Abstract
Using current diagnostic criteria, primary mediastinal B cell lymphoma (PMBL) cannot be distinguished from other types of diffuse large B cell lymphoma (DLBCL) reliably. We used gene expression profiling to develop a more precise molecular diagnosis of PMBL. PMBL patients were considerably younger than other DLBCL patients, and their lymphomas frequently involved other thoracic structures but not extrathoracic sites typical of other DLBCLs. PMBL patients had a relatively favorable clinical outcome, with a 5-yr survival rate of 64% compared with 46% for other DLBCL patients. Gene expression profiling strongly supported a relationship between PMBL and Hodgkin lymphoma: over one third of the genes that were more highly expressed in PMBL than in other DLBCLs were also characteristically expressed in Hodgkin lymphoma cells. PDL2, which encodes a regulator of T cell activation, was the gene that best discriminated PMBL from other DLBCLs and was also highly expressed in Hodgkin lymphoma cells. The genomic loci for PDL2 and several neighboring genes were amplified in over half of the PMBLs and in Hodgkin lymphoma cell lines. The molecular diagnosis of PMBL should significantly aid in the development of therapies tailored to this clinically and pathogenetically distinctive subgroup of DLBCL.
Keywords: gene expression profiling, microarray, outcome prediction, PMBL, DLBCL
Introduction
Primary mediastinal B cell lymphoma (PMBL) has been recognized as a subtype of diffuse large B cell lymphoma (DLBCL) based on its distinctive clinical and morphological features (1). In many patients given this diagnosis, the only site of lymphoma involvement is the mediastinum, but the lymphoma can extend locally to involve other thoracic structures and occasionally disseminate to distinctive extranodal sites such as the kidney and the brain (2). PMBL patients tend to be young, with a median age of 30–35 yr at diagnosis, which contrasts with other DLBCL patients who have a median age of >60 yr at diagnosis. Pathologically, PMBL tumors are frequently highly sclerotic and are characterized by a diffuse proliferation of large cells, often with a clear cytoplasm.
Immunohistochemical analysis has confirmed the B cell origin of this lymphoma, although the expression of immunoglobulin genes is notably low (1). The relationship of PMBL to other DLBCL subgroups is uncertain. One DLBCL subgroup, termed germinal center B cell (GCB)–like DLBCL, maintains the gene expression program of normal germinal center B cells, whereas another DLBCL subgroup, termed activated B cell (ABC)–like DLBCL, expresses genes characteristic of activated B cells and plasma cells (3–5). PMBLs have low expression of CD10, a marker of the germinal center stage of B cell differentiation, prompting speculation that PMBL may be distinct from the GCB DLBCL subgroup (1).
Comparative genomic hybridization and fluorescence in situ hybridization analyses have supported the notion that PMBL is a pathogenetically distinct subgroup of DLBCL. Gains of chromosome arm 9p have been detected in over half of the PMBL cases, and this karyotypic abnormality is only occasionally detected in other DLBCLs (6, 7). Chromosome 9p gains can also be accompanied by amplification of the JAK2 gene, and interestingly, both 9p gains and JAK2 amplification have also been detected in Hodgkin lymphoma (8). Some patients with Hodgkin lymphoma have been noted to develop PMBL within 1 yr after treatment, and some “grey zone” lymphomas can have histological features that are intermediate between Hodgkin lymphoma and PMBL (9, 10). These observations have led to speculation that PMBL and Hodgkin lymphoma may be pathogenetically related (1, 11).
Clinically, PMBL is an aggressive lymphoma, and its relative responsiveness to treatment is controversial (1). Some studies concluded that PMBL patients have a relatively poor prognosis (12, 13), but another study showed a 5-yr overall survival rate of 46% with anthracycline-based chemotherapy, similar to that of other DLBCLs (14). A more recent study that combined chemotherapy with radiotherapy reported an 82% overall survival at 3 yr, a rate much higher than in other DLBCLs (15).
Imprecision in the diagnosis of PMBL may account for some of the heterogeneity in reported clinical responses. In particular, other DLBCLs that may originate by chance in the mediastinal region may be confused with PMBL. Currently, no molecular tests are routinely available for the diagnosis of PMBL. Two genes, MAL and FIG1 , are expressed frequently in PMBLs, but these markers may not identify all PMBL cases, and FIG1 is also expressed in some DLBCLs (16, 17).
Figure 1.

Identification of a PMBL gene expression signature. (A) Hierarchical clustering identified a set of 23 PMBL signature genes that were more highly expressed in most lymphomas with a clinical diagnosis of PMBL than in lymphomas assigned to the GCB or ABC DLBCL subgroups. Each row presents gene expression measurements from a single Lymphochip microarray feature representing the genes indicated. Each column represents a single lymphoma biopsy sample. Relative gene expression is depicted according to the color scale shown. (B) Hierarchical clustering of the lymphoma biopsy samples based on expression of the PMBL signature genes identified in A. A “core” cluster of lymphoma cases was identified that highly expressed the PMBL signature genes.
We undertook a gene expression profiling study of PMBL to establish a molecular diagnosis of this disease. We identified a gene expression signature of PMBL that distinguished this subgroup from other DLBCLs and showed that PMBL patients have distinctive clinical features and a favorable overall survival rate after therapy. The PMBL signature genes revealed an extraordinarily robust gene expression relationship between PMBL and Hodgkin lymphoma, strongly supporting a pathogenetic relationship between these two lymphoma types.
Materials and Methods
Analysis of Gene Expression and Clinical Data.
Pretreatment lymphoma biopsy samples were studied according to a protocol approved by the NCI Institutional Review Board. Lymphoma biopsies were reviewed by a panel of hematopathologists and were found to be DLBCLs morphologically. A “training” set of cases consisted of 36 biopsy specimens from 35 patients for whom the diagnosis of PMBL was considered. These patients all had mediastinal masses of at least 6 cm at presentation. These samples were profiled for gene expression using Lymphochip DNA microarrays comprised of 15,133 cDNA elements as described (3), and the data are available at http://llmpp.nih.gov/PMBL. A “validation” set of 274 lymphoma samples was previously profiled using Lymphochip DNA microarrays comprised of 12,196 cDNA elements (4); data for these samples were obtained from http://llmpp.nih.gov/DLBCL. All patients were treated with anthracycline-containing multiagent chemotherapy protocols with some patients additionally receiving radiation therapy.
