Abstract
Dendritic cells (DCs) and macrophages are professional antigen-presenting cells (APCs) that play key roles in both innate and adaptive immunity. ChemR23 is an orphan G protein–coupled receptor related to chemokine receptors, which is expressed specifically in these cell types. Here we present the characterization of chemerin, a novel chemoattractant protein, which acts through ChemR23 and is abundant in a diverse set of human inflammatory fluids. Chemerin is secreted as a precursor of low biological activity, which upon proteolytic cleavage of its COOH-terminal domain, is converted into a potent and highly specific agonist of ChemR23, the chemerin receptor. Activation of chemerin receptor results in intracellular calcium release, inhibition of cAMP accumulation, and phosphorylation of p42–p44 MAP kinases, through the Gi class of heterotrimeric G proteins. Chemerin is structurally and evolutionary related to the cathelicidin precursors (antibacterial peptides), cystatins (cysteine protease inhibitors), and kininogens. Chemerin was shown to promote calcium mobilization and chemotaxis of immature DCs and macrophages in a ChemR23-dependent manner. Therefore, chemerin appears as a potent chemoattractant protein of a novel class, which requires proteolytic activation and is specific for APCs.
Keywords: orphan receptor, dendritic cells, macrophages, chemotaxis, proteolytic processing
Introduction
DCs and macrophages display phagocytic properties and are activated by various microorganism-derived molecules through the family of Toll-like receptors, which trigger the release of costimulatory molecules, cytokines, and chemokines. Therefore, they play an essential role in innate immunity. These cell populations also constitute the professional APCs, which are responsible for the mounting of adaptive immune responses (1, 2). DCs and macrophages are attracted to infection and inflammatory sites by a variety of factors, among which chemokines constitute the largest group so far (3). It has been shown that tremendous functional, morphological, and metabolic diversity exists among these cell populations. One of these functional differences is the expression of differential sets of chemoattractant receptors, which is responsible for the selective recruitment of specific cell subpopulations according to their lineage, origin, and maturation state (2).
Many tumor types have been demonstrated to attract macrophages and DCs through the direct or indirect production of chemoattractant factors (4, 5). These include a number of CC chemokines, such as MCP-1. APC recruitment is expected to contribute to the development of an antitumoral immune response, and such an effect has been demonstrated in a number of animal studies in which the forced expression of chemokines, such as RANTES, by tumor cell lines resulted in a decreased pathogenicity of these cells in vivo. However, a number of studies have demonstrated that the infiltration of tumors by macrophages and DCs contributed to their aggressive phenotype by supplying different classes of factors necessary for tumor cell proliferation and invasiveness, such as growth and angiogenic factors, as well as proteolytic enzymes (4, 5). Chemoattractant factors for DCs and macrophages are therefore frequently expressed in the tumor microenvironment, and these factors can play dual roles in the context of tumor progression.
Due to their key roles in immune responses and their involvement in tumor pathogenesis, there is a strong interest in the identification of new factors contributing to the recruitment of macrophages and DCs. They might indeed on one hand help to understand better the development of innate and adaptive immune responses, and on the other hand constitute tools or targets in therapeutic strategies aiming at modulating the immune responses in situations such as cancer, inflammatory diseases, and graft rejection (6, 7).
We have previously described ChemR23 as a human orphan G protein–coupled receptor expressed in macrophages and immature DCs (8). ChemR23, also known as Dez in mouse (9), was shown to constitute a minor coreceptor for a subset of human immunodeficiency viruses (10), but no ligand, either natural or synthetic, has been so far identified for this receptor. ChemR23 is structurally related to receptors for chemokines and other chemoattractant molecules and was therefore expected, given its distribution, to specifically attract APCs in response to an unknown mediator. However, most chemokines and chemoattractant molecules are characterized by a relatively poor specificity, as they usually attract a diverse set of leukocyte subpopulations (7, 11). Therefore, ChemR23, by its specific expression in macrophages and immature DCs, appeared as a particularly attractive candidate receptor involved in the initiation and early regulation of immune responses. We have therefore developed an experimental strategy to identify the ligand of this receptor. Given its expression pattern, we reasoned that the cognate ligand should be expressed in inflammatory situations and selected, as sources of potential agonists, a number of pathological situations in which such ligands can be expected to be generated in large amounts.
Materials and Methods
Expression of Human ChemR23.
The human coding sequence (Y14838, 8) was cloned into the pcDNA3 (Invitrogen) and pEFIN3 vectors (Euroscreen) and sequenced. The pEFIN3 construct was transfected using Fugene 6 into CHO-K1 cells expressing or not Gα16 and apoaequorin, and G418-resistant clones were characterized for ChemR23 (chemerinR) expression by Northern blotting. A functional assay based on the luminescence of mitochondrial aequorin (12) was performed as previously described (13). Results were expressed as relative light units or as percentage of the endogenous response to 20 μM ATP.
Purification of Human Native Chemerin.
