||||
| Chlamydia trachomatis | L1/440/LN | 15 | Human | − | No | − | No |
| Chlamydia suis | DC-6 | K. Sachse | Porcine | − | ND | − | ND |
| Chlamydia muridarum | MoPn | VR-123 | Murine | − | ND | − | ND |
Abbreviations and symbols: S, sensitive to infection; −, not sensitive to infection; B, binding; No, no binding; ND, not done.
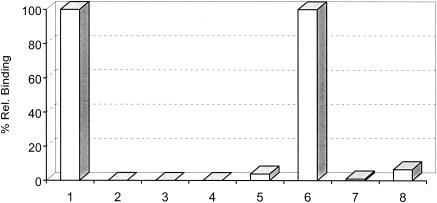
Previously it had been shown that host restriction of Chp2 was mediated through the ability of this bacteriophage to bind to a chlamydial surface protein-based receptor (4). In the present host challenge study we monitored for bacteriophage replication by immunofluorescence assay and showed that φCPAR39 is unable to infect C. felis and Chp3 is unable to infect C. pneumoniae. Quantitative cell binding assays (Fig. 1) confirmed that φCPAR39 cannot bind to C. felis and Chp3 cannot bind to C. pneumoniae but that φCPAR39 binds to C. pneumoniae and Chp3 binds to C. felis, demonstrating that host restriction is due solely to receptor recognition on a susceptible host. These studies were extended to include an example of each chlamydial species (Table 1), and the results confirmed that the ability of φCPAR39 or Chp3 to bind to elementary bodies (EBs) correlated directly with their ability to replicate and cause infection.
FIG. 1.
Binding of Chp3 and φCPAR39 with susceptible and nonsusceptible hosts. Shown are relative binding of C. felis EBs by Chp3 (column 1) and φCPAR39 (column 2) and the effects on such binding by Chp3 (column 3) and φCPAR39 (column 4) following treatment of EBs with proteinase K. Columns 5 to 8 show relative binding to C. pneumoniae EBs by Chp3 (column 5) and φCPAR39 (column 6) and the effects on such binding by Chp3 (column 7) and φCPAR39 (column 8) following treatment of EBs with proteinase K.
We have shown that the Chp2 receptor is a protease-sensitive component of the chlamydial outer membrane (4). To test whether the receptor(s) for φCPAR39 and Chp3 was also protease sensitive, EBs of both C. felis and C. pneumoniae were treated with proteinase K and then binding assays were performed as previously described (4). These analyses (Fig. 1) indicate that treatment of EBs with protease completely eliminates binding to susceptible host bacteria by phages φCPAR39 and Chp3.
Chlamydiaphages do not encode an external scaffold or major spike protein (10). Alignment of the structural proteins of Chp2 and Chp3 with those of φCPAR39 showed very little variation except in the coat protein VP1, which has an area of significant divergence between amino acids 216 and 299. The related microviruses φMH2K and SpV4 also have this distinctive region of sequence variation within their coat proteins (1). For SpV4, cryoimage reconstruction shows a large insertion loop that forms spikes at the threefold axis of symmetry (3). These spikes are thought to be formed by trimerization of the insertion loop (IN5) at each threefold axis of symmetry of the capsid and are a relic either of the external scaffolding protein or of a major spike protein (1). It has been speculated that in chlamydiaphages similar insertion loops are the viral antireceptors (12). Our observations are consistent with this proposal, as Chp2 and Chp3 have an identical cell tropism which is different from that of φCPAR39 and the predicted IN5 equivalent sequences of both Chp2 and Chp3 are highly conserved, with only two amino acid changes from each other, whereas the IN5 region of φCPAR39 is 15 amino acids shorter and highly divergent (12). The simplest explanation is that Chp2-Chp3 and φCPAR39 bind to different outer membrane protein receptors via the IN5 domain. This hypothesis is supported by the observation that saturation of the susceptible host C. abortus by Chp2 and Chp3 does not prevent binding with φCPAR39 (data not shown). Thus, φCPAR39 and Chp3 infect both C. abortus and C. pecorum, because their respective individual receptors are both present in these species.
Studies of bacteriophages that infect chlamydiae are of great interest, because they may provide the foundation for a genetic transfer system. Surface proteins, such as bacteriophage receptors, may also be involved in host-pathogen interactions, and while many chlamydial outer envelope proteins have been identified, many more remain to be discovered and the functions of most remain obscure. The observation that chlamydiaphages have different but overlapping cell tropisms is an important and significant discovery because it affords the opportunity to create recombinant chlamydiaphages by coinfection using host species of chlamydiae susceptible to both groups of chlamydiaphages. Future work will focus on producing the necessary monoclonal antibodies that will allow the selection and isolation of recombinant chlamydiaphages. These will then be used to map antireceptor binding domains to explore in fine detail the role of extracellular factors on cell tropism.
Acknowledgments
This work was supported by grant no. 063882 from the Wellcome Trust. S.A.G. was supported by an MRC research studentship.
