Abstract
Human natural killer (NK) cells express a series of activating receptors and coreceptors that are involved in recognition and killing of target cells. In this study, in an attempt to identify the cellular ligands for such triggering surface molecules, mice were immunized with NK-susceptible target cells. On the basis of a functional screening, four mAbs were selected that induced a partial down-regulation of the NK-mediated cytotoxicity against the immunizing target cells. As revealed by biochemical analysis, three of such mAbs recognized molecules of ∼70 kD. The other mAb reacted with two distinct molecules of ∼65 and 60 kD, respectively. Protein purification followed by tryptic digestion and mass spectra analysis, allowed the identification of the 70 kD and the 65/60 kD molecules as PVR (CD155) and Nectin-2 δ/α (CD112), respectively. PVR-Fc and Nectin-2-Fc soluble hybrid molecules brightly stained COS-7 cells transfected with the DNAM-1 (CD226) construct, thus providing direct evidence that both PVR and Nectin-2 represent specific ligands for the DNAM-1 triggering receptor. Finally, the surface expression of PVR or Nectin-2 in cell transfectants resulted in DNAM-1–dependent enhancement of NK-mediated lysis of these target cells. This lysis was inhibited or even virtually abrogated upon mAb-mediated masking of DNAM-1 (on NK cells) or PVR or Nectin-2 ligands (on cell transfectants).
Keywords: natural killer cells, tumors, activating receptors, cellular ligands, protein sequencing
Introduction
In humans the function of NK cells is regulated by a series of surface receptors that either inhibit or enhance the NK-mediated cytolytic activity. When NK cells encounter potential target cells, clonally distributed killer inhibitory Ig-like receptors (KIR) and/or the CD94/NKG2A heterodimer sense the expression of HLA class I molecules at the cell surface (1–4). When appropriate inhibitory receptor/HLA class I ligand interactions occur, the receptors deliver negative signals that inhibit the NK-mediated killing. On the other hand, in the absence of such interactions, as it may occur in tumor or virus infected target cells, (non-HLA class I–specific) activating receptors are allowed to deliver triggering signals to NK cells resulting in target cell killing. Several non-HLA class I–specific activating receptors have been identified including NKp46, NKp30, and NKp44 (collectively termed “natural cytotoxicity receptors”, NCR) and NKG2D (5, 6). While the expression of NCR is restricted to NK cells, NKG2D is also expressed by virtually all TCR γ/δ+ as well as by CD8+ TCR α/β+ T cells. The combined mAb-mediated masking of both NCR and NKG2D results in strong inhibition of the NK cytotoxicity against most target cells. This suggests that NCR and NKG2D play a predominant role in target cell recognition and killing by NK cells. However, additional surface molecules have been implicated in this process. These include 2B4 (CD244), NTB-A, and NKp80 that appear to function as coreceptors since their triggering function is dependent on the simultaneous engagement of NCR (7–9). Another surface molecule that has been shown to participate in the induction phase of NK cell activation is the leukocyte adhesion molecule DNAX accessory molecule-1 (DNAM-1; CD226; references 10 and 11). DNAM-1 is expressed by virtually all NK cells, T cells, and monocytes. Encoded on human chromosome 18, DNAM-1 is characterized by two Ig-like domains in the extracellular portion and a cytoplasmic tail containing three tyrosine residues. Cross-linking of DNAM-1 results in Fyn-mediated tyrosine phosphorylation and triggering of cytotoxicity in NK cells (11). Remarkably, DNAM-1 function is dependent on the surface expression of lymphocytes function-associated antigen 1 (LFA-1; CD18/CD11a). Indeed, it is defective in NK cells from patients with leukocyte adhesion deficiency syndrome 1 (LAD1), who lack LFA-1 but express DNAM-1. In this case the function could be rescued upon genetic reconstitution of LFA-1 (12).
Although functional data would indicate that the various activating NK receptors and coreceptors recognize ligands expressed at the target cell surface, so far only few of these ligands have been identified. For example NKG2D recognizes the stress-inducible MHC class I–related chain molecules (MIC)A/B and UL16-binding protein (ULBP) molecules (13–15) while 2B4 specifically interacts with CD48 (16, 17). On the other hand, NCR, NTB-A, NKp80, and DNAM-1 are orphan receptors since their cellular ligands are still unknown. In an attempt to identify such ligands, we generated mAbs by immunizing mice with NK-susceptible target cells. These mAbs were screened for their ability to interfere with the NK-mediated cytotoxicity against the immunizing target cells. Using this functional approach we were able to identify two molecules, poliovirus receptor (PVR) (18) and Nectin-2 (19, 20), that specifically induce NK cell activation by interacting with DNAM-1.
Materials and Methods
Monoclonal Antibodies.
M5A10 and M2C24 or L95 and L14 mAbs were obtained by immunizing a 5-week-old BALB/c mouse with M14 (human melanoma) or 293T (embryonic fibroblast) cell lines respectively as described previously (8). The following mAbs, selected in our lab, were used in this study: GN18, KRA236, and FS123 (IgG3, IgG1, and IgG2a, respectively, anti-DNAM-1), BAB281 and KL247 (IgG1 and IgM, respectively, anti-NKp46), Z231 and KS38 (IgG1 and IgM, respectively, anti-NKp44), Z25 and F252 (IgG1 and IgM, respectively, anti-NKp30), PP35 (IgG1, anti-2B4), BAT221 (IgG1, anti-NKG2D), c127 (IgG1, anti-CD16), c218 (IgG1, anti-CD56), MA152 (IgG1, anti-NKp80), MA127 (IgG1, anti-NTB-A), BAM195 (IgG1, anti-MICA), and JT3A (IgG2a, anti-CD3).
D1.12 (IgG2a, anti-HLA-DR) mAb was provided by Dr. R.S. Accolla (University of Insubria, Varese, Italy). HP2.6 (IgG2a, anti-CD4) mAb was provided by Dr. P. Sanchez-Madrid (Universidad Autonoma de Madrid, Madrid, Spain). M295 (IgG1, anti-ULBP1), M310 (IgG1, anti-ULBP2), and M551 (IgG1, anti-ULBP3) were provided by Immunex, Seattle, WA. TU145 (IgM, anti-CD48) was purchased from Becton Dickinson.
Polyclonal NK and T Cell Populations and NK Cell Lines.
PBMCs, derived from healthy donors, were isolated on Ficoll-Hypaque gradients and PBLs were obtained by depleting PBMCs of plastic-adherent cells. Enriched NK cells were isolated by incubating PBLs with anti-CD3 (JT3A), anti-CD4 (HP2.6), and anti-HLA-DR (D1.12) mAbs (30 min at 4°C) followed by goat anti–mouse coated Dynabeads (Dynal; 30 min at 4°C) and immunomagnetic depletion (8). CD3− 4− DR− cells were cultured on irradiated feeder cells in the presence of 100 U/ml rIL-2 (Proleukin; Chiron Corp.) and 1.5 ng/ml PHA (GIBCO BRL) in order to obtain polyclonal NK cell populations. NK92 and YT NK cell lines were provided by E.O. Long (NIAID, NIH, Rockville, MD) and E.D. Carosella (Institut Universitaire d'Hematologie, Paris, France), respectively.
PHA blasts were obtained by culturing for one day PBL with 1.5 ng/ml PHA (GIBCO BRL). After this period of time cells were washed and cultured in medium supplemented with 100 U/ml of rIL2.
Cytolytic Activity and Flow Cytofluorimetric Analysis.
NK cells were tested for cytolytic activity against the indicated target cells in a 4-h 51Cr-release assay as previously described (8, 15). The concentrations of the various mAbs added for masking experiments were 10 μg/ml. The E/T ratios are indicated in the text.
In some cytolytic assays, as indicated in the text, to avoid the involvement of NKG2D-dependent lysis, effector cells were first incubated with BAT 221 (anti-NKG2D) mAb (10 μg/ml) for 15 min at room temperature (15).