The Bayesian statistical procedure used to create the gene expression-based PMBL predictor has been described (5). In the training set of cases, a Bayesian PMBL predictor was constructed from the 46 genes shown in Fig. 2 A. Since cases in the validation set were profiled on Lymphochip microarrays that lacked some of these genes, we constructed another Bayesian PMBL predictor using the 26 discriminating genes that were represented on these microarrays. After demonstrating that this predictor performed identically to the 46-gene predictor on the training set (not shown), it was then used to classify cases in the validation set of cases without reoptimization of the model parameters (Fig. 2 B).
Figure 2.

Development of a gene expression-based molecular diagnosis of PMBL. (A) A PMBL predictor was created based on the expression of the 46 genes shown. Relative gene expression for each lymphoma biopsy sample is presented according to the color scale shown in Fig. 1. The probability that each sample is PMBL or DLBCL based on gene expression is shown at the top. See Results for details. (B) Validation of the PMBL predictor. The PMBL predictor was used to classify 274 lymphoma samples from an independent cohort of patients (4) as PMBL or DLBCL. Some patients had been diagnosed as PMBL based on current diagnostic criteria (PMBL clinical diagnosis), whereas others had not been given this diagnosis (DLBCL clinical diagnosis). The prediction results are summarized on the right, and the relative gene expression for each case that was classified by the predictor as PMBL is shown on the left. In addition, the average expression of each gene in the samples classified as DLBCL is shown. Shown are the 20 genes from the PMBL predictor (A) that were more highly expressed in PMBL than in DLBCL and that were represented on the Lymphochip microarrays used to profile this set of lymphoma samples (4). Not shown are eight genes from the PMBL predictor that were more highly expressed in DLBCL than in PMBL.
Survival probabilities were estimated using the Kaplan-Meier method. P values for survival differences were evaluated using a log-rank test. P values for differences in age group and gender were generated using a chi-squared test. P values for differences in age as a continuous variable were computed using an ANOVA test.
Genomic Copy Number Analysis.
Quantitative PCR assays were used to assess the genomic copy number of the PDL2 gene relative to the control PRKCQ gene, as described (18). Control samples of genomic DNA from peripheral blood mononuclear cells of normal volunteers yielded a PDL2 to PRKCQ ratio of 0.99 with a SD of 0.08. A threshold PDL2 to PRKCQ ratio for gain/amplification of the PDL2 genomic locus was set at 1.31, which is four SDs above the mean. A biopsy specimen comprised of 100% malignant cells would be expected to yield a PDL2/PRKCQ ratio of 1.5 if the malignant cells had a gain of a single chromosome copy. In some of the DLBCL tumor biopsies studied, up to 40% of the cells were reactive normal cells; in such biopsies, a gain of a single chromosome copy in the malignant cells would yield a PDL2 to PRKCQ ratio of 1.30. PCR primers for PDL2 amplification were 5′-CTGGCCAAACGTCAGCGT-3′ and 5′-TGACCTGGTAGAGGCCTTCAG-3′, and the fluorescent Taqman probe was 5′-CCTGCCAACACCAGCCACTCCAG-3′. PCR primers for PRKCQ amplification were 5′-TCGCCATTTCTTCGGATTG-3′ and 5′-GCCTCGCCCTGACAAGACT-3′, and the fluorescent Taqman probe was 5′-TGTCCAACTTTGACTGCGGGTCCTG-3′. JAK2 and SMARCA2 genomic copy numbers were assessed relative to the CDKN2C gene. In control peripheral blood mononuclear cell samples, the JAK2 to CDKN2C ratio was 0.91 ± 0.08, and the SMARCA2 to CDKN2C ratio was 0.99 ± 0.13. PCR primers for JAK2 amplification were 5′-GTCCTAATGATCTCTTAGCTAGGATGTG-3′ and 5′-AATATGCTCATGATCCCAGATTTTC-3′, and the fluorescent Taqman probe was 5′-TTTATGTTGACAACAGATTTGAAT-AACTCGAGCAAA-3′. PCR primers for SMARCA2 amplification were 5′-CGAGTTTTGCCTTGTGGACTG-3′ and 5′-CCTTATCCCCTTTGGGTTAGTGA-3′, and the fluorescent Taqman probe was 5′-AAGGTTCAAGTTTGCTAGTCCGCAGATTGC-3′. PCR primers for CDKN2C amplification were 5′-TCAGGAGTCGGGAGGAATAAAA-3′ and 5′-CGGCATGACCGTAGAGACAA-3′, and the fluorescent Taqman probe was 5′-AATTTTCTAATCAGAGCTCAGCTGCAGTGTC-3′.
Expression of PMBL Signature Genes in Primary Hodgkin Reed-Sternberg Cells.
A case of nodular sclerosis classical Hodgkin lymphoma with easily recognizable Hodgkin Reed-Sternberg (HRS) cells was identified and 4-μm sections were cut from the frozen tissue block embedded in OCT and applied to plastic film–coated slides (Leica). The section was briefly stained in freshly prepared hematoxylin and eosin after fixation in 70% ethyl alcohol. Microdissection was performed using a Leica AS LMD. The cutting nitrogen laser used in this microdissection method destroyed reactive cells that surrounded the HRS cells. About 700 HRS cells were dissected from the tissue sections and collected in 400 μl of Trizol reagent (Invitrogen). An equivalent number of cells from the Hodgkin cell line L428, the PMBL cell line K1106, and the GCB DLBCL cell line OCI-Ly19 was also collected in Trizol. RNA was precipitated from each sample and resuspended in 190 μl of DEPC-treated water. A 5-μl aliquot of RNA was used in a Taqman one-step quantitative RT-PCR assay performed according to the manufacturer's protocol (Applied Biosystems) using Assays-on-Demand reagents for the following genes: CCL17/TARC (Hs00171074), SNFT (Hs00232744), MAL (Hs00242749), and TNFRSF6/Fas (Hs00236330) relative to ACTB/β-actin (Hs99999903_m1). Immunoperoxidase staining for MAL protein in a case of classical Hodgkin lymphoma was performed as described (16).
Online Supplemental Material.
Fig. S1 shows the expression of “lymph node” signature genes in PMBLs, and Fig. S2 shows the expression of PMBL signature genes in Hodgkin lymphoma cell lines. Figs. S1 and S2 are available at http://www.jem.org/cgi/content/full/jem.20031074/DC1.