We are grateful to K. Sachse for the kind gift of Chlamydia suis.
REFERENCES
- 1.Brentlinger, K. L., S. Hafenstein, C. R. Novak, B. A. Fane, R. Borgon, R. McKenna, and M. Agbandje-McKenna. 2002. Microviridae, a family divided: isolation, characterization and genome sequence of ΦMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovirus. J. Bacteriol. 184:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cello, R. M. 1967. Ocular infections in animals with PLT (Bedsonia) group agents. Am. J. Ophthalmol. 63:1270-1273. [DOI] [PubMed] [Google Scholar]
- 3.Chipman, P. R., M. Agbandje-McKenna, J. Renaudin, T. S. Baker, and R. McKenna. 1998. Structural analysis of the spiroplasma virus, SpV4: implications for evolutionary variation to obtain host diversity among the Microviridae. Structure 6:135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Everson, J. S., S. A. Garner, B. Fane, B.-L. Liu, P. R. Lambden, and I. N. Clarke. 2002. Biological properties and cell tropism of Chp2, a bacteriophage of the obligate intracellular bacterium Chlamydophila abortus. J. Bacteriol. 184:2748-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francis, T. J., and T. P. Magill. 1938. An unidentified virus producing acute meningitis and pneumonitis in experimental animals. J. Exp. Med. 68:147-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garner, S., S. Everson, B. Fane, P. R. Lambden, and I. N. Clarke. 2002. Discovery and molecular characterisation of a bacteriophage infecting C. pecorum, p. 25-28. In J. Schachter, G. Christiansen, I. N. Clarke, M. R. Hammerschlag, B. Kaltenboeck, C.-C. Kuo, R. G. Rank, G. L. Ridgway, P. Saikku, W. E. Stamm, R. S. Stephens, J. T. Summersgill, P. Timms, and P. B. Wyrick (ed.), Proceedings of the Tenth International Symposium on Human Chlamydial Infections. GRAFMAT Basin ve Reklam Sanayi Tic. Ltd. Şti, Antalya, Turkey.
- 7.Hsia, R.-C., H. Ohayon, P. Gounton, and P. M. Bavoil. 1996. Phage infection of Chlamydia psittaci GPIC, p. 48. In A. Stary (ed.), Proceedings of the Third European Society for Chlamydia Research. Societa Editrice Esculapio, Bologna, Italy.
- 8.Hsia, R. C., H. Ohayon, P. Gounon, A. Dautry-Varsat, and P. M. Bavoil. 2000. Phage infection of the obligate intracellular bacterium, Chlamydia psittaci strain guinea pig inclusion conjunctivitis. Microbes Infect. 2:761-772. [DOI] [PubMed] [Google Scholar]
- 9.Hsia, R. C., L. M. Ting, and P. M. Bavoil. 2000. Microvirus of Chlamydia psittaci strain Guinea pig inclusion conjunctivitis: isolation and molecular characterization. Microbiology 146:1651-1660. [DOI] [PubMed] [Google Scholar]
- 10.Liu, B. L., J. S. Everson, B. Fane, P. Giannikopoulou, E. Vretou, P. R. Lambden, and I. N. Clarke. 2000. Molecular characterization of a bacteriophage (Chp2) from Chlamydia psittaci. J. Virol. 74:3464-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Read, T. D., C. M. Fraser, R. C. Hsia, and P. M. Bavoil. 2000. Comparative analysis of Chlamydia bacteriophages reveals variation localized to putative receptor binding domain. Microb. Comp. Genomics 5:223-231. [DOI] [PubMed] [Google Scholar]
- 13.Richmond, S., P. Stirling, and C. Ashley. 1982. Chlamydia have phage too, p. 41-44. In P.-A. Mardh, K. K. Holmes, J. D. Oriel, P. Piot, and J. Schachter (ed.), Fifth International Symposium on Human Chlamydial Infection, Elsevier Biomedical Press, Amsterdam, The Netherlands.
- 14.Richmond, S. J., P. Stirling, and C. Ashley. 1982. Virus infecting the reticulate bodies of an avian strain of Chlamydia psittaci. FEMS Microbiol. Lett. 14:31-36. [Google Scholar]
- 15.Schachter, J., and K. F. Meyer. 1969. Lymphogranuloma venereum II. Characterization of some recently isolated strains. J. Bacteriol. 99:636-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stamp, J. T., A. D. McEwen, J. A. A. Watt, and D. I. Nisbet. 1950. Enzootic abortion in ewes. I. Transmission of the disease. Vet. Rec. 62:251-256. [DOI] [PubMed] [Google Scholar]
- 17.Storey, C. C., M. Lusher, and S. J. Richmond. 1989. Analysis of the complete nucleotide sequence of Chp1, a phage which infects avian Chlamydia psittaci. J. Gen. Virol. 70:3381-3390. [DOI] [PubMed] [Google Scholar]