For one-color cytofluorimetric analysis (FACSCalibur™; Becton Dickinson) cells were stained with the appropriate mAbs followed by PE- or FITC-conjugated isotype-specific goat anti–mouse second reagent (Southern Biotechnology Associates, Inc.; reference 9). PVR-Fc and Nectin-2-Fc molecules were used at the concentration of 20 μg/ml followed by PE-conjugated isotype-specific goat anti–human second reagent (Southern Biotechnology Associates, Inc.)
Biochemical Characterization of PVR and Nectin-2 Molecules.
20 × 106 cells were incubated (15' at 20°C) in 1 ml PBS pH 8.0 containing 250 μg of EZ-LINK SULFO-NHS-LC-LC-BIOTIN (Pierce Chemical Co.) and washed three times in Washing Buffer (10 mM TRIS, pH 8, 0.14 M NaCl). Cells were then lysed in 1% NP-40 and immunoprecipitated with Sepharose-PA- (Amersham Biosciences) coupled mAbs. Samples were analyzed by discontinuous SDS-PAGE either undigested or digested with N-glycosidase F (Boehringer) and transferred to Immobilon P (Millipore). After staining with Neutravidin (Pierce Chemical Co.) the Renaissance Chemiluminescence Kit (NEN Life Science Products) was used for detection (8). For Western blotting experiments mAbs, purified using AKTA PRIME (Amersham Biosciences), were labeled with Biotin (Pierce Chemical Co.). Neutravidin (Pierce Chemical Co.) was used as second reagent.
PVR and Nectin-2 Purification.
To obtain 293T cell membranes, cells were washed twice in PBS pH 7.5 and resuspended at 108 cells/ml in ice-cold hypotonic swelling buffer (10 mM Tris-HCl, pH 7.6, 0.5 mM MgCl2, 10 mM 1AA, with protease inhibitors) for 10' at 4°C before their disruption with Polytron 3100 (Kinematica AG). Tonicity was restored by adding 10 mM Tris-HCl, pH 7.6, 0.5 mM MgCl2, 1.5 M NaCl with protease inhibitors and suspension was centrifuged 5′ at 500 g at 4°C to remove the nuclear fraction. Supernatant, upon addition of EDTA (at final concentration of 5 mM), was centrifuged 45' at 150,000 g at 4°C. Pellet was resuspended in 1 ml 1% NP-40 lysis buffer with protease inhibitors and stored at –80°C. Membrane lysates from 5 × 109 cells were precleared twice with Sepharose-PA and incubated with Sepharose CnBr-coupled mAbs O/N at 4° under rotation. After extensive washes, specific proteins were eluted with 0.1 M glycine, 150 mM NaCl, pH 2.8, highly concentrated with Amicon Ultra (Millipore) and analyzed by discontinuous SDS-PAGE under nonreducing conditions. To detect the purified protein polyacrylamide gel was stained using Simply Blue Safestain (Invitrogen) 1 h at room temperature.
In-gel Enzymatic Digestion.
After the staining procedure, the gel was washed three times with water for 60 min. The band of interest was excised using a sterile blade, placed in a 1.5 ml microtube and cut into pieces. In-gel digestion was performed as described by Ha et al. (21) modified as follows: 30 μl of 100 mM ammonium bicarbonate, 1 mM CaCl2, pH 8.9, and 5 μl of trypsin solution (200 μg/ml, Promega) were added to the gel particles. After 10', 30 μl of 60% acetonitrile in 100 mM ammonium bicarbonate, 1 mM CaCl2 pH 8.9 (final concentration: 30%) were added to the mixture. After overnight incubation at 37°C, the supernatant was recovered and dried in a vacuum centrifuge (Savant Instruments) until the volume was reduced to 30 μl; 30 μl of 0.25% formic acid was then added. The sample was filtered using a 0.02 μm Anodisc 13 filter (Whatman) in a MicroFilter system (ProteinSolutions).
LC/ESI-MS/MS Analysis of Tryptic Peptides.
An automated LCQ-DECA MS/MS ion trap mass spectrometer coupled to a HPLC Surveyor (Thermo Finnigan) and equipped with a Hypersil BDS, C18 column, 1 × 100 mm (ThermoHypersil) were used. Peptides were eluted from the column using an acetonitrile gradient, 5% B for 3 min followed by 5 to 90% B within 52 min (eluent A: 0.25% formic acid in water; eluent B: 0.25% formic acid in acetonitrile) at a flow-rate of 50 μl/min. The capillary of the ion trap was kept at 200°C and the voltage at 30 V. Spray voltage was 5.0 kV. Spectra were acquired in automated MS/MS mode: each full MS scan (in the range 400–2,000 m/z) was followed by three MS/MS of the most abundant ions, using relative collision energy of 35%. Computer analysis of peptide MS/MS spectra was performed using the version 1.2 of the TurboSEQUEST software (University of Washington, licensed to ThermoFinnigan Corp.) and searched against the National Center for Biotechnology Information (NCBI) human protein database.
CHO-K Cell Transfectants and Chimeric Molecules.
CHO-K cells were stably transfected with either the pSV2-PVRα (gift of Dr. Akio Nomoto, University of Tokyo, Tokyo, Japan) or the pLX2S.B1 (Nectin-2α) or the pLX2L.C12 (Nectin-2δ) vectors (20, 22)
Construction, production, and purification of soluble Nectin-2-Fc, PVR-Fc, and Nectin-4tr-Fc (a truncated form of Nectin-4 ectodomain) were already described (20, 23). Briefly, Nectin ectodomains were fused to the Fc portion of human IgG1 using the pCOS Fc Link vector (Smith Kline Beecham). Proteins were produced in COS cells with FuGENE6 according to the recommendations of the manufacturer (Roche Diagnostics). The proteins were purified from supernatants on Affi-Gel protein A. Purification was controlled by Coomassie Blue-stained SDS-PAGE.
RT-PCR Amplification of cDNA Encoding for DNAM-1 and Transient Cell Transfection.
Total RNA extracted using peQGold RNA pure (peQLab) from a polyclonal NK cell population was reverse transcribed using oligodT priming. Primers used for cDNA amplification of complete DNAM1-ORF (1124 bp) were the following: 5′ AGA ACC AGC CTT TCA AAC AG (DNAM-frw) and 5′ CTT GGG TAG TGG AAA AAA ATT G (DNAM rev). Amplification was performed for 30 cycles (30 s at 94°C, 30 s at 55°C, 30 s at 72°C), followed by 7 min incubation at 72°C, using AmpliTAQ (PerkinElmer/Applied Biosystems). The cDNA obtained was subcloned in VR1012 expression vector (Vical Inc.) and sequenced.
COS7 cells were transiently transfected with VR1012/DNAM-1 construct using Fugene 6 (Roche). Briefly, cells were seeded at 5 × 105/plate, 24 h later they were incubated with 6 μg of plasmid and 10 μl of FuGENE6 in DMEM/10% FCS. After 48 or 72 h, transfected cells were used for cytofluorimetric analysis. Cell transfectants were stained with GN18 or FS123 anti-DNAM-1 mAbs followed by PE-conjugated goat anti–mouse IgG second reagent (Southern Biotechnology Associates, Inc.). In parallel, DNAM-1 transfected cells were stained with PVR-Fc, Nectin-2-Fc, or Nectin-4tr-Fc soluble chimeric molecules (from 0.2 to 1 μg/sample) followed by PE-conjugated goat-anti-human IgG antibody (Southern Biotechnology Associates, Inc.) and analyzed by flow cytometry using a FACSort™ (Becton Dickinson).
Results
Isolation of mAbs against NK-susceptible Target Cells.