Results
Development of a PMBL Predictor.
To discover a gene expression signature of PMBL, we first identified 35 DLBCL patients whose lymphomas involved the mediastinum prominently and for whom the diagnosis of PMBL was entertained. Since the clinical diagnosis of PMBL is imprecise, we anticipated that this group of patients would be heterogeneous, including some cases of “true” PMBL and other cases with standard DLBCL that happened to involve the mediastinum. We used Lymphochip DNA microarrays to profile gene expression in tumor biopsies from these patients and in 25 biopsies from patients with DLBCL who had previously been assigned to either the ABC or GCB subgroup (4). A hierarchical clustering algorithm (19) was used to organize the genes by their expression patterns across these lymphoma samples. We noted a large group of genes that were tightly clustered together in the resulting dendrogram and that were more highly expressed in the lymphomas with mediastinal involvement than in the other DLBCLs (Fig. 1 A). This gene cluster included two genes that had previously been shown to be highly expressed in PMBL cases, MAL and FIG1 (16, 17), along with many other genes that had not previously been associated with PMBL.
Some of the lymphomas with mediastinal involvement did not express this set of putative PMBL signature genes, and we suspected that these cases were more likely to be conventional DLBCLs than PMBL. We used hierarchical clustering to organize the cases according to their expression of the PMBL signature genes, which resulted in two major clusters of cases (Fig. 1 B). One cluster contained 21 samples that were designated as “PMBL core” samples by virtue of their high expression of PMBL signature genes. The other cluster contained some samples that had virtually no expression of these genes and other samples that did express these genes, albeit at lower levels than in the PMBL core cases.
We next developed a gene expression-based method to distinguish the PMBL core cases from the GCB and ABC DLBCL cases. This PMBL predictor used a Bayesian algorithm to assign a probability that an individual case belonged to the PMBL core group versus the DLBCL group (5). Briefly, this algorithm begins with the selection of a set of differentially expressed genes between two cancer groups, A and B. For each biopsy sample, the expression levels of these genes are combined linearly to derive a single “linear predictor score.” Each cancer group will have a different distribution of linear predictor scores. Based on these distributions, Bayes rule can be used to estimate the probability that a particular sample belongs to cancer group A or cancer group B. An arbitrary probability cutoff of ≥90% is used to classify a sample into a cancer group.
Given the heterogeneity of DLBCL, it was challenging to select an optimal set of differentially expressed genes to be used in the PMBL predictor. As a first step, we chose genes that were differentially expressed between the PMBL core group and both the GCB and ABC DLBCL subgroups (P < 0.001). This set of genes included all of the PMBL signature genes that had been identified by hierarchical clustering (Fig. 1 A) and a large number of additional genes. Many of the genes in this set belonged to the so-called “lymph node” gene expression signature, which was previously identified as a variable feature of DLBCL tumors that appears to reflect a host response to the malignant cells (3, 4). The lymph node signature includes genes encoding extracellular matrix components and genes that are characteristically expressed in macrophage, NK, and T cells. It is well known that PMBL tumors are often intensely fibrotic (20), and indeed, most of the biopsy samples in the PMBL core group had high expression of the lymph node signature genes (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20031074/DC1). We chose to exclude these genes from our PMBL predictor since they might cause some DLBCLs with high expression of the lymph node signature genes to be misclassified as PMBLs. Therefore, we refined our list of PMBL distinction genes by requiring that they also be differentially expressed between the PMBL core group and subgroup of six DLBCLs with high expression of the lymph node signature genes (P < 0.001).
The resulting list of PMBL/DLBCL distinction genes was used to create a PMBL predictor, which included 35 genes that were more highly expressed in PMBL and 11 genes that were more highly expressed in DLBCL (Fig. 2 A). All of the PMBL core samples were classified as PMBL according to this predictor as were six of the other lymphomas with mediastinal involvement. However, nine of the lymphomas with mediastinal involvement were classified as DLBCL by the predictor, as were all of the GCB and ABC DLBCLs.
We next tested the performance of the PMBL predictor on an independent set of 274 DLBCL cases that we had previously analyzed by gene expression profiling (4). In this validation set, 11 cases were identified on clinical grounds as being consistent with a diagnosis of PMBL, and the PMBL predictor classified 9 of these as PMBL (Fig. 2 B). Interestingly, 12 of the remaining 263 DLBCL samples were classified as PMBL by the predictor. Fig. 2 B shows that these cases were indistinguishable by gene expression from the 9 cases given a diagnosis of PMBL on clinical grounds. As expected, the average expression of the PMBL predictor genes in the 249 samples classified as DLBCL was notably lower than in the 22 PMBL cases (Fig. 2 B). Thus, PMBL represents a third subgroup of DLBCL that can be distinguished by gene expression profiling from GCB DLBCL and ABC DLBCL.
Previously, we developed a Bayesian predictor of the distinction between the ABC and GCB DLBCL subgroups (21). Of the 48 cases assigned to the PMBL subgroup, this ABC/GCB predictor classified 42 as GCB DLBCL and none as ABC DLBCL, whereas 8 cases remained unclassified. Although the PMBL subgroup was somewhat more related to the GCB subgroup than the ABC subgroup, the GCB and PMBL subgroups were easily distinguishable using the PMBL predictor.
The PMBL Subgroup Is Clinically Distinct from Other DLBCL Subgroups.
Table I compares the clinical parameters of patients assigned to the PMBL, ABC, and GCB subgroups of DLBCL by our gene expression-based predictor. PMBL patients were significantly younger than other DLBCL patients, with a median age at diagnosis of 33 yr compared with 66 and 61 yr for ABC and GCB DLBCL patients, respectively. Although there was no significant difference in the gender distributions among the DLBCL subgroups, young women (age <35 yr) accounted for 35% of the PMBL patients, which was significantly higher than in the other DLBCL subgroups. Young men (age <35 yr) were also more frequently represented in the PMBL subgroup, accounting for 19% of the patients. Correspondingly, older men and women (age >60) were significantly underrepresented in the PMBL subgroup. These clinical characteristics were observed in both the training and validation sets of PMBL cases, demonstrating that the PMBL predictor reproducibly identified a clinically distinct subgroup of DLBCL patients.