1 liter of ascitic fluid was filtered and loaded (50 ml/run) onto a reverse phase column (10 × 100 mm; Poros 20 R2 beads; Applied Biosystems). A 5–95% CH3CN gradient (6%/min) in 0.1% TFA was applied and 5 ml fractions were collected and assayed for ChemR23 activation. Active fractions were adjusted to pH 4.8 and applied to a cation-exchange HPLC column (Polycat 9.6 × 250 mm; Vydac) in the presence of 10% CH3CN, eluted with a 0–1 M NaCl gradient (10%/min) in acetate buffer, pH 5. Active fractions were desalted (Ultrafree, cut-off: 10 kD; Millipore), loaded onto a second cation-exchange column (Polycat 2.1 × 250 mm; Vydac), and eluted with the same buffer (2%/min NaCl gradient). 0.5 ml active fractions (desalted) were pooled, diluted eightfold with 0.1% H3PO4, and loaded onto a C18 column (4.6 × 250 mm; Vydac). A 5–95% CH3CN gradient (0.3%/min) in 0.1% H3PO4 was applied and individual UV absorption peaks (214 nm) were collected manually and assayed. The active fractions were loaded onto a second C18 column (2.1 × 250 mm; Vydac; 5–95% CH3CN in 0.1% TFA, 0.3%/min). The peaks were collected manually and analyzed by mass spectrometry. The use of human material collected for diagnostic or therapeutic purposes was approved by the ethical committee of the Université Libre de Bruxelles Medical School.
Cloning and Expression of Human Tazarotene-induced Gene (Tig)-2, Renamed Chemerin.
Human (Q99969) Tig-2 cDNA was amplified by PCR from liver cDNA (CLONTECH Laboratories, Inc.) using human sense (5′-CAGGAATTCAGCATGCGACGGCTGCTGA-3′) and antisense (5′-GCTCTAGATTAGCTGCGGGGCAGGGCCTT-3′) primers in standard conditions (94°C for 3 min; 94°C for 90 s, 58°C for 1 min, and 72°C for 90 s, 35 cycles; 72°C for 10 min). The resulting 500-bp fragment was cloned in pEFIN3 and sequenced. The plasmid was transfected in CHO-K1 cells and G418-resistant clones were characterized by the activity of conditioned medium on chemerinR. For recombinant chemerin production, cells were grown to 70% confluence and further incubated with serum-free DMEM-F12, which was collected after 3 d. The recombinant protein was purified by filtration through 0.45 μm Millex filters (Millipore) and separation through a cation-exchange HPLC column (Polycat 9.6 × 250 mm; Vydac; 0–1 M NaCl gradient in acetate buffer, pH 5). The protein concentration in active fractions was determined after SDS/PAGE by comparison with glutathione S-transferase and lysozyme standards after silver staining.
Mass Spectrometry Analysis.
The active fractions were vacuum dried, resuspended in 10 μl of 100 mM Tris-HCl, pH 8.7, heated for 15 min at 95°C, incubated overnight at 37°C with 250 ng trypsin (Promega), and purified by solid phase extraction (C18 ZipTip; Millipore). The digested peptides were eluted in 1.5 μl of 70% CH3CN/0.1% TFA onto a metallic MALDI target, dried, and then mixed in 1.5 μl of matrix mix (2 mg/ml 2,5-dihydroxybenzoic acid and 10 mg/ml α-cyano-4-hydroxycinnamic acid, 2 mM fucose, 5 mM ammonium acetate). For proteic samples excised from SDS/acrylamide gels, the samples were processed as previously described (14). For determination of the NH2 terminus of the recombinant protein, the digested peptides were first separated onto a C18 column (250 mm; Vydac, 5–95% CH3CN in 0.1% TFA, 2%/min) and each HPLC fraction was analyzed separately. Mass spectrometry analysis was performed on a Q-TOF Ultima Global mass spectrometer equipped with a MALDI source (Micromass) and calibrated using the monoisotopic masses of tryptic and chymotryptic peptides from bovine serum albumin. Ionization was achieved using a nitrogen laser (337 nm beam, 10 Hz) and acquisitions were performed in a V mode reflectron position. Microsequencing was performed by argon-induced fragmentation after selection of the parent ion.
Quantitative RT-PCR.
ChemerinR and chemerin transcripts were detected by quantitative RT-PCR (TaqMan) in total or polyA+ RNA samples from human tissues and blood cell populations obtained commercially (CLONTECH Laboratories, Inc. and Ambion) or prepared locally (RNeasy Mini Kit; QIAGEN). Primers were: 5′-GCAGACAAGCTGCCGGA-3′ forward, 5′-AGTTTGATGCAGGCCAGGC-3′ reverse, and 5′-AACCCGAGTGCAAAGTCAGGCCC-3′ as probe for chemerin; 5′-GTCCCAGAACCACCGCAG-3′ forward, 5′-AAGAAAGCCAGGACCCAGATG-3′ reverse, and 5′-TTCGCCTGGCTTACATGGCCTGC-3′ as probe for chemerinR; and 5′-GAAGGTGAAGGTCGGAGTC-3′ forward, 5′-GAAGATGGTGATGGGATTTC-3′ reverse, and 5′-AGCTCTCCCGCC- GGCCTCTG-3′ as probe for the reference housekeeping gene GAPDH. Standard curves were run systematically for the three genes and the transcript copy number of prochemerin and chemerinR was normalized to the GAPDH transcript copy number for each sample.
Production of Monoclonal Antibodies.
BALB/c mice were injected with 100 μg pcDNA-chemerinR as previously described (15), or with the prochemerin COOH-terminal octapeptide FSKALPRS. Sera were tested by FACS® on the CHO-chemerinR cell line or by Elisa for the prochemerin peptide, and immune mice were used to generate monoclonal antibodies by standard hybridoma technology, using the NSO myeloma cell line (15). The Ig class of selected hybridomas was determined with a mouse monoclonal antibody isotyping kit (IsoStrip; Boehringer).
Binding Assay.