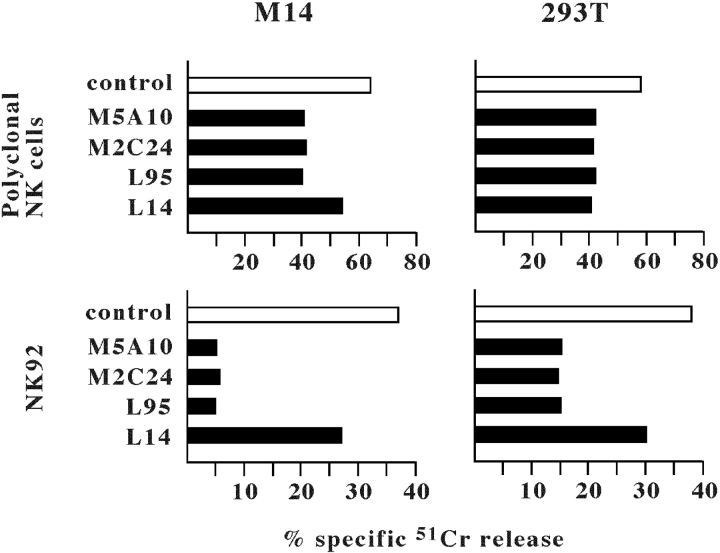
In an attempt to identify novel cell surface ligands recognized by orphan triggering NK receptors, mice were immunized with the NK-susceptible M14 (human melanoma) or 293T (human embryonic fibroblasts) cell lines (15). After cell fusion, the mAbs obtained were screened for their capability of inhibiting the NK cytotoxicity against the immunizing cells. Both target cells did not express CD48 (i.e., the 2B4 ligand) while they did express the NKG2D ligands. In particular, M14 expressed ULBP2 while 293T expressed MICA, ULBP2, and ULBP3 (not shown). Thus, in order to select mAbs directed to ligands different from MICA and ULBPs, the cytolytic assay was performed in the presence of an anti-NKG2D mAb to block NKG2D on the effector NK cells (15). These were represented either by polyclonal NK cell populations or by the NK92 NK cell line (surface phenotype: CD16−, NKp46−, NKp30+, NKp44+, NKG2D+, 2B4+, NTB-A+, DNAM-1+) (8). Using this experimental approach, four mAbs were selected that inhibited the cytotoxicity residual after NKG2D blocking (Fig. 1) . In particular, M5A10 and M2C24 mAbs were isolated from M14 immunization, while L95 and L14 mAbs were selected from 293T immunization. It is of note that the inhibitory effect of the four selected mAbs was higher when the NK92 cell line was used, instead of normal polyclonal NK cells, as a source of effector cells. This is likely to reflect differences in the expression and/or surface density of the various triggering NK receptors in the two different effector cells. Indeed, normal polyclonal NK cells, unlike the NK92 cell line, express NKp46, a major triggering NK receptor that is also involved in recognition and killing of both M14 and 293T cells (24). The inhibition mediated by L14 mAb was lower as compared with the other mAbs particularly when the effector cells were represented by NK92. This suggested that the molecule(s) recognized by L14 mAb might be different from those recognized by the other mAbs.
Figure 1.
Inhibition of NK-mediated cytotoxicity by M5A10, M2C24, L95, and L14 mAbs. A polyclonal NK cell population (top panels) or the NK92 NK cell line (bottom panels) pretreated with anti-NKG2D mAb (BAT221; for 15 min at room temperature) were analyzed for cytolytic activity against the indicated target cell lines in the presence of either a control mAb (white bars) or one or another of the indicated mAbs (black bars). The effector/target ratio used was 10:1 for polyclonal NK cells and 20:1 for NK92. The results are representative of three independent experiments; the standard deviation of the mean of the triplicates was <5%.
Altogether, the above data suggested that the selected mAbs could recognize surface structures functioning as cellular ligands for triggering receptors other than NKG2D. Thus, we next evaluated, by indirect immunofluorescence and cytofluorimetric analysis, the distribution of the surface molecules reactive with the selected mAbs. As shown in Table I, all mAbs brightly stained not only the immunizing cells but also a large panel of tumor cell lines of different histotype. Notably, the cellular distribution of the mAbs-reactive molecule(s) was mostly overlapping although minor differences could be detected between the reactivity of L14 and that of the other mAbs (see for example the MOLT4 cell line in Table I). Finally, the analysis of normal leukocytes revealed that all four mAbs did not react with resting NK, B, and T cells or with in vitro activated NK cells. Low reactivity could be detected in monocytes and granulocytes (with all four mAbs), while in vitro–activated T cells were weakly stained by L95, M5A10, and M2C24 mAbs but not by L14 mAb.
Table I.
Surface Reactivity of the Selected mAbs on Different Cell Types
| Cells | Histotype | L95, M5A10, M2C24 mAbs |
L14mAb |
|---|---|---|---|
| Resting NK cells | − | − | |
| Activated NK cells | − | − | |
| Resting T cells | − | − | |
| PHA Blasts | +/− | − | |
| Resting B cells | − | − | |
| Monocytes | +/− | +/− | |
| Granulocytes | +/− | +/− | |
| YT | NK cell line | − | − |
| NK92 | NK cell line | − | − |
| CEM | T leukemia | − | − |
| MOLT4 | T leukemia | + | − |
| Jurkat | T leukemia | + | + |
| H9 | T leukemia | ++ | +/− |
| HSB2 | T leukemia | + | + |
| Raji | Burkitt lymphoma | − | − |
| DAUDI | Burkitt lymphoma | − | − |
| Namalwa | Burkitt lymphoma | − | +/− |
| LCL 721.221 | EBV cell line | − | − |
| LCL C1R | EBV cell line | − | − |
| RPMI 18226 | EBV cell line | + | +/− |
| K562 | Erythroleukemia | ++ | + |
| MM6 | Promyelocytic leukemia | ++ | +/− |
| U937 | Myeloid leukemia | + | +/− |
| 293T | Embryonic fibroblasts | ++ | ++ |
| M14 | Melanoma | ++ | + |
| MEL15392 | Melanoma | ++ | +/− |
| FO-1 | Melanoma | ++ | + |
| Me 1386 | Melanoma | ++ | + |
| Me 18105 | Melanoma | ++ | ++ |
| A549 | Lung carcinoma | ++ | + |
| SMMC | Hepatoma | + | + |
| HELA | Cervical carcinoma | + | + |
| IGROV-1 | Ovarian carcinoma | + | + |
| SKNEP | Kidney carcinoma | + | + |
| A172 | Glioblastoma | ++ | + |
| HT29 | Colon carcinoma | ++ | + |
| SW480 | Colon carcinoma | ++ | ++ |
| SK-N-BE | Neuroblastoma | +/− | +/− |
| LAN5 | Neuroblastoma | ++ | + |
| HTLA260 | Neuroblastoma | + | +/− |
| YAC-1 | Murine thymoma | − | − |
| BW1502 | Murine thymoma | − | − |
| P815 | Murine mastocytoma | − | − |
| COS-7 | Monkey kidney fibroblast | + | + |
| CHO-K | Chinese hamster, ovarian carcinoma | − |
− |
Normal cells and tumor cell lines of different histotype were analyzed by indirect immunofluorescence and FACS® analysis for reactivity with the indicated mAbs. Cells are of human origin unless otherwise specified. Mean fluorescence intensity (MFS): overlapping the negative control (−)(MFI set on 5); MFI below 20 (+/−); MFI between 20 and 100 (+); MFI over 100 (++). Immunizing cells (M14 and 293T) and MOLT4 are in bold.
Biochemical Characterization of the Surface Molecules Recognized by the Selected mAbs.