Table I.
Clinical Characeristics of PMBL and Other DLBCL Patients
| PMBL | PMBL | PMBL | ||||
|---|---|---|---|---|---|---|
| ABC DLBCL (n = 86) |
GCB DLBCL (n = 103) |
Training set (n = 18) |
Validation set (n = 25) |
All cases (n = 43) |
P value | |
| Median age | 66 | 61 | 33 | 33 | 33 | 4.4E-16 |
| Age <35 | 5% | 10% | 52% | 56% | 53% | 7.2E-14 |
| Age 35–60 | 29% | 38% | 44% | 28% | 37% | |
| Age >60 | 66% | 52% | 4% | 17% | 9% | |
| Gender = male | 59% | 53% | 44% | 50% | 47% | 0.38 |
| Female <35 | 2% | 3% | 32% | 39% | 35% | 1.1E-12 |
| Male <35 | 2% | 7% | 20% | 17% | 19% | |
| Female 35–60 | 6% | 18% | 24% | 6% | 16% | |
| Male 35–60 | 23% | 19% | 20% | 22% | 21% | |
| Female >60 | 33% | 25% | 0% | 6% | 2% | |
| Male >60 | 34% | 27% | 4% | 11% | 7% |
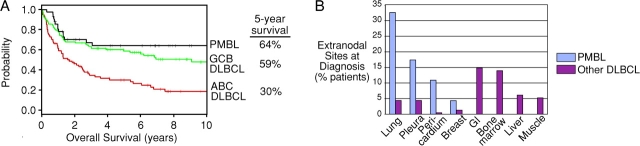
The PMBL subgroup defined by the PMBL predictor had a relatively favorable overall survival rate after therapy (Fig. 3 A). PMBL patients had a 5-yr survival rate of 64%, which was superior to the rate of 46% for all DLBCL patients (P = 0.0067). The survival of the PMBL subgroup was significantly better than the 30% 5-yr survival rate of the ABC DLBCL subgroup (Fig. 3 A; P = 5.8 E-5), but only marginally better than the 59% 5-yr survival rate of the GCB DLBCL subgroup (P = 0.18). In our series, 14 PMBL patients received radiation therapy in addition to multiagent chemotherapy, but their survival was not significantly different from those that did not (not shown). Notably, no patients in the PMBL subgroup died after 37 mo, suggesting that those PMBL patients who survive more than 3 yr have a high probability of being cured by multiagent chemotherapy.
Figure 3.
Clinical characteristics of PMBL patients. (A) Kaplan-Meier plot of overall survival of PMBL, GCB DLBCL, and ABC DLBCL patients after chemotherapy. (B) Distribution of extranodal sites of disease involvement at diagnosis for PMBL and other DLBCL patients.
Finally, the lymphomas of PMBL patients characteristically involved other thoracic structures in addition to the mediastinum (Fig. 3 B). Lung, pleura, pericardium, and breast were more frequently involved in PMBL than in other DLBCLs. Conversely, the gastrointestinal tract, bone marrow, liver, and muscle were more frequently involved in other DLBCLs than in PMBLs. This distinctive tissue distribution of the PMBLs further supports their classification as a separate DLBCL subgroup.
Chromosome 9p Aberrations in PMBL.
Previous studies demonstrated that gains of chromosome arm 9p are frequent in PMBL, and one case of PMBL was found to have a high level amplification of the JAK2 gene, which resides at chromosome band 9p24 (6–8). We noted that JAK2 and three other genes located near JAK2 on chromosome band 9p24 (PDL1, PDL2, and SMARCA2) were all expressed at significantly higher levels in PMBLs than in other DLBCLs (Figs. 1 and 2). Indeed, PDL2 was the overall best PMBL distinction gene represented on the microarray, with a 5.6-fold higher expression, on average, in PMBLs than in other DLBCLs (P = 7.55 E-15) (Fig. 2 A).
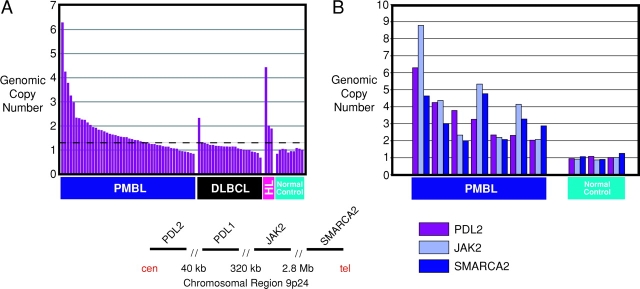
To directly assess chromosome 9p gains and amplifications in our set of PMBLs, we designed a quantitative PCR assay for PDL2 in which the genomic copy number of this gene was compared with that of a control gene on chromosome band 10p15 (PRKCQ). Using control genomic DNA from peripheral blood mononuclear cells of 10 normal donors, the average PDL2 to PRKCQ ratio was 0.99, as expected (Fig. 4 A). More than half of the PMBLs tested (30 out of 48; 62.5%) had elevated PDL2 to PRKCQ ratios (see Materials and Methods), consistent with a chromosome 9p gain and/or amplification of the PDL2 locus. By contrast, only 1 out of 23 DLBCL samples had a similarly elevated PDL2 to PRKCQ ratio. Seven of the PMBLs (23%) had PDL2 to PRKCQ ratios in excess of two, consistent with an amplification of the PDL2 locus. Quantitative PCR assays for the JAK2 and SMARCA2 genes detected coamplification of these genes in the seven cases with a twofold PDL2 amplification (Fig. 4 B), and no PMBL cases were found with only amplification of the JAK2 gene (not shown). The preferential gain/amplification of this chromosomal region in PMBL underscores the unique pathogenesis of this DLBCL subgroup.
Figure 4.
Amplification of genes on chromosome band 9p24 in PMBL. (A) The genomic copy number of the PDL2 gene on chromosome arm 9p is compared with that of the control PRKCQ gene. A PDL2 to PRKCQ ratio above the threshold indicated by the dashed line was taken as evidence of a gain/amplification of the PDL2 genomic locus (see Materials and Methods for details). HL, Hodgkin lymphoma cell lines L428, HDLM2, and L540. Normal control is defined as peripheral blood mononuclear cell samples from normal volunteers. (B) Coamplification of the PDL2, JAK2, and SMARCA2 genes in PMBL. The genomic copy numbers of the JAK2 and SMARCA2 genes relative to the control CDKN2C gene are shown for seven PMBL cases with greater than twofold amplification of PDL2 (A) and for three normal control samples. The structure of the chromosome 9p24 region near the PDL2 gene is shown. cen, centromere; tel, telomere.