ChemR23-expressing CHO-K1 cells were collected from plates with PBS supplemented with 5 mM EDTA, gently pelleted for 2 min at 1,000 g, and resuspended in binding buffer (50 mM Hepes, pH 7.4, 1 mM CaCl2, 5 mM MgCl2, 0.5% BSA). Competition binding assays were performed in Minisorb tubes (Nunc) using the 125I-YHSFFFPGQFAFS peptide as tracer (specific activity: 600 Ci/mmol, 50,000 cpm/tube), variable concentrations of competitors, and 500,000 cells in a final volume of 0.1 ml. Total binding was measured in the absence of competitor and nonspecific binding was measured in the presence of a 100-fold excess of unlabeled ligand. Samples were incubated for 90 min at 27°C and then bound tracer was separated by filtration through GF/B filters presoaked in 0.5% BSA. Filters were counted in a β-scintillation counter. Binding parameters were determined with the PRISM software (Graphpad Software) using nonlinear regression applied to a one-site competition model.
Intracellular Cascade Assays.
GTPγ[35S] binding assays were performed as previously described (16). Functional parameters were determined with the PRISM software (Graphpad Software) using nonlinear regression applied to a sigmoidal dose-response model. ERK1/2 activation was assayed by Western blotting, using an anti–phospho-p42/44 monoclonal antibody (E10; Cell Signaling Technology) as previously described (13). All assays were run with or without overnight pretreatment with 100 ng/ml pertussis toxin. It was shown that such pertussis toxin pretreatment did not inhibit the functional response to ATP in these cells.
Chemotaxis and Ca2+ Mobilization Assays on Primary Cells.
Monocyte-derived DCs were generated by 50 ng/ml GM-CSF and 20 ng/ml IL-13 stimulation as previously described (17). Maturation of DCs was achieved after stimulation with 100 ng/ml LPS. Macrophages were obtained by incubating monocytes in Petriperm dishes (Haereus) for 6 d in RPMI supplemented with 10% FCS and 10 ng/ml macrophage colony-stimulating factor. Cell migration was evaluated using a 48-well microchemotaxis chamber technique as previously described (18). For Ca2+ mobilization assays, monocyte-derived DCs or macrophages (107 cells/ml in HBSS without phenol red but containing 0.1% BSA) were loaded with 5 μM FURA-2 (Molecular Probes) for 30 min at 37°C in the dark. The loaded cells were washed twice, resuspended at 106 cells/ml, kept for 30 min at 4°C in the dark with or without 10 μg/ml of the blocking 4C7 monoclonal antibody, and transferred into the quartz cuvette of a luminescence spectrometer (LS50B; PerkinElmer). Ca2+ mobilization in response to recombinant chemerin was measured by recording the ratio of fluorescence emitted at 510 nm after sequential excitation at 340 and 380 nm.
Results
Isolation and Characterization of Chemerin as the ChemR23 Ligand.
We developed, as a screening assay, CHO-K1 cell lines coexpressing human ChemR23, apoaequorin, and Gα16, allowing to test fractions from human inflammatory fluids as well as various porcine and human lymphoid organs. A biological activity, specific for ChemR23-expressing cell lines, was detected in fractions resulting from the reverse phase HPLC fractionation of human ascitic fluids (secondary to ovary or liver neoplasms) in arthritic synovial fluid as well as in extracts from human spleen (unpublished data). The activity was purified to homogeneity from a single batch of ascitis by five successive steps of liquid chromatography (Fig. 1) , and the active compound was digested by trypsin and analyzed by mass spectrometry (Fig. 2, A and B) . Eight peptides were predicted to derive from the product of the human Tig-2 (Fig. 2 B), covering 91 amino acids out of the 143 amino acid–long sequence of the Tig-2 gene product (after removal of the predicted signal peptide; 19). However, the COOH-terminal peptide (peptide 8) was not tryptic, lacking the last six amino acids of the predicted protein. This observation indicated that the active compound might result from the proteolytic processing of the encoded precursor. The active protein (137 amino acids, 15,876 D) was named chemerin and ChemR23, the chemerin receptor (chemerinR). The cDNA sequence of mouse chemerin was reconstructed from expressed sequence tags present in the public databases and the predicted primary structure (Fig. 2 C) shares 65% identity with the human polypeptide.
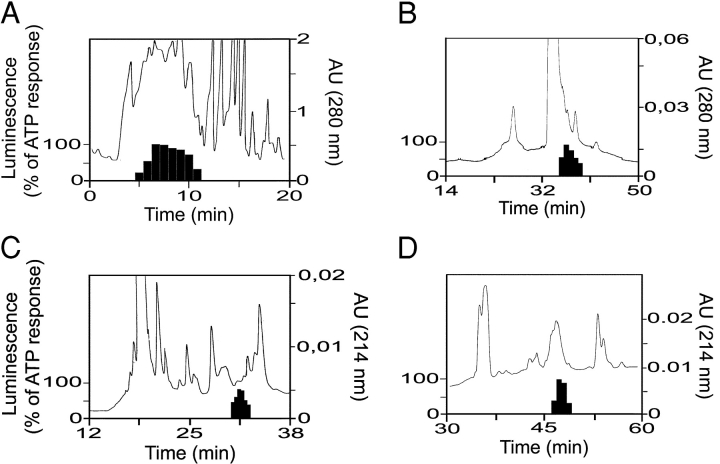
Figure 1.
Purification of the natural ligand of the ChemR23 receptor from human inflammatory fluid. (A) First step HPLC fractionation (Poros column) of human ascitic fluid. The absorbance (AU) and biological activity on ChemR23 (luminescence in an aequorin-based assay, normalized to the ATP response, solid bars) are shown. (B) Third step (cation-exchange column). (C) Fourth step (C18 column). (D) Last step purification of the active compound (C18 column). The x axis is magnified to focus on the region of interest.