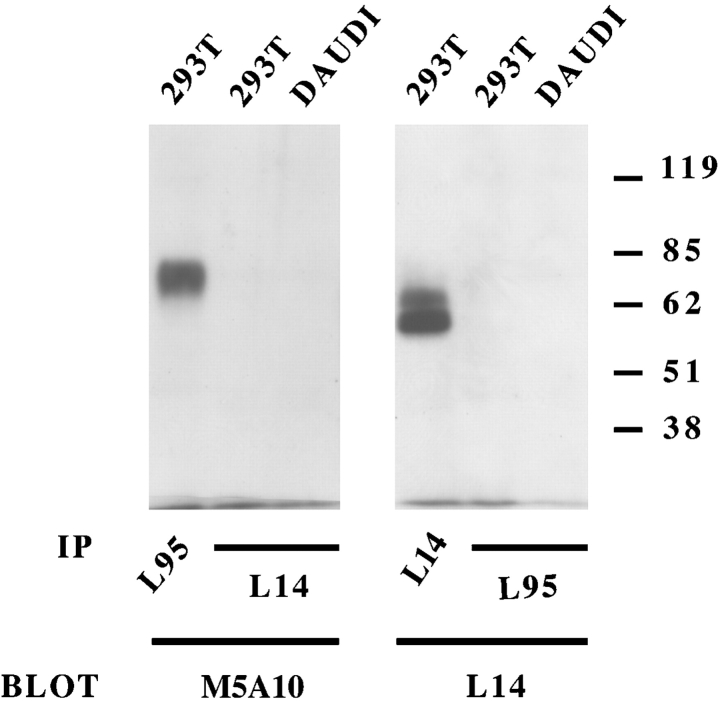
Different cell lines were surface labeled with biotin and cell lysates were immunoprecipitated with one or another mAb. In all instances, M5A10, M2C24, and L95 mAbs immunoprecipitated a surface molecule of ∼70 kD both under reducing (Fig. 2 A) and nonreducing conditions (not depicted). The protein backbone remaining after treatment with N-glycosidase F, displayed a molecular mass of ∼40–45 kD. L14 mAb immunoprecipitated two surface molecules of ∼65 and 60 kD, respectively, both under-reducing (Fig. 2 B) and nonreducing conditions (not shown). After treatment with N-glycosidase F these molecules displayed protein backbones of ∼60 and 55 kD, respectively. These results, together with data on cell surface distribution and cytotoxicity suggested that L14 mAb may recognize surface molecule(s) distinct from that reactive with the other mAbs. To gain further information, 293T cells were immunoprecipitated with each of the selected mAbs and, after blotting, probed either with the same mAb used for immunoprecipitation or with a different mAb. M5A10 (Fig. 3 , left panel) and M2C24 (not shown) reacted with the molecule immunoprecipitated by L95 mAb thus confirming that they recognized the same molecule. On the other hand, they did not recognize the molecule immunoprecipitated by L14 mAb and vice versa. It is of note that both the 65 and 60 kD surface molecules were recognized by L14 mAb also by Western blot (Fig. 3, right panel), suggesting that they might represent alternative forms of a single molecule.
Figure 2.
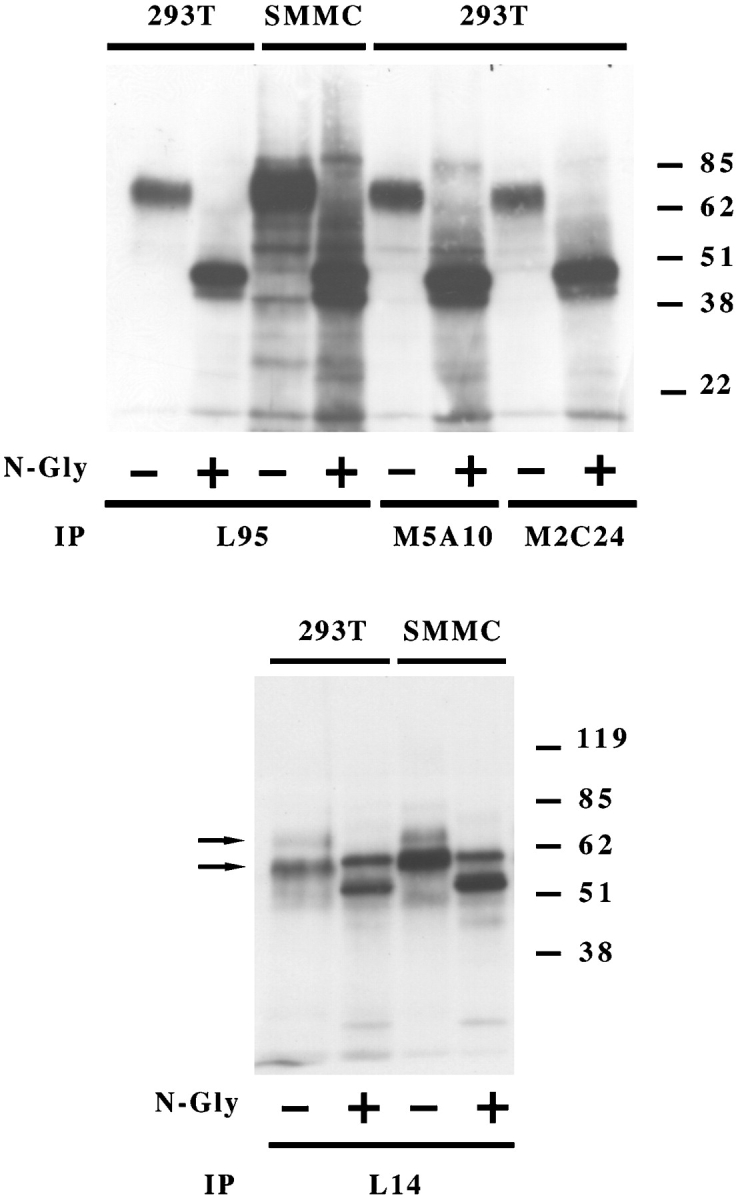
Biochemical characterization of the surface molecules recognized by M5A10, M2C24, L95, and L14 mAbs. The indicated cell lines were surface labeled with biotin and immunoprecipitated with L95, M5A10, M2C24 (top panel), and L14 (bottom panel) mAbs. Samples were treated (+) or not (−) with N-glycosidase F and analyzed in an 8% SDS-PAGE under reducing conditions. Molecular weight markers (kD) are indicated.
Figure 3.
Western blot analysis of M5A10, L95, and L14 mAbs. The indicated cell lines were immunoprecipitated with L95 and L14 mAbs. Samples were analyzed in an 8% SDS-PAGE under nonreducing conditions and probed with biotin-labeled M5A10 and L14 mAbs. Molecular weight markers (kD) are indicated. DAUDI cell lines were used as negative control.