Similarities Between PMBL and Hodgkin Lymphoma.
Two of the PMBL distinction genes, CD30 and TARC, are both characteristically expressed in the malignant HRS cells of Hodgkin lymphoma (22, 23). This led us to investigate whether other PMBL distinction genes might be expressed in Hodgkin lymphoma. Given the rarity of HRS cells in primary Hodgkin lymphoma biopsies, we addressed this question by profiling gene expression in three cell lines derived from Hodgkin lymphoma, L428, HDLM2, and L540. Previous gene expression profiling studies of Hodgkin lymphoma cell lines (including L428 and HDLM2) have demonstrated that they reflect the gene expression program of primary HRS cells (24). For comparison, we profiled gene expression in one cell line derived from PMBL, K1106 (25), and six DLBCL cell lines that resemble the GCB DLBCL subgroup in gene expression.
For this analysis, we selected a set of PMBL signature genes that were more highly expressed in primary PMBL than in other DLBCLs (P < 0.001). A set of Hodgkin lymphoma signature genes was chosen such that at least two of the Hodgkin lymphoma cell lines had more than twofold greater expression than the average expression in the GCB DLBCL cell lines. Of the 348 PMBL signature genes, a remarkably large subset of 118 (34%) were also Hodgkin lymphoma signature genes (Fig. 5 A). Since Hodgkin lymphoma signature genes represented only 13% of all Lymphochip microarray genes, the set of PMBL signature genes were 2.6-fold enriched in Hodgkin lymphoma signature genes (Fig. 5 D). For comparison, we identified a set of genes that were more highly expressed (greater than or equal to twofold) in the PMBL cell line K1106 than in the GCB DLBCL cell lines and found that 153 of these genes were also PMBL signature genes. This represents a 2.75-fold enrichment of K1106 high genes within the set of PMBL signature genes, which is similar to the enrichment of Hodgkin lymphoma signature genes (Fig. 5 D). Of the 118 genes that were both PMBL and Hodgkin lymphoma signature genes, 69% were also more highly expressed in Hodgkin lymphoma cell lines than in two ABC DLBCL cell lines, OCI-Ly3 and OCI-Ly10, demonstrating that these genes also distinguish PMBL and Hodgkin lymphoma from ABC DLBCL. Recently, gene expression data from four Hodgkin lymphoma cell lines and five GCB DLBCL cell lines were generated using Affymetrix microarrays (24). Of the 235 PMBL signature genes represented on these arrays, 86 (37%) were twofold more highly expressed in greater than or equal to three Hodgkin lymphoma cell lines than in the GCB DLBCL cell lines (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20031074/DC1), confirming the gene expression relationship between PMBL and Hodgkin lymphoma.
Figure 5.

Relationship of PMBL to Hodgkin lymphoma. Relative gene expression is shown in primary PMBLs (average of all biopsy samples), the PMBL cell line K1106, three Hodgkin lymphoma (HL) cell lines, and six GCB DLBCL cell lines, according to the color scale shown in Fig. 1. (A) PMBL signature genes that are also expressed at high levels in Hodgkin lymphoma cell lines compared with GCB DLBCL cell lines. (B) PMBL signature genes not expressed in Hodgkin lymphoma cell lines. (C) Mature B cell markers expressed in PMBL and GCB DLBCL but not in Hodgkin lymphoma. (D) Enrichment within the set of PMBL signature genes of genes highly expressed in Hodgkin lymphoma cell lines or in the K1106 PMBL cell line relative to GCB DLBCL cell lines. See Results for details.
Notably, two of the previously described PMBL signature genes, MAL and FIG1 , were also highly expressed in Hodgkin lymphoma cell lines. In addition, a number of cytokines (IL-15, CSF-1, and TRAIL) and chemokines (TARC, RANTES, and Fractalkine) were also expressed by both PMBL and Hodgkin lymphoma cell lines. Many of the genes in this list are inducible by various stimuli, including interferon-responsive genes (STAT1, GBP1, IFP35, AIM2, IFIT2, IFI-6–16, PML, and IRF-1) and targets of the NF-κB transcription factor (A20, jun-B, IκBα, NFκB2, TRAF1, IL-15, CSF-1, and RANTES), possibly indicating that PMBL and Hodgkin lymphoma have activated a similar set of signaling pathways.
We confirmed the expression of four PMBL signature genes in primary HRS cells that we microdissected from a case of classical Hodgkin lymphoma (Fig. 6 A). Using quantitative RT-PCR assays, CCL17/TARC, SNFT, MAL, and TNFRSF6/Fas were all found to be expressed as mRNA in primary HRS cells at levels comparable to those observed in the L428 Hodgkin lymphoma cell line and the K1106 PMBL cell line. By contrast, the GCB DLBCL cell line OCI-Ly19 did not detectably express these PMBL signature genes. In addition, using immunohistochemistry, we could detect MAL protein expression in primary HRS cells of some cases of classical Hodgkin lymphoma (Fig. 6 B).
Figure 6.
Expression of PMBL signature genes in primary HRS cells. (A) Quantitative RT-PCR measurement of mRNA levels for CCL17/TARC, SNFT, MAL, and TNFRSF6/Fas in the indicated cell types. mRNA expression for each gene is presented relative to the expression of ACTB/β-actin. (B) Immunohistochemical staining of MAL protein in a malignant HRS cell from a case of nodular sclerosis classical Hodgkin lymphoma.
Using the quantitative PCR assay for PDL2 genomic alterations, all three Hodgkin lymphoma cell lines had PDL2 to PRKCQ ratios above two (Fig. 4 A). This result is in keeping with previous reports of chromosome 9p24 gains and amplification in Hodgkin lymphoma (8) and provides a further connection between PMBL and Hodgkin lymphoma.