Figure 2.

Identification of chemerin as the natural ligand of ChemR23, the chemerin receptor. (A) Monoisotopic peptide mass fingerprinting of the active fraction on a Maldi Q-TOF mass spectrometer after trypsin digestion. (B) Sequences corresponding to selected major peaks of the Maldi Q-TOF mass spectrometer spectrum after trypsin digestion. Peptides 1–7 correspond to tryptic peptides derived from the Tig-2 gene product (prochemerin), whereas peptide 8 is not tryptic and corresponds to the COOH-terminal end of the purified protein. The position of the peptides within this sequence is given. The sequence of peptides in peaks 3, 5, 7, and 8 was confirmed by microsequencing. (C) Amino acid sequence alignment of human and mouse (Protein Data Bank accession no. AK002298) preprochemerin and human cathelicidin FALL39 precursor. Amino acid identities as compared with human preprochemerin are boxed. The signal peptides (predicted for mouse preprochemerin) are in bold lowercase characters and cysteines are in bold. Cleaved COOH-terminal peptides are in bold italics and underlined (predicted by analogy for mouse prochemerin). The location of introns, which interrupt the gene coding sequences between codons, are indicated by arrowheads.
Expression and Pharmacological Characterization of Human Chemerin.
Human Tig-2 cDNA was cloned and expressed in CHO-K1 cells. The bioactive recombinant protein was purified to homogeneity from conditioned medium and analyzed by mass spectrometry and SDS/PAGE, which confirmed COOH-terminal truncation after serine 157 (unpublished data). A monoclonal antibody generated against a peptide (FSKALPRS) corresponding to the predicted COOH-terminal sequence of the gene product was used to purify to homogeneity from CHO-K1–conditioned medium, an unprocessed form of the protein (prochemerin), which was confirmed by mass spectrometry to retain the six COOH-terminal amino acids (unpublished data). The amount of purified recombinant chemerin (Fig. 3 A) and prochemerin (unpublished data) was determined by comparison with protein standards following SDS/PAGE and silver staining. It was inferred that >90% of prochemerin released by CHO-K1 cells was enzymatically processed into chemerin. Comparison of the biological activity of the two purified proteins assayed in parallel on CHO-K1 cells expressing human chemerinR (Fig. 3 B) demonstrated that processed chemerin was about 100-fold more active (EC50: 4.5 ± 0.7 nM, mean ± SEM for seven independent experiments) than unprocessed prochemerin (EC50: 393 ± 116 nM, mean ± SEM for three independent experiments). The NH2 terminus of prochemerin and chemerin was determined by mass spectrometry. A tryptic peptide (ELTEAQR, 845.45 D) corresponding to amino acids 21–27 of preprochemerin was identified by sequencing (unpublished data), confirming the signal peptide cleavage site predicted by the SignalP software from Expasy (http://www.cbs.dtu.dk/services/SignalP/).
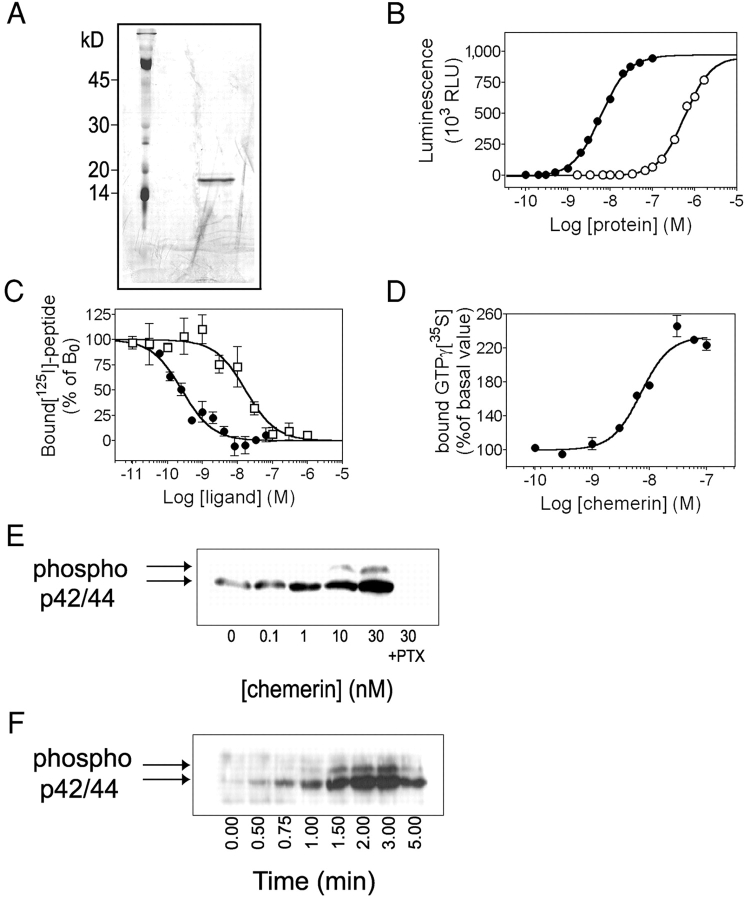
Figure 3.