Purification and Sequence Analysis of the Molecules Recognized by the Selected mAbs
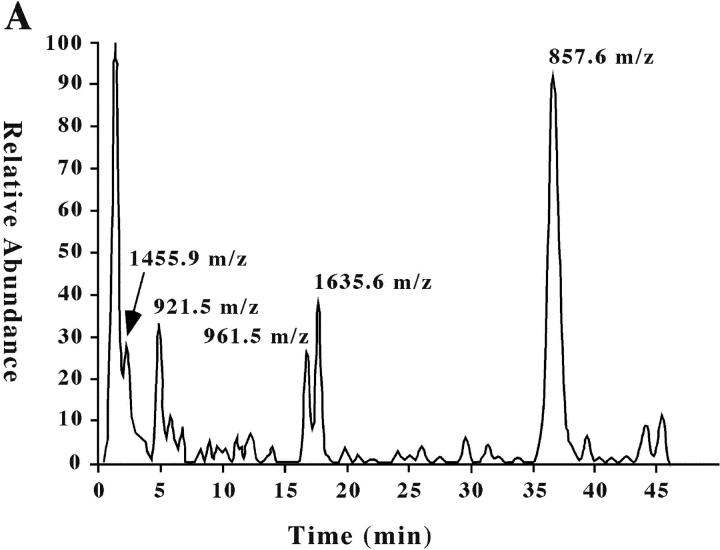
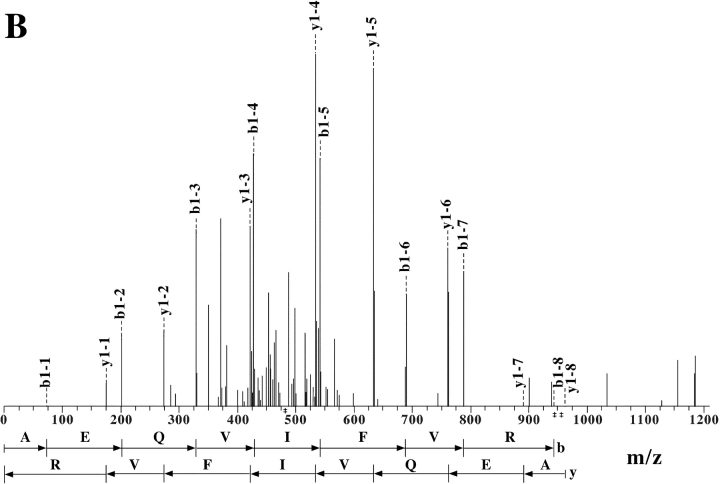
To ascertain the identity of the surface molecules identified by the four selected mAbs, cell membranes were prepared from 293T cells. After lysis, molecules were purified by affinity chromatography using L95 or L14 mAbs covalently bound to sepharose beads. Eluted proteins were concentrated and analyzed in SDS-PAGE under nonreducing conditions. The quality of the purification process was verified by the Western blot analysis of the eluted molecules with the corresponding mAb (not shown). After running, polyacrylamide gels were stained with coomassie blue. The band corresponding to the L14-specific molecules(s) were excised from the gel for the in-gel tryptic digestion and analyzed on a LC-MS/MS system. Fig. 4 A shows the ion chromatogram displaying the base peaks of the tryptic digest of the L14-specific molecule(s). Analysis of the mass spectra (full MS and MS/MS) using the TurboSEQUEST software showed five peptides with m/z (mass-to-charge ratio) of 1455.9, 921.5, 857.6, 961.5, and 1635.6. Fig. 4 B shows a representative fragmentation spectrum of peptide 961.5 m/z, from which its sequence (AEQVIFVR) was deduced. Similar spectra were obtained for the other four peptides of the same protein (see Table II). These results identified the L14-reactive protein(s) as Nectin-2 (δ and α isoforms; CD112). It is of note that the Nectin-2 δ and α isoforms are characterized by identical extracellular portions but different transmembrane and cytoplasmic regions (long and short, respectively). This was in agreement with our biochemical data showing that L14 mAb specifically recognized two surface molecules of different molecular mass (see above).
Figure 4.
Extract ion chromatogram of the L14-antigen tryptic digest and representative fragmentation spectrum for ion m/z 961. (A) Ion chromatogram displaying the base peaks of the tryptic digest of L14-specific molecule analyzed on a LC-MS/MS system as described in Materials and Methods. (B) MS/MS of the peptide with molecular ion 961.55 m/z from which the amino acid sequence AEQVIFVR was obtained in the automated system from NH2 to COOH terminal and vice versa.
Table II.
Amino Acid Sequence and m/z of Peptides Obtained by Tryptic Digestion of L14- or L95 mAb-defined Antigens
| Identified peptides | m/z | |
|---|---|---|
| Nectin-2 δa and αb | MGPSFPSPKPGSER | 1,455.9 |
| VQVLPEVR | 921.5 | |
| FTLVPSGR | 857.6 | |
| AEQVIFVR | 961.5 | |
| Nectin-2 αb | SPGGAGGGASGDGGFYDPK | 1,635.6 |
| PVR δc and αd | VLAKPQNTAEVQK | 1,425.8 |
| QAELTVQVK | 1,015.6 | |
| VQLTGEPVPPMAR | 1,297.7 | |
| HGESGSMAVFHQTQGPSYSESK | 2,351.0 | |
| RLEFVAAR | 961.6 |
Peptides identified from L14 specific protein, corresponding to Nectin-2δ (long) or α (short) isoforms, and from L95 specific protein, corresponding to PVRδ and α are shown. The bands, from SDS-PAGE, corresponding to the L14 or L95-specific molecules, were trypsin digested and analyzed on a LC-MS/MS system as described in Materials and Methods. The m/z (mass-to-charge ratio) of each peptide is reported.
(NCBI, gi 12643789, Nectin-2δ).
(NCBI, gi 2118920, Nectin-2α).
(NCBI, gi 319893, PVRδ).
(NCBI, gi 319892, PVRα).
Similar experiments were performed for the molecule recognized by the M5A10, M2C24, and L95 mAbs. In this case, the search in the NCBI human protein database of the MS/MS spectra analysis matched with the Poliovirus Receptor δ and α isoforms (PVR, CD155; for detected ions and amino acid sequences see Table II). The predicted molecular masses of identified proteins (57743 and 51358 kD for Nectin-2δ and α, respectively, and 42912 and 45351 kD for PVR δ and α, respectively) were in agreement with those obtained from SDS electrophoresis (see Fig. 2).
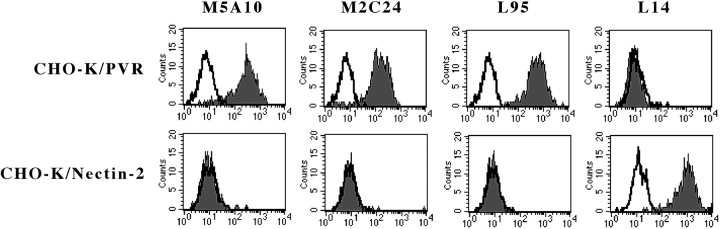
To unequivocally confirm that the two molecules corresponded to PVR (CD155) and Nectin-2 (CD112), respectively, hamster CHO-K cells either untransfected or transfected with PVR or Nectin-2 constructs were analyzed for their surface reactivity with the four mAbs. In all instances, no reactivity could be detected in untransfected CHO-K cells (not shown). On the other hand, M5A10, M2C24, and L95 mAbs specifically recognized PVRα cell transfectants while Nectin-2δ and Nectin-2α transfectants were uniquely stained by L14 mAb (Fig. 5) .
Figure 5.
Surface reactivity of M5A10, M2C24, L95 and L14 mAbs with CHO-K cells transfected with the PVR or Nectin-2 constructs CHO-K cells transfected with the PVRα or Nectin-2δ (or Nectin-2α, data not shown) constructs were stained with the indicated mAbs followed by PE-conjugated goat anti–mouse isotype-specific second reagent and analyzed by flow cytometry. White profiles indicate cells incubated with the second reagent only.
Identification of DNAM-1 as the Surface NK Receptor Specific for PVR and Nectin-2.
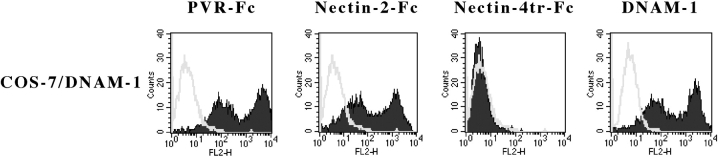
To identify the receptor(s) expressed by NK cells that specifically recognize PVR or Nectin-2 on target cells, chimeric molecules formed by the extracellular domain of PVR or Nectin-2 molecules and by the Fc portion of human IgG1 were analyzed for their surface reactivity with COS-7 cells transiently transfected with a panel of activating NK receptors. PVR-Fc and Nectin-2-Fc did not react with untransfected cells or with COS-7 cells transfected with NKp46 and NKp30 (not shown). On the contrary, COS-7 cells transfected with DNAM-1 (CD226) were brightly stained by both PVR-Fc and Nectin-2-Fc molecules (Fig. 6) . Nectin-4tr-Fc molecule, used as control, did not display surface reactivity with either untransfected or transfected COS-7 cells. In conclusion, all these data demonstrate that PVR and Nectin-2 represent specific cell surface ligands recognized by DNAM-1 receptor.
Figure 6.
Surface reactivity of PVR-Fc or Nectin-2-Fc molecules with DNAM-1 cell transfectants COS-7 cells transfected with DNAM-1 construct were stained with PVR-Fc, Nectin-2-Fc, or Nectin-4tr-Fc (negative control) molecules, followed by PE-conjugated goat anti–human second reagent, or with GN18 (anti-DNAM-1) mAb, followed by PE-conjugated goat anti–mouse isotype-specific second reagent. Samples were analyzed by flow cytometry. White profiles indicate cells incubated with the second reagent only.
