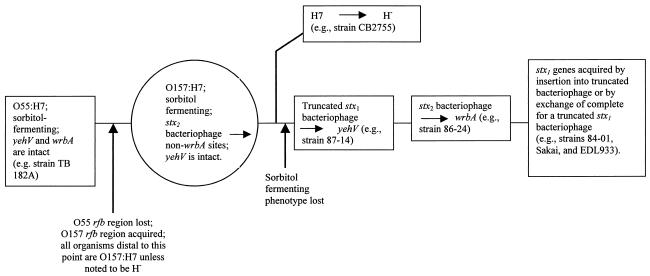
Volume 185, no. 12, p. 3596-3605. Page 3597: Table 1 erroneously designated wrbA as being occupied by a bacteriophage in the sorbitol-fermenting strains CB2755 and 493/89, whereas it is, in fact, intact (as demonstrated in Fig. 1). Therefore, the stx2 bacteriophage integrated into a site or sites other than wrbA in these isolates. Thus, our assumptions that these sorbitol-fermenting organisms diverged from other members of the STEC1 lineage after the stx2 bacteriophage integrated into wrbA and that integration of the stx2 bacteriophage into wrbA is an ancestral event are not supported by the data. The corrected data, which suggest that the stx2 bacteriophage inserted into wrbA at a later point in the evolution of the STEC1 clade after a truncated bacteriophage was acquired by Escherichia coli O157:H7, lead to a simpler evolutionary scenario which obviates the need to invoke lineages A, B, and C (see the revised Fig. 4 shown below).
FIG. 4.
Proposed evolutionary scenario for the STEC1 clade. Boxes represent organisms known to exist today. The circle represents a proposed intermediate organism between E. coli O55:H7 and E. coli O157:H7.
Page 3600: The stx1A and stx1B labels in Fig. 2 were reversed.
Page 3603: Figure 4 and its legend should appear as shown below. .