Pharmacology of the chemerin receptor. (A) SDS/PAGE of human recombinant chemerin expressed in CHO-K1 cells and purified by HPLC. The gel was silver stained and the major band corresponds to a protein of 18 kD. Mass spectrometry analysis demonstrated the cleavage of the six COOH-terminal amino acids in this biologically active protein. (B) Biological activity on chemerinR of human recombinant chemerin (•) and prochemerin (○), using the aequorin assay. (C) Competition binding assay using as tracer an iodinated peptide derived from the chemerin COOH terminus. Competition was performed with the unlabeled peptide (□) or human recombinant chemerin (•). (D) Concentration-action curve of human chemerin in a GTPγ[35S] binding assay, using membranes of CHO/chemerinR cells. (E) Immunodetection of phosphorylated ERK1/2 in CHO/chemerinR cells after stimulation by human recombinant chemerin for 2 min. (F) Kinetics of ERK1/ ERK2 activation after stimulation by 10 nM human chemerin. Each experiment was repeated at least three times.
ChemerinR is structurally and evolutionary related to the C5a and C3a receptors, the prostaglandin D2 receptor CRTH2, and the orphan GPR1 receptor (20). These receptors, as well as a large set of other characterized and orphan receptors, including most chemokine receptors, were shown to be totally unreactive to purified human chemerin (unpublished data). The activation of chemerinR by a set of >200 bioactive molecules, including all currently available chemokines, C5a, C3a, fMLP, bradykinin, PAF, and leukotrienes, was also tested. All of these agents were unable to promote receptor activation even at concentrations significantly higher (100 nM or 1 μM) than those reported to activate their own receptors. Therefore, chemerin and its receptor appear as a specific signaling system, in contrast to the situation prevailing with inflammatory chemokines and their respective receptors.
To investigate whether proteolytic activation of prochemerin is performed intracellularly in the secretory pathway or is an extracellular process potentially regulated by the activation of extracellular proteases, we tested the activation of human-purified prochemerin in the medium of cultured cells and conditioned media. We could show that human prochemerin can be fully converted into a form active on chemerinR during the incubation (at 100 nM) in the culture medium of hamster CHO-K1 cells, simian Cos-7 cells, or human HEK293 cells (unpublished data), as well as in conditioned media from these cells (Fig. 4 A). These data indicate that prochemerin processing is performed extracellularly and the active chemerin product is not degraded further by the proteolytic activity and is therefore stable in extracellular medium. Although the protease responsible for this processing is not known, the regulation of this enzyme activity is expected to control the extracellular generation of active chemerin in vivo.
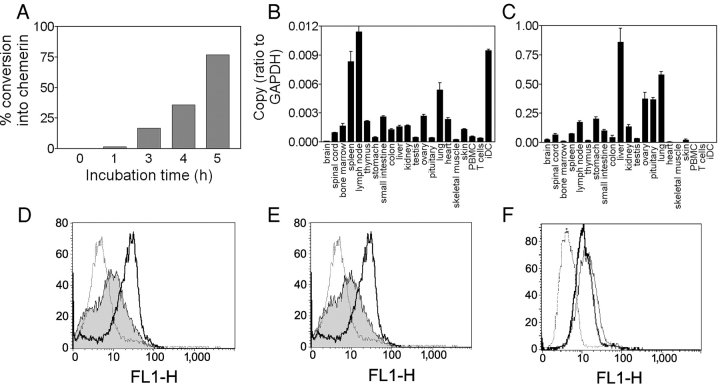
Figure 4.
Expression of human chemerin and its receptor. (A) Conversion of 100 nM human recombinant prochemerin in conditioned medium from hamster CHO-K1 cells. Conversion rate was estimated by comparing the biological activity with that of the same molar amount of purified processed chemerin. (B and C) Transcripts encoding human chemerinR (B) and prochemerin (C) were amplified by quantitative RT-PCR in a set of human tissues and cell populations. PBMC, peripheral blood mononuclear cells; iDC, immature DCs. (D and E) The expression of chemerinR was analyzed by FACS® in immature (solid line) and mature DCs (gray area) after stimulation by LPS (D) or CD40L (E), using the 1H2 monoclonal antibody (IgG2A). Control labeling (dotted line) was made with an antibody of the same isotype. (F) ChemerinR expression on macrophages was monitored using the 1H2 (thick solid line) and 4C7 (thin solid line) monoclonal antibodies. Control labeling (dotted line) was made with an antibody of the same isotype.
Pharmacology and Intracellular Signaling of the Chemerin Receptor.
Attempts to develop a binding assay based on the iodination of full-size chemerin were unsuccessful. However, structure function analysis of peptides derived from the COOH terminus of chemerin (unpublished data) allowed the design of a bioactive peptide (YHSFFFPGQFAFS, EC50 of 28 nM on chemerinR-expressing CHO-K1 cells, using the aequorin-based assay) that could be labeled on its NH2 terminus tyrosine for binding studies. This iodinated peptide was used in a competition binding assay, using the unlabeled peptide or recombinant chemerin as competitors (Fig. 3 C). The Ki values were estimated to 2.5 ± 1.2 nM (pKi: 8.82 ± 0.38) for recombinant chemerin and 12.1 ± 4.97 nM (pKi: 7.95 ± 0.18) for the peptide (mean ± SEM for three independent experiments). The signaling pathways activated by chemerinR were next investigated in CHO-K1 cells expressing the human receptor, but not Gα16 or apoaequorin (CHO/chemerinR cells). Receptor activation was tested in a GTPγ[35S] binding assay, using membranes from CHO/chemerinR cells (EC50: 7.8 ± 0.4 nM, mean ± SEM for four independent experiments; Fig. 3 D). Stimulation of these cells by human chemerin at low nanomolar concentrations resulted in the release of intracellular calcium and inhibition of cAMP accumulation (unpublished data), as well as phosphorylation of the p42 and p44 MAP kinases (Fig. 3, E and F). All of these effects were inhibited by pertussis toxin pretreatment, demonstrating the involvement of Gi family members. No activity of recombinant chemerin or prochemerin was obtained in any of these assays on wild-type CHO-K1 cells (unpublished data).