Despite these links between PMBL and Hodgkin lymphoma, these two lymphoma types were clearly distinguishable by the expression of other genes. A subset of PMBL signature genes were highly expressed in the K1106 PMBL cell line but not in the Hodgkin lymphoma or GCB DLBCL cell lines (Fig. 5 B). Another clear difference between PMBL and Hodgkin lymphoma was in the expression of mature B cell genes. Hodgkin lymphoma cells extinguish expression of much of the mature B cell gene expression program by an unknown mechanism (24). Accordingly, several mature B cell genes (CD19, CD20, CD22, CD79A, CD79B, and Oct-2) were not detectably expressed in the Hodgkin lymphoma cell lines, whereas these genes were expressed in PMBLs and in GCB DLBCLs (Fig. 5 C). Thus, PMBL shares some, but not all, of the Hodgkin lymphoma gene expression program.
Discussion
We have used gene expression profiling to establish a molecular diagnosis of PMBL. The lymphoma cases identified by our predictor as PMBL have several clinical and molecular features in common with PMBL cases identified by current diagnostic methods (1). First, 53% of the patients predicted to be PMBL were younger than 35 at diagnosis, and in this age group women outnumbered men 1.8 to 1. Second, the lymphomas of these PMBL patients frequently extended to other thoracic structures besides the mediastinum but did not involve many of the extrathoracic sites typical of other DLBCL subgroups. Third, two of the molecular markers previously associated with PMBL, MAL (16), and FIG1 (17) were expressed in the majority of PMBL cases identified by our predictor. Finally, over half of the PMBLs identified by our predictor had a gain/amplification of a region of chromosome 9p24, which is a hallmark genomic abnormality of PMBL (6, 7).
We propose that the molecular diagnosis of PMBL by gene expression yields a more precise definition of this DLBCL subgroup than is provided by current diagnostic methods. Of the 46 patients for whom the diagnosis of PMBL was considered, 35 (76%) were classified as PMBL by gene expression. Of the 11 cases that were not classified as PMBL by our predictor, 7 were classified as GCB DLBCL and 4 were classified as ABC DLBCL by gene expression (unpublished data), which roughly reflects the distribution of these subgroups among all DLBCL cases (4). Therefore, it appears likely that these cases represent other forms of DLBCL that happened to predominantly involve the mediastinum at presentation.
Gene expression profiling identified PMBL as a subgroup of DLBCL with a relatively favorable survival rate after therapy: the 5-yr survival rate of PMBL was 64% compared with 59% and 30% for the GCB and ABC DLBCL subgroups, respectively. In our series, four patients with predominant mediastinal involvement had tumors that were not classified as PMBL by our predictor but instead were found to be ABC DLBCL; these patients had rapid disease progression and died within 2 yr of diagnosis. This example highlights how molecular diagnosis can provide valuable prognostic information that could help guide the management of DLBCL patients with mediastinal masses.
The gene expression signature of PMBL revealed a fascinating relationship between this lymphoma subgroup and Hodgkin lymphoma. Over one third of all PMBL signature genes were also more highly expressed in Hodgkin lymphoma cell lines than in GCB DLBCL cell lines. Five of the PMBL signature genes that we identified (MAL, SNFT, TNFRSF6, TARC, and CD30) have been shown to be expressed as proteins in primary HRS cells (Fig. 6 and references [22, 23]). Further, we found that over half of the PMBLs and three Hodgkin lymphoma cell lines shared gains/amplifications in a region of chromosome 9p, in accord with previous studies (6–8). The PMBLs identified in our study shared many clinical and pathological features with classical Hodgkin lymphomas, especially those with nodular sclerosis histology (26). Both lymphoma types are prevalent in younger patients, especially women, and frequently involve the mediastinum and other thoracic structures. As the name implies, nodular sclerosis Hodgkin lymphomas are characterized by a fibrotic stromal cell reaction, which is also observed frequently in PMBLs (Fig. S1). Further, there are reports of Hodgkin lymphoma patients who developed PMBL as a second malignancy within one year after treatment (9, 10). These findings strongly support the hypothesis that there is a pathogenetic overlap between PMBL and some forms of Hodgkin lymphoma (1, 11).
Two general possibilities could account for the striking gene expression similarities between PMBL and Hodgkin lymphoma. First, the PMBL signature genes that are also expressed by Hodgkin lymphoma may be the downstream targets of a signaling pathway or transcription factor that is active in both lymphoma types. Many of these genes are known to be activated by the NF-κB or interferon signaling pathways, but the actual pathways responsible for their expression in these lymphomas remain to be elucidated. A second possibility is that PMBL and some forms of Hodgkin lymphoma may originate from a thymic B cell (16, 27, 28). Indeed, 60% of Hodgkin lymphomas involve the mediastinum at presentation and can involve the thymus (29). In this scenario, the gene expression program that is shared by PMBL and Hodgkin lymphoma might reflect the normal characteristics of thymic B cells (16, 27, 28) or might reflect an oncogenic mechanism that is required for a malignant B cell to develop in this anatomical site.
It is intriguing that the chromosome 9p region that is gained/amplified in PMBL and Hodgkin lymphoma contains two regulators of T cell responses, PDL1 and PDL2. These genes encode members of the B7 family that are ligands for the PD-1 receptor on T cells (30–34) but may also bind other T cell surface molecules (35). PDL1 and PDL2 have been reported to have both negative and positive effects on T cell responses (30–35), which may be explained by the existence of more than one receptor for these ligands on T cells (35). Expression of PDL1 on tumor cells has been shown to inhibit tumor immunity (34, 36), and therefore PDL1 expression might allow a malignant B cell arising in the thymus to evade T cell recognition. Alternatively, the ability of PDL2 to costimulate T cells might lead to local cytokine production that is beneficial to the tumor cells. It is important to emphasize that PDL1 and PDL2 were highly expressed in most PMBLs, even those that lacked evidence of gains or amplifications of this genomic region. Thus, elevated transcription of these genes appears to be a characteristic feature of PMBL that may be augmented by genomic copy number gains. Besides PDL1 and PDL2, the amplicon on chromosome 9p in PMBL includes JAK2, which encodes a tyrosine kinase, and SMARCA2, which encodes a putative chromatin regulator. Functional studies will be needed to elucidate the relative contributions of each of these chromosome 9p genes to the pathogenesis of PMBL and Hodgkin lymphoma.