The Interaction between DNAM-1 and PVR or Nectin-2 Promotes Human NK Cell Triggering and Cytotoxicity.
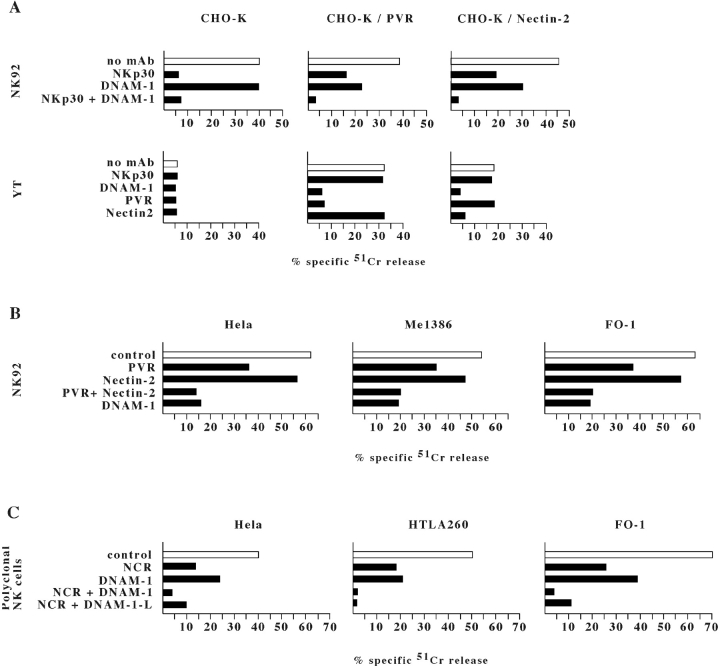
In the first set of experiments NK cells were assessed for cytolytic activity against the CHO-K hamster cell line either untransfected or transfected with PVR or Nectin-2 constructs. As shown in Fig. 7 A, NK92 cells killed untransfected CHO-K cells. Cytotoxicity was mostly dependent on NKp30–mediated recognition of undefined xenogeneic ligand(s) expressed on hamster cells since it was sharply inhibited by mAb-mediated masking of NKp30. On the contrary, in agreement with the lack of human PVR or Nectin-2 ligands on the CHO-K cells, mAb-mediated masking of DNAM-1 had no inhibitory effect. On the other hand, the NK92-mediated lysis of CHO-K cells that had been transfected with either PVR or Nectin-2 constructs was only partially inhibited by anti-NKp30 mAb. In these cell transfectants, cytotoxicity was partially inhibited also by anti-DNAM-1 mAb. Remarkably, the simultaneous masking of NKp30 and DNAM-1 virtually abrogated cytotoxicity (Fig. 7 A). These results were further substantiated by experiments in which the effector NK cells were represented by the YT NK cell line. YT express DNAM-1 and 2B4 but lack CD16, NKp30, NKp46, NKp44, and NKG2D molecules. Thus, due to the lack of the major triggering NK receptors, it is conceivable that in these cells DNAM-1 may play a predominant role in inducing cytotoxicity. As shown in Fig. 7 A, YT cells did not kill untransfected CHO-K cells. However, they lysed both PVR and Nectin-2 CHO-K cell transfectants. Moreover, mAb-mediated masking of either DNAM-1 or PVR or Nectin-2 virtually abrogated target cell lysis.
Figure 7.
mAb-mediated disruption of DNAM-1/ligands interaction inhibits the NK-mediated lysis of PVR or Nectin-2 cell transfectants and of tumor cell targets. (A) NK92 or YT NK cell lines were analyzed for cytolytic activity against the CHO-K cells either untransfected or transfected with PVRα or Nectin-2δ constructs. Cytolytic assays were performed either in the absence of mAb (white bars) or in the presence of F252, GN18, L14, and L95 mAbs to the indicated molecules (black bars). The effector/target ratio used was10:1 for NK92 and 40:1 for YT. Similar results were obtained with Nectin-2α transfectants (data not shown). (B) NK92 NK cell line pretreated with anti-NKG2D mAb (for 15 min at room temperature) was analyzed for cytolytic activity against the indicated tumor target cells in the presence of either a control mAb (white bars) or mAbs (as in panel A) to the indicated molecules (black bars). The effector/target ratio used was 10:1. (C) A polyclonal NK cell population pretreated with anti-NKG2D mAb was analyzed for cytolytic activity against the indicated tumor target cells in the presence of either a control mAb (white bars) or mAbs to NCR (F252 and KL247), DNAM-1 (KRA236), or DNAM-1-Ligands (L95 and L14). These mAbs were used alone or in combination as indicated. The effector/target ratio used was 4:1. The results are representative of four independent experiments; the standard deviation of the mean of the triplicates was <4%.
In most experiments, the YT-mediated lysis of PVR transfectants was slightly higher than that of Nectin-2 transfectants. As, in CHO-K transfectants the expression of Nectin-2 was similar or even higher than that of PVR the above difference may reflect an intrinsic disparity between the two ligands in the ability to promote DNAM-1–dependent NK cell activation.
DNAM-1 Contributes to NK-mediated Lysis of Tumor Cells.
As our data demonstrated that DNAM-1 recognizes two different ligands on target cells we next analyzed the role of the interactions between DNAM-1 and PVR and Nectin-2 in the NK-mediated cytotoxicity against a panel of tumor target cells. In a first set of experiments effector cells were represented by the NK92 NK cell line. As target cells expressed one or another NKG2D ligands, experiments were performed in the presence of anti-NKG2D mAb (see above). Although not shown, we could not detect any DNAM-1 involvement in NK-mediated lysis of target cells that lack PVR and/or Nectin-2. Lysis of PVR+, Nectin-2+ targets (see the representative HELA, Me1386, and FO-1 in Fig. 7 B) was partially inhibited by mAb-mediated masking of PVR or Nectin-2. Again, PVR appeared to play a predominant role in inducing DNAM-1–dependent NK cell triggering as compared with Nectin-2. Importantly however, the combined masking of both PVR and Nectin-2 resulted in inhibition of lysis comparable to that obtained by mAb-mediated masking of DNAM-1. Altogether, these data suggest that PVR and Nectin-2 cooperate in the induction of DNAM-1–dependent killing of target cells. In addition, they suggest that they represent the major cellular ligands of DNAM-1.
In another set of experiments we evaluated the role of DNAM-1-mediated recognition of PVR and Nectin-2 using polyclonal NK populations as a source of effector cells. These experiments were performed in order to compare the relative contribution of DNAM-1 versus NCR in the process of NK-mediated killing of different tumors including carcinomas, melanomas, and neuroblastomas.
It is of note that, at variance with NK92 or YT cells, the cytolytic activity of polyclonal NK cells is induced via multiple triggering receptors. These may display a more or less predominant involvement depending upon the expression of their cellular ligands at the target cell surface. Fig. 7 C shows the results of cytotoxicity experiments in which polyclonal NK cells were analyzed against three representative tumor cell lines. Lysis of HELA carcinoma was substantially inhibited by the use of anti-NCR mAbs although some inhibition was also evident in the presence of anti-DNAM-1 mAb; only the combined use of both reagents could abrogate lysis. In the case of FO1 melanoma, again a predominant role of NCR could be detected although blocking of DNAM-1 also resulted in potent inhibition. This was confirmed also by the finding that blocking both NCR and DNAM-1 (or its ligands) could virtually abrogate cytotoxicity. Finally, in the case of HTLA260 neuroblastoma, blocking of DNAM-1 resulted in an inhibitory effect comparable to that induced by anti-NCR mAbs. Although not shown, blocking of PVR alone on HTLA260 resulted in inhibition comparable to that mediated by anti-DNAM-1 mAb. This data is consistent with the finding that HTLA260 express PVR but very little Nectin-2.