Expression of Chemerin and Its Receptor.
We investigated the presence of prochemerin and chemerinR transcripts in various human tissues and leukocyte populations by real-time RT-PCR (TaqMan). In addition to immature DCs, chemerinR transcripts were found primarily in spleen, lymph nodes, and lung, and at lower levels in a number of other tissues (Fig. 4 B). Abundant chemerin transcripts were found in liver, lung, pituitary, and ovary (Fig. 4 C), and lower levels could be detected in most tissues. Interestingly, however, no expression of chemerin was found in peripheral blood leukocyte populations. Monoclonal antibodies generated against human chemerinR by genetic immunization (15) and characterized by FACS® on CHO-K1 cell lines expressing the receptor (unpublished data) were used to confirm the presence of the receptor at the surface of DCs and macrophages. High levels of chemerinR immunoreactivity were found on monocyte-derived immature DCs, and chemerinR was downmodulated after maturation of the cells as a result of LPS or CD40L stimulation (Fig. 4, D and E). Similarly, chemerinR immunoreactivity was observed at the surface of monocyte-derived human macrophages (Fig. 4 F). The ability of anti-chemerinR antibodies to block receptor activation by chemerin was investigated using the aequorin assay on chemerinR-expressing CHO-K1 cells. We found that two antibodies (4C7 and 1H2) were able to efficiently inhibit calcium mobilization promoted by recombinant chemerin in a concentration-dependent manner (Fig. 5 A).
Figure 5.
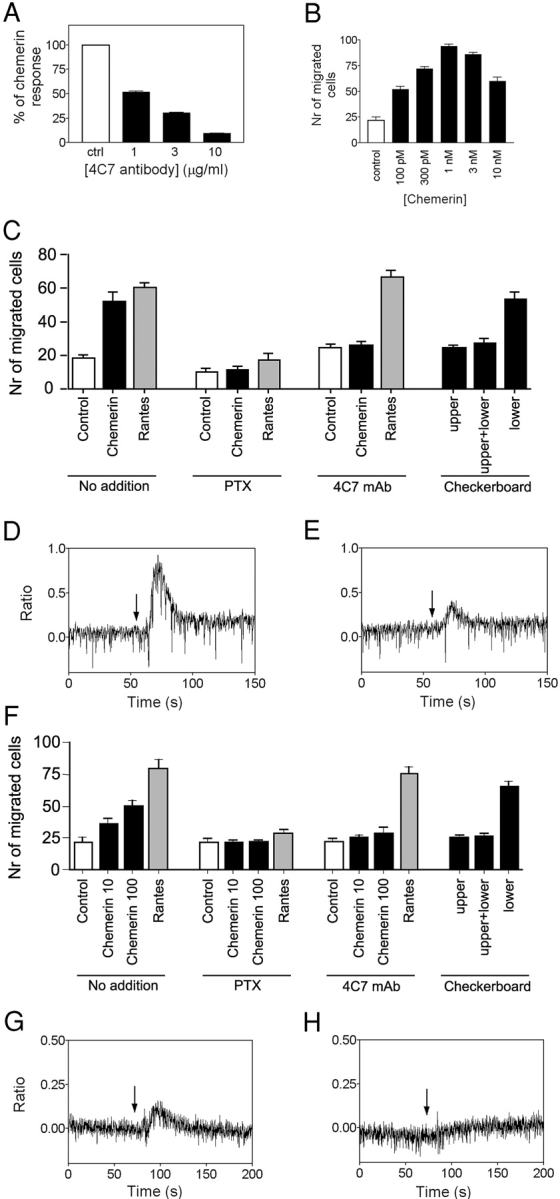
Biological activity of chemerin on primary cells. (A) Inhibition of the functional response of CHO-K1 cells expressing the chemerinR (aequorin assay) by the 4C7 anti-chemerinR monoclonal antibody. The cells were preincubated for 30 min at room temperature with various amounts of the 4C7 antibody before stimulation by 10 nM recombinant chemerin. The data were normalized according to the response in the absence of antibody (100%) and in the absence of agonist (0%). (B) Chemotaxis of human immature DCs by recombinant chemerin. Results are expressed as the mean ± SD (n = 3) and are representative of three donors. (C) 10 pM chemerin-induced DC migration was inhibited by 3 μg/ml pertussis toxin pretreatment of the cells, as well as by preincubation of the cells with 10 μg/ml of the 4C7 monoclonal antibody. Checkerboard analysis investigates chemotactic versus chemokinetic effects of chemerin on DCs. 10 pM human chemerin was added to the lower and/or upper chamber of the chemotaxis device. 10 nM of the chemokine RANTES was used as a positive control in the experiments. (D) Ca2+ flux in monocyte-derived DCs in response to 30 nM recombinant chemerin (arrow). (E) The same experiment after 30 min preincubation of the cells with 10 μg/ml of the 4C7 monoclonal antibody. (F) Chemerin-induced macrophage migration (10 and 100 pM) and its inhibition by 3 μg/ml pertussis toxin pretreatment and 10 μg/ml 4C7 monoclonal antibody. Checkerboard analysis investigates chemotactic versus chemokinetic effects of chemerin on macrophages. (G) Ca2+ flux in macrophages in response to 30 nM recombinant chemerin (arrow). (H) The same experiment after 30 min preincubation of the cells with 10 μg/ml of the 4C7 monoclonal antibody.