Despite these striking parallels between PMBL and Hodgkin lymphoma, there are important differences. A subset of the PMBL signature genes was not expressed highly in Hodgkin lymphoma cell lines, and the down-regulation of mature B cell genes that is characteristic of Hodgkin lymphoma was not observed in PMBLs. In addition, the intense immune cell infiltration that is characteristic of Hodgkin lymphoma is not observed in most PMBLs. Interestingly, “grey zone” lymphomas have been described that have histological features of both PMBL and Hodgkin lymphoma (1, 11). Such cases may indicate that there is a spectrum of lymphomas between PMBL and Hodgkin lymphoma, a prospect that can be tested by gene expression profiling and other molecular analyses.
Finally, our studies emphasize that DLBCL is a heterogeneous diagnostic category that harbors at least three molecularly and clinically distinct subgroups: GCB DLBCL, ABC DLBCL, and PMBL. Clearly, clinical trials in DLBCL must incorporate gene expression profiling so that these three disease entities can be recognized. The molecular diagnosis of these DLBCL subgroups is the first step toward understanding the oncogenic mechanisms that cause these diseases, which will ultimately lead to rational and disease-specific treatments.
Acknowledgments
This work was supported by an NCI Director's Challenge grant (UO1-CA84967) and by the Center for Cancer Research, NCI, and was performed under the auspices of the Lymphoma/Leukemia Molecular Profiling Project of the NCI.
Abbreviations used in this paper: ABC, activated B cell; DLBCL, diffuse large B cell lymphoma; GCB, germinal center B cell; HRS, Hodgkin Reed-Sternberg; PMBL, primary mediastinal B cell lymphoma.
References
- 1.Barth, T.F., F. Leithauser, S. Joos, M. Bentz, and P. Moller. 2002. Mediastinal (thymic) large B-cell lymphoma: where do we stand? Lancet Oncol. 3:229–234. [DOI] [PubMed] [Google Scholar]
- 2.Bishop, P.C., W.H. Wilson, D. Pearson, J. Janik, E.S. Jaffe, and P.C. Elwood. 1999. CNS involvement in primary mediastinal large B-cell lymphoma. J. Clin. Oncol. 17:2479–2485. [DOI] [PubMed] [Google Scholar]
- 3.Alizadeh, A.A., M.B. Eisen, R.E. Davis, C. Ma, I.S. Lossos, A. Rosenwald, J.C. Boldrick, H. Sabet, T. Tran, X. Yu, et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 403:503–511. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwald, A., G. Wright, W.C. Chan, J.M. Connors, E. Campo, R.I. Fisher, R.D. Gascoyne, H.K. Muller-Hermelink, E.B. Smeland, J.M. Giltnane, et al. 2002. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 346:1937–1947. [DOI] [PubMed] [Google Scholar]
- 5.Wright, G., B. Tan, A. Rosenwald, E.H. Hurt, A. Wiestner, and L.M. Staudt. 2003. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B Cell lymphoma. Proc. Natl. Acad. Sci. USA. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joos, S., M.I. Otano-Joos, S. Ziegler, S. Bruderlein, S. du Manoir, M. Bentz, P. Moller, and P. Lichter. 1996. Primary mediastinal (thymic) B-cell lymphoma is characterized by gains of chromosomal material including 9p and amplification of the REL gene. Blood. 87:1571–1578. [PubMed] [Google Scholar]
- 7.Bentz, M., T.F. Barth, S. Bruderlein, D. Bock, M.J. Schwerer, M. Baudis, S. Joos, A. Viardot, A.C. Feller, H.K. Muller-Hermelink, et al. 2001. Gain of chromosome arm 9p is characteristic of primary mediastinal B-cell lymphoma (MBL): comprehensive molecular cytogenetic analysis and presentation of a novel MBL cell line. Genes Chromosomes Cancer. 30:393–401. [DOI] [PubMed] [Google Scholar]
- 8.Joos, S., M. Kupper, S. Ohl, F. von Bonin, G. Mechtersheimer, M. Bentz, P. Marynen, P. Moller, M. Pfreundschuh, L. Trumper, and P. Lichter. 2000. Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res. 60:549–552. [PubMed] [Google Scholar]
- 9.Zarate-Osorno, A., L.J. Medeiros, D.L. Longo, and E.S. Jaffe. 1992. Non-Hodgkin's lymphomas arising in patients successfully treated for Hodgkin's disease. A clinical, histologic, and immunophenotypic study of 14 cases. Am. J. Surg. Pathol. 16:885–895. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez, C.L., L.J. Medeiros, and E.S. Jaffe. 1991. Composite lymphoma. A clinicopathologic analysis of nine patients with Hodgkin's disease and B-cell non-Hodgkin's lymphoma. Am. J. Clin. Pathol. 96:81–89. [DOI] [PubMed] [Google Scholar]
- 11.Jaffe, E.S., and K. Muller-Hermelink. 1999. Relationship between Hodgkin's disease and non-Hodgkin's lymphomas. Hodgkin's Disease. P.M. Mauch, J.O. Armitage, V. Diehl, R.T. Hoppe, and L.M. Weiss, editors. Lippincott Williams & Wilkens, Philadelphia, PA. 181–191.