In conclusion, these experiments further indicate that killing of different tumor targets by normal NK cells is the result of the engagement of multiple triggering receptors. The involvement of DNAM-1 can be appreciated in mAb-dependent blocking experiments against various tumors of different histotype. However we could not identify target cells that were lysed in a strictly DNAM-1–dependent manner.
The most prominent inhibition with anti-DNAM-1 antibodies was detected against the neuroblastoma cell line although this may simply reflect a reduced surface density of the NCR ligands on this target.
Discussion
In this study, mAbs specific for molecules expressed at the cell surface of NK-susceptible target cells were selected on the basis of their ability to down-regulate the NK-mediated cytolytic activity. As revealed by the LC-MS/MS amino acid sequence analysis, the surface molecules recognized by the selected mAbs were PVR and Nectin-2, i.e., two members of the Nectin family (23). Analysis of the surface reactivity of PVR-Fc and Nectin-2-Fc soluble hybrid molecules indicated that both molecules represent specific ligands of the DNAM-1 activating receptor (10, 11). The functional interaction between DNAM-1 and PVR or Nectin-2 was substantiated by cytolytic assays in which NK cells were used as effectors and hamster CHO-K cells transfected with PVR or Nectin-2 as target cells. Indeed, in these experiments mAb-mediated masking of DNAM-1 or PVR or Nectin-2 resulted in inhibition or even in virtual abrogation of cytotoxicity.
PVR (18) and Nectin-2 (19, 20) are closely related molecules encoded by genes located in the 19q13 chromosomal region. Alternative splicing of PVR (PVRα and PVRδ) and Nectin-2 (Nectin-2α and Nectin-2δ) yield to surface molecules characterized by identical extracellular portions, formed by three Ig-like domains in the order V/C/C (18, 19). Alternative splicing sites of PVR and Nectin-2 isoforms are located in the cytoplasmic and in the extracellular regions respectively. Differences in length of the Nectin-2α and δ cytoplasmic tails result in a short or long isoform, respectively. Importantly, PVR and Nectin-2, are highly expressed in tumor cell lines of epithelial or neuronal origin such as carcinomas, melanomas and neuroblastomas (see Table I).
DNAM-1 is a transmembrane glycoprotein, involved in lymphocyte adhesion and signaling, characterized by two extracellular Ig-like domains and by a cytoplasmic portion containing three tyrosine residues (11). It is expressed by virtually all human NK cells, T cells, monocytes, and by a small subset of B lymphocytes. Upon mAb-mediated cross-linking DNAM-1 transduces activating signals resulting in tyrosine phosphorylation of DNAM-1 itself, enhancement of cytotoxicity and cytokine production in both T and NK cells. Interestingly, the role played by DNAM-1 in both CTL- and NK-mediated killing varied upon the target cells analyzed. Thus, mAb-mediated blocking of DNAM-1 resulted in inhibition of cytotoxicity against colon or ovarian carcinomas, K562, Jurkat, and U937 cell lines. In contrast, lysis of the Epstein-Barr virus (EBV)-transformed LCL 721.221 B cell lines was unaffected (11). These data suggested that cell surface antigen(s) that are broadly expressed in both nonhematopoietic and hematopoietic tissues could represent the DNAM-1 ligand(s). Our present identification of PVR and Nectin-2 as DNAM-1–specific cellular ligands, is in agreement with this concept. Indeed, carcinomas and hematopoietic cell lines such as K562, Jurkat, and U937 express PVR and Nectin-2. Moreover, in accordance with the inability of anti-DNAM-1 mAb to inhibit T- or NK-mediated lysis of B-EBV cells, the majority of B-EBV cell lines analyzed did not express both molecules (see Table I). The involvement of DNAM-1 in the NK-mediated lysis of various tumor cell lines analyzed consistently correlated with the expression of PVR and Nectin-2 on target cells suggesting that they may represent major ligands for DNAM-1. It is of note that the involvement of DNAM-1 in NK cytotoxicity could be better appreciated when NK92 or YT were used as effectors instead of normal NK cells. This data is explained by the fact that normal NK cells express several activating receptors and coreceptors that cooperate in target cell recognition and killing (5). As a consequence, mAb-mediated blocking of individual receptors may not significantly inhibit cytotoxicity since most target cells express multiple ligands for the various receptors and coreceptors. On the contrary, the NK cell lines NK92 and YT lack one or more major activating receptor involved in NK cell cytotoxicity (8). Thus, the mAb-mediated disruption of a single receptor/ligand interaction (namely DNAM-1/PVR or Nectin-2) was sufficient to significantly inhibit cytolytic activity.
Our data shows that DNAM-1, upon engagement by the specific ligands (i.e., PVR and Nectin-2), cooperates with other triggering NK receptors in the induction of NK-mediated cytotoxicity. In this context, the analysis of polyclonal NK populations that express the whole set of triggering NK receptors indicated that the induction of cytotoxicity against the carcinoma, melanoma, and neuroblastoma cell lines analyzed is consequent to the engagement of both NCR and DNAM-1 (Fig. 7 C). Most tumor target cells overexpressing DNAM-1 ligands displayed a reduced expression of HLA class I molecules. Thus, in the absence of effective HLA class I/inhibitory receptors interactions, the engagement of DNAM-1 and other triggering NK receptors, resulted in induction of cytolytic activity. In the case of normal target cells, however, DNAM-1/ligands interactions should not result in activation of NK-mediated killing. Although some normal cells may express the DNAM-1 ligands they also express normal levels of HLA class I molecules that block the NK cell function upon interaction with inhibitory NK receptors. In this context, it is of note that normal endothelial or epithelial cells express members of the Nectin family that appear to be localized to intercellular borders (20). This specific localization may have relevant implication in the process of leukocyte recirculation. Leukocyte recirculation from the blood to lymphoid organs is critical for the induction of inflammatory responses. To recirculate, leukocytes must pass between endothelial cells, a process called extravasation. While LFA-1 molecules are crucial for the firm adhesion of leukocytes to endothelium (25), the final step, i.e., leukocyte migration between endothelial cells is largely unknown. Given the particular endothelial localization of nectins and the fact that DNAM-1 is expressed by different leukocytes, it is conceivable that Nectin/DNAM-1 interactions may also play a role in the interaction between leukocytes and endothelial cells in the process of leukocyte recirculation.
It is of note that DNAM-1 and LFA-1 molecules are physically and functionally associated in normal cells (12). Accordingly, in the absence of LFA-1, as in the case of LAD1 patients, the function of DNAM-1 was defective. Importantly, in normal NK cells not only the mAb-mediated cross-linking of DNAM-1 itself but also that of LFA-1 induces tyrosine phosphorylation of DNAM-1. These data support the notion that LFA-1/CAMs interactions may precede and modulate DNAM-1/Nectins interactions in target cell recognition and, possibly, in the process of extravasation.
Acknowledgments
We are grateful to Dr. Luciano Zardi for fruitful discussions and to Dr. Pier Luigi Mauri (Istituto Tecnologie Biomediche - CNR, Milano) for his valuable support in mass spectrometry analysis. We thank Ms. Tiziana Baffi for secretarial assistance.
This work was supported by grants awarded by Associazione Italiana per la Ricerca sul Cancro (A.I.R.C.), Istituto Superiore di Sanità (I.S.S.), Ministero dell'Istruzione, dell'Università e della Ricerca (M.I.U.R.), Consiglio Nazionale delle Ricerche, Progetto Finalizzato Biotecnologie, EU QLRT-2000-01495 Project “Engineering human antibody derivatives, which specifically recognize and ablate new blood vessels, for the therapy of angiogenesis-related pathologies”, INSERM, the Ligue Nationale Française Contre le Cancer (LNFCC), and the Association pour la Recherche contre le Cancer (ARC). Also the financial support of Fondazione Compagnia di San Paolo, Torino, Italy, is gratefully acknowledged. Roberta Castriconi is recipient of a fellowship awarded by Fondazione Italiana per la Ricerca sul Cancro (F.I.R.C.).