To investigate whether chemerin is frequently generated in pathological situations in humans, we fractionated a set of inflammatory fluids and assayed the chemerin content by measuring the biological activity of the fractions on chemerinR as compared with a standard curve made with purified recombinant chemerin. Significant levels of active chemerin, well within the active range (33–358 ng/ml, corresponding to 2–23 nM), were found in the majority of ascitic fluids resulting from ovary cancer, but also in ascitic fluids resulting from a liver cancer and from an ovary hyperstimulation syndrome, as well as in a pool of synovial fluids from arthritic patients (Table I). Interestingly, active chemerin was not detected in synovial fluids pooled from patients with osteoarthritis (Table I), nor in fractions from human hemofiltrate (unpublished data), demonstrating that its presence is linked to inflammatory situations.
Table I.
Bioactive Chemerin Concentration in Human Samples
| Sample | Pathology | Chemerin (ng/ml) |
|---|---|---|
| 1 | Ovary Carcinoma | 74 |
| 2 | Ovary Carcinoma | 73 |
| 3 | Ovary Carcinoma | 104 |
| 4 | Ovary Carcinoma | 92 |
| 5 | Ovary Carcinoma | n.d. (<10) |
| 6 | Ovary Carcinoma | 82 |
| 7 | Ovary Carcinoma | 103 |
| 8 | Ovary Carcinoma | 43 |
| 9 | Ovary Carcinoma | 87 |
| 10 | Ovary Carcinoma | n.d. (<10) |
| 11 | Ovary Carcinoma | 90 |
| 12 | Ovary Carcinoma | 33 |
| 13 | Ovary Carcinoma | 57 |
| 14 | Ovary Carcinoma | 87 |
| 15 | Ovary Carcinoma | 62 |
| 16 | Ovary Carcinoma | 37 |
| 17 | O.H.S. | 116 |
| 18 | Arthritis | 358 |
| 19 | Osteoarthritis | n.d. (<1) |
The amount of chemerin in ascitic (samples 1-17) and synovial (samples 18 and 19) fluids was estimated after two fractionation steps, by assaying the fractions on chemerinR-expressing cells using the aequorin-based assay and a standard curve made with purified recombinant human chemerin. Synovial fluids from arthritis and osteoarthritis patients were pooled for measurement after centrifugation. O.H.S., ovarian hyperstimulation syndrome; n.d., not detectable (the limit of detection in the assay conditions is given).
Biological Activity of Chemerin.
The biological function of chemerin was further investigated on leukocyte populations. In accordance to the coupling of human chemerinR through the Gi class of G proteins, its structural relatedness to chemoattractant receptors, and its expression in APCs, we showed that chemerin acted as a chemotactic factor for these cells. DCs and macrophages were differentiated in vitro from human monocytes. Human recombinant chemerin promoted in vitro migration of macrophages and immature DCs (Fig. 5, B, C, and F), whereas no chemotaxis of mature DCs was observed (unpublished data). Maximal chemotactic responses were obtained for concentrations of 100 pM to 1 nM, according to the batch of recombinant chemerin. Such bell-shaped chemotactic response, with a maximum corresponding to concentrations below the EC50 observed in other functional assays, is typically observed for other chemotactic factors such as chemokines. The effect was completely abolished after treatment with pertussis toxin (Fig. 5, C and F), demonstrating the involvement of the Gi class of G proteins. Migration of macrophages and DCs was also inhibited by the anti-chemerinR monoclonal antibody 4C7 (Fig. 5, C and F) without affecting RANTES-induced cell migration, demonstrating that the effect is specifically mediated by the chemerinR. A checkerboard analysis showed that when equal concentrations of chemerin were present in both the lower and upper wells, no significant increase in cell migration was observed (Fig. 5, C and F). Thus, the migration of macrophages and immature DCs induced by chemerin is essentially a chemotactic effect rather than chemokinesis. We also investigated whether recombinant chemerin could induce Ca2+ mobilization in APCs. As expected, intracellular Ca2+ levels increased in immature DCs in response to recombinant chemerin (Fig. 5 D), whereas the 4C7 antibody inhibited the Ca2+ response (Fig. 5 E). Similar observations were made for macrophages (Fig. 5, G and H).
Discussion
Orphan G protein–coupled receptors have proved over the past few years to constitute key tools allowing the identification of new extracellular pathways. Our work represents one of the few examples so far, in which the natural ligand of an orphan receptor has been purified from a human natural source, using the receptor as a bioassay. This approach has led to the discovery of new ligands such as nociceptin/orphanin FQ, orexins, or ghrelin, which have since taken a growing importance in various aspects of physiology (21–23). We have focused in this work on ChemR23 because of its relatedness to chemoattractant receptors and its unique expression pattern among leukocyte populations, being specific to macrophages and immature DCs, the two major classes of APCs involved both in innate and adaptive immune responses.