- 12.Haioun, C., P. Gaulard, F. Roudot-Thoraval, M. Divine, H. Jouault, J.P. Lebourgeois, M. Kuentz, J.P. Farcet, and F. Reyes. 1989. Mediastinal diffuse large-cell lymphoma with sclerosis: a condition with a poor prognosis. Am. J. Clin. Oncol. 12:425–429. [DOI] [PubMed] [Google Scholar]
- 13.Lavabre-Bertrand, T., D. Donadio, N. Fegueux, D. Jessueld, J. Taib, D. Charlier, T. Rousset, J.M. Emberger, P. Baldet, and M. Navarro. 1992. A study of 15 cases of primary mediastinal lymphoma of B-cell type. Cancer. 69:2561–2566. [DOI] [PubMed] [Google Scholar]
- 14.Abou-Elella, A.A., D.D. Weisenburger, J.M. Vose, J.P. Kollath, J.C. Lynch, M.A. Bast, P.J. Bierman, T.C. Greiner, W.C. Chan, and J.O. Armitage. 1999. Primary mediastinal large B-cell lymphoma: a clinicopathologic study of 43 patients from the Nebraska Lymphoma Study Group. J. Clin. Oncol. 17:784–790. [DOI] [PubMed] [Google Scholar]
- 15.Zinzani, P.L., M. Martelli, M. Magagnoli, E. Pescarmona, L. Scaramucci, F. Palombi, M. Bendandi, M.P. Martelli, S. Ascani, G.F. Orcioni, et al. 1999. Treatment and clinical management of primary mediastinal large B-cell lymphoma with sclerosis: MACOP-B regimen and mediastinal radiotherapy monitored by (67)Gallium scan in 50 patients. Blood. 94:3289–3293. [PubMed] [Google Scholar]
- 16.Copie-Bergman, C., A. Plonquet, M.A. Alonso, M.L. Boulland, J. Marquet, M. Divine, P. Moller, K. Leroy, and P. Gaulard. 2002. MAL expression in lymphoid cells: further evidence for MAL as a distinct molecular marker of primary mediastinal large B-cell lymphomas. Mod. Pathol. 15:1172–1180. [DOI] [PubMed] [Google Scholar]
- 17.Copie-Bergman, C., M.L. Boulland, C. Dehoulle, P. Moller, J.P. Farcet, M.J. Dyer, C. Haioun, P.H. Romeo, P. Gaulard, and K. Leroy. 2003. Interleukin 4-induced gene 1 is activated in primary mediastinal large B-cell lymphoma. Blood. 101:2756–2761. [DOI] [PubMed] [Google Scholar]
- 18.Rosenwald, A., G. Wright, A. Wiestner, W.C. Chan, J.M. Connors, E. Campo, R.D. Gascoyne, T.M. Grogan, H.K. Muller-Hermelink, E.B. Smeland, et al. 2003. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 3:185–197. [DOI] [PubMed] [Google Scholar]
- 19.Eisen, M.B., P.T. Spellman, P.O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anagnostopoulos, I., F. Dallenbach, and H. Stein. 2001. Diffuse large cell lymphomas. Neoplastic Hematopathology. D.M. Knowles, editor. Lippincott Williams & Wilkins, Philadelphia, PA. 855–905.
- 21.Wright, G., B. Tan, A. Rosenwald, E.H. Hurt, A. Wiestner, and L.M. Staudt. 2003. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. USA. 100:9991–9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein, H., J. Gerdes, U. Schwab, H. Lemke, V. Diehl, D.Y. Mason, H. Bartels, and A. Ziegler. 1983. Evidence for the detection of the normal counterpart of Hodgkin and Sternberg-Reed cells. Hematol. Oncol. 1:21–29. [DOI] [PubMed] [Google Scholar]
- 23.Peh, S.C., L.H. Kim, and S. Poppema. 2001. TARC, a CC chemokine, is frequently expressed in classic Hodgkin's lymphoma but not in NLP Hodgkin's lymphoma, T-cell-rich B-cell lymphoma, and most cases of anaplastic large cell lymphoma. Am. J. Surg. Pathol. 25:925–929. [DOI] [PubMed] [Google Scholar]
- 24.Kuppers, R., U. Klein, I. Schwering, V. Distler, A. Brauninger, G. Cattoretti, Y. Tu, G.A. Stolovitzky, A. Califano, M.L. Hansmann, and R. Dalla-Favera. 2003. Identification of Hodgkin and Reed-Sternberg cell-specific genes by gene expression profiling. J. Clin. Invest. 111:529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nacheva, E., M.J. Dyer, C. Metivier, D. Jadayel, G. Stranks, R. Morilla, J.M. Heward, T. Holloway, S. O'Connor, P.C. Bevan, et al. 1994. B-cell non-Hodgkin's lymphoma cell line (Karpas 1106) with complex translocation involving 18q21.3 but lacking BCL2 rearrangement and expression. Blood. 84:3422–3428. [PubMed] [Google Scholar]
- 26.Mauch, P.M., L.A. Kalish, M. Kadin, C.N. Coleman, R. Osteen, and S. Hellman. 1993. Patterns of presentation of Hodgkin disease. Implications for etiology and pathogenesis. Cancer. 71:2062–2071. [DOI] [PubMed] [Google Scholar]
- 27.Addis, B.J., and P.G. Isaacson. 1986. Large cell lymphoma of the mediastinum: a B-cell tumour of probable thymic origin. Histopathology. 10:379–390. [DOI] [PubMed] [Google Scholar]
- 28.Isaacson, P.G., A.J. Norton, and B.J. Addis. 1987. The human thymus contains a novel population of B lymphocytes. Lancet. 2:1488–1491. [DOI] [PubMed] [Google Scholar]
- 29.Jaffe, E.S., N.L. Harris, H. Stein, and J.W. Vardiman. 2001. Tumours of Haematopoietic and Lymphoid Tissues. IARC Press, Lyon, France. 351 pp.
- 30.Freeman, G.J., A.J. Long, Y. Iwai, K. Bourque, T. Chernova, H. Nishimura, L.J. Fitz, N. Malenkovich, T. Okazaki, M.C. Byrne, et al. 2000. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 192:1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latchman, Y., C.R. Wood, T. Chernova, D. Chaudhary, M. Borde, I. Chernova, Y. Iwai, A.J. Long, J.A. Brown, R. Nunes, et al. 2001. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2:261–268. [DOI] [PubMed] [Google Scholar]
- 32.Dong, H., G. Zhu, K. Tamada, and L. Chen. 1999. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 5:1365–1369. [DOI] [PubMed] [Google Scholar]
- 33.Dong, H., S.E. Strome, D.R. Salomao, H. Tamura, F. Hirano, D.B. Flies, P.C. Roche, J. Lu, G. Zhu, K. Tamada, et al. 2002. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 8:793–800. [DOI] [PubMed] [Google Scholar]
- 34.Curiel, T.J., S. Wei, H. Dong, X. Alvarez, P. Cheng, P. Mottram, R. Krzysiek, K.L. Knutson, B. Daniel, M.C. Zimmermann, et al. 2003. Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nat. Med. 9:562–567. [DOI] [PubMed] [Google Scholar]
- 35.Liu, X., J.X. Gao, J. Wen, L. Yin, O. Li, T. Zuo, T.F. Gajewski, Y.X. Fu, P. Zheng, and Y. Liu. 2003. B7DC/PDL2 promotes tumor immunity by a PD-1–independent mechanism. J. Exp. Med. 197:1721–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong, H., and L. Chen. 2003. B7-H1 pathway and its role in the evasion of tumor immunity. J. Mol. Med. 81:281–287. [DOI] [PubMed] [Google Scholar]