Abbreviations used in this paper: DNAM-1, DNAX accessory molecule-1; LFA-1, lymphocytes function-associated antigen 1; MIC, MHC class I–related chain molecules; NCR, natural cytotoxicity receptor; PVR, poliovirus receptor; ULBP, UL16-binding protein.
C. Bottino, R. Castriconi, and D. Pende contributed equally to this work.
References
- 1.Moretta, A., C. Bottino, M. Vitale, D. Pende, R. Biassoni, M.C. Mingari, and L. Moretta. 1996. Receptors for HLA-class I molecules in human natural killer cells. Annu. Rev. Immunol. 14:619–648. [DOI] [PubMed] [Google Scholar]
- 2.Long, E.O. 1999. Regulation of immune responses through inhibitory receptors. Annu. Rev. Immunol. 17:875–904. [DOI] [PubMed] [Google Scholar]
- 3.Ravetch, J.V., and L.L. Lanier. 2000. Immune inhibitory receptors. Science. 290:84–89. [DOI] [PubMed] [Google Scholar]
- 4.Shilling, H.G., N. Young, L.A. Guethlein, C.N. Wheng, C.M. Gardiner, D. Tyan, and P. Parham. 2002. Genetic control of human NK cell repertoire. J. Immunol. 169:239–247. [DOI] [PubMed] [Google Scholar]
- 5.Moretta, A., C. Bottino, M. Vitale, D. Pende, C. Cantoni, M.C. Mingari, R. Biassoni, and L. Moretta. 2001. Activating receptors and co-receptors involved in human natural killer cell-mediated cytolysis. Annu. Rev. Immunol. 19:197–223. [DOI] [PubMed] [Google Scholar]
- 6.Cerwenka, A., and L.L. Lanier. 2001. Ligands for natural killer cell receptors: redundancy or specificity. Immunol. Rev. 181:158–169. [DOI] [PubMed] [Google Scholar]
- 7.Sivori, S., S. Parolini, M. Falco, E. Marcenaro, R. Biassoni, C. Bottino, L. Moretta, and A. Moretta. 2000. 2B4 functions as a co-receptor in human natural killer cell activation. Eur. J. Immunol. 30:787–793. [DOI] [PubMed] [Google Scholar]
- 8.Bottino, C., M. Falco, S. Parolini, E. Marcenaro, R. Augugliaro, S. Sivori, E. Landi, R. Biassoni, L. Notarangelo, L. Moretta, and A. Moretta. 2001. NTB-A, a novel SH2D1A-associated surface molecule contributing to the inability of natural killer cells to kill Epstein-Barr virus-infected B cells in X-linked lymphoproliferative diseases. J. Exp. Med. 194:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vitale, M., M. Falco, R. Castriconi, S. Parolini, R. Zambello, G. Semenzato, R. Biassoni, C. Bottino, L. Moretta, and A. Moretta. 2001. Identification of NKp80, a novel triggering molecule expressed by human natural killer cells. Eur. J. Immunol. 31:233–242. [DOI] [PubMed] [Google Scholar]
- 10.Burns, G.F., T. Triglia, J.A. Werkmeister, C.G. Begley, and A.W. Boyd. 1985. TLiSA1, a human T lineage-specific activation antigen involved in the differentiation of cytotoxic T lymphocytes and anomalous killer cells from their precursors. J. Exp. Med. 161:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibuya, A., D. Campbell, C. Hannum, H. Yssel, K. Franz-Bacon, T. McClanahan, T. Kitamura, J. Nicholl, G.R. Sutherland, L.L. Lanier, and J.H. Phillips. 1996. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity. 4:573–581. [DOI] [PubMed] [Google Scholar]
- 12.Shibuya, K., L.L. Lanier, J.H. Phillips, H.D. Ochs, K. Shimizu, E. Nakayama, H. Nakauchi, and A. Shibuya. 1999. Physical and functional association of LFA-1 with DNAM-1 adhesion molecule. Immunity. 11:615–623. [DOI] [PubMed] [Google Scholar]
- 13.Bauer, S., V. Groh, J. Wu, A. Steinle, J.H. Phillips, L.L. Lanier, and T. Spies. 1999. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 285:727–729. [DOI] [PubMed] [Google Scholar]
- 14.Cosman, D., J. Mulberg, C. Sutherland, W. Chin, R. Armitage, W. Fanslow, M. Kubin, and J. Chalupny. 2001. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 14:123–133. [DOI] [PubMed] [Google Scholar]
- 15.Pende, D., P. Rivera, S. Marcenaro, C.C. Chang, R. Biassoni, R. Conte, M. Kubin, D. Cosman, S. Ferrone, L. Moretta, and A. Moretta. 2002. Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumor cell lines of different histotypes: analysis of tumor susceptibility to NKG2D-dependent natural killer cell cytotoxicity. Cancer Res. 62:6178–6186. [PubMed] [Google Scholar]
- 16.Kubin, M.Z., D.L. Parsley, W. Din, J.Y. Waugh, T. Davis-Smith, C.A. Smith, B.M. Macduff, R.J. Armitage, W. Chin, L. Cassiano, et al. 1999. Molecular cloning and biological characterization of NK cell activation-inducing ligand, a counterstructure for CD48. Eur. J. Immunol. 29:3466–3477. [DOI] [PubMed] [Google Scholar]
- 17.Nakajima, H., M. Cella, H. Langen, A. Friedlein, and M. Colonna. 1999. Activating interactions in human NK cell recognition: the role of 2B4-CD48. Eur. J. Immunol. 29:1676–1683. [DOI] [PubMed] [Google Scholar]
- 18.Mendelsohn, C.L., E. Wimmer, and V.R. Racaniello. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 56:855–865. [DOI] [PubMed] [Google Scholar]
- 19.Eberle, F., P. Dubreuil, M.G. Mattei, E. Devilard, and M. Lopez. 1995. The human PRR2 gene, related to the human poliovirus receptor gene (PVR), is the true homolog of the murine MPH gene. Gene. 159:267–272. [DOI] [PubMed] [Google Scholar]
- 20.Lopez, M., M. Aoubala, F. Jordier, D. Isnardon, S. Gomez, and P. Dubreuil. 1998. The human poliovirus receptor related 2 protein is a new hematopoietic/endothelial homophilic adhesion molecule. Blood. 92:4602–4611. [PubMed] [Google Scholar]
- 21.Ha, G.H., S.U. Lee, D.G. Kang, N.Y. Ha, S.H. Kim, J. Kim, J.M. Bae, J.W. Kim, and C.W. Lee. 2002. Proteome analysis of human stomach tissue: separation of soluble proteins by two-dimensional polyacrylamide gel electrophoresis and identification by mass spectrometry. Electrophoresis. 23:2513–2524. [DOI] [PubMed] [Google Scholar]
- 22.Koike, S., H. Horie, I. Ise, A. Okitsu, M. Yoshida, N. Iizuka, K. Takeuchi, T. Takegami, and A. Nomoto. 1990. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 9:3217–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reymond, N., S. Fabre, E. Lecocq, J. Adelaide, P. Dubreuil, and M. Lopez. 2001. Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with nectin1/PRR1 through V domain interaction. J. Biol. Chem. 276:43205–43215. [DOI] [PubMed] [Google Scholar]
- 24.Sivori, S., D. Pende, C. Bottino, E. Marcenaro, A. Pessino, R. Biassoni, L. Moretta, and A. Moretta. 1999. NKp46 is the major triggering receptor involved in the natural cytotoxicity of fresh or cultured human natural killer cells. Correlation between surface density of NKp46 and natural cytotoxicity against autologous, allogeneic or xenogeneic target cells. Eur. J. Immunol. 29:1656–1666. [DOI] [PubMed] [Google Scholar]
- 25.Springer, T.A. 1994. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 76:301–314. [DOI] [PubMed] [Google Scholar]