We have identified a novel protein, chemerin, as the natural ligand of ChemR23, renamed the chemerin receptor or chemerinR. Chemerin is encoded by a poorly characterized gene (Tig-2), previously shown to be overexpressed in nonlesional psoriatic skin and at lower levels in psoriatic lesions, and to be up-regulated by the synthetic retinoid tazarotene in these lesions (19). Induction of this gene was also shown in the bone stromal cell line ST2 after stimulation by 1,25-dihydroxy-vitamin D3 and dexamethasone (24). However, no specific function has been proposed so far for this gene, which was not predicted previously to encode a secreted protein, although the presence of a signal peptide is unambiguously predicted by classical algorithms. We have demonstrated here that prochemerin is indeed secreted, but that its activation into chemerin requires a specific proteolytic processing. Therefore, the generation of chemerin appears to be regulated at two levels. First, it is transcriptionally regulated, and this regulation appears to vary according to cell types and tissues. Second, the processing of prochemerin is mediated by a so far uncharacterized extracellular protease, and this proteolytic activity is likely regulated as well, according to the extracellular matrix remodeling status of the tissue.
The COOH-terminal processing of prochemerin, i.e., proteolytic cleavage after serine 157, is reminiscent of the situation in other proteins that share a similar fold. Indeed, database searches identified cathelicidins, cystatins, and related proteins such as the bradykinin precursor kininogen, as the closest structural relatives of prochemerin. The sequence of the human cathelicidin FALL-39 precursor is displayed in Fig. 2 C. Although primary structure similarity is low, a pattern of conserved cysteines is shared across the various proteins. In addition, the number and location of introns is strictly conserved within the gene structure of prochemerin, cathelicidins, cystatins, and kininogens, further demonstrating their evolutionary relationship. This is illustrated for the FALL-39 gene in Fig. 2 C. Prochemerin is therefore likely to adopt the same fold as chicken cystatin (Protein Data Bank accession no. 1CEW). However, prochemerin contains six cysteines instead of four in procathelicidins and cystatins, suggesting the presence of a third disulfide bond. Cathelicidins are secreted as propeptides that require proteolytic processing to generate a COOH-terminal peptide endowed with bactericidal properties (25). Both the COOH-terminal peptide and the structured NH2-terminal (cystatin-like) domain have been shown to display chemotactic properties for leukocytes, properties which involve G protein–coupled receptors (26, 27). In addition, bradykinin, a well known G protein–coupled receptor agonist, is also released by the COOH-terminal processing of its precursor, kininogen, which is made of three cystatin-like modules (28). Therefore, chemerin belongs to a large family of evolutionary related protein precursors, which after proteolytic processing, generate peptides and proteins involved in innate and adaptive immune responses.
Chemerin is a potent chemoattractant of immature DCs and macrophages, acting at concentrations similar to that of most chemokines (10 pM to 1 nM). The use of neutralizing antibodies allowed the demonstration of the involvement of the chemerinR in this process. Nanomolar concentrations of active chemerin were found in a large proportion of ascitic fluids secondary to ovarian carcinomas (Table I). Such ascitic fluids are known to display a strong inflammatory component, and bioactive concentrations of various chemokines have been described in the same samples (29). Bioactive chemerin was also recovered in other samples well known to contain high levels of inflammatory mediators. The expression pattern suggests that this new system is involved in many human diseases characterized by strong inflammatory responses. Previous demonstrations of prochemerin expression suggest an involvement in psoriasis (18) as well as in the control of bone remodeling by the macrophage-related osteoclast population (24). Therefore, this novel protein and its receptor constitute interesting candidates as therapeutic targets for future development. The development of knockout models for the chemerin and its receptor gene, currently in progress, will help further delineate the role of this system.
Acknowledgments
We thank Dominique Revets for his expert technical assistance and Raymond Lecocq for his advice. We are grateful to Drs. S. Steinfeld and A. Delbaere for providing human clinical samples.
This work was supported by AIRC (Associazione Italiana Ricerca sul Cancro), the Actions de Recherche Concertées of the Communauté Française de Belgique, the Belgian program on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming, the Cell Factory program of the European Community, the Fonds de la Recherche Scientifique Médicale of Belgium, Télévie, and the Fondation Médicale Reine Elisabeth to M. Parmentier. The scientific responsibility is assumed by the authors. V. Wittamer is the recipient of a grant from the FIRST-Industrie program of the Walloon Region. I. Migeotte and C. Blanpain are Aspirant and D. Communi is Research Associate of the Belgian Fonds National de la Recherche Scientifique.
M. Parmentier and D. Communi contributed equally to this work.
Abbreviation used in this paper: Tig, tazarotene-induced gene.
References
- 1.Mellman, I., and R.M. Steinman. 2001. Dendritic cells: specialized and regulated antigen processing machines. Cell. 106:255–258. [DOI] [PubMed] [Google Scholar]
- 2.Lanzavecchia, A., and F. Sallusto. 2001. Regulation of T cell immunity by dendritic cells. Cell. 106:263–266. [DOI] [PubMed] [Google Scholar]
- 3.Caux, C., B. Vanbervliet, C. Massacrier, S. Ait-Yahia, C. Vaure, K. Chemin, M.C. Dieu-Nosjean, and A. Vicari. 2002. Regulation of dendritic cell recruitment by chemokines. Transplantation. 73:S7–S11. [DOI] [PubMed] [Google Scholar]
- 4.Coussens, L.M., and Z. Werb. 2002. Inflammation and cancer. Nature. 420:860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vicari, A.P., and C. Caux. 2002. Chemokines in cancer. Cytokine Growth Factor Rev. 13:143–154. [DOI] [PubMed] [Google Scholar]
- 6.Baggiolini, M. 2001. Chemokines in pathology and medicine. J. Intern. Med. 250:91–104. [DOI] [PubMed] [Google Scholar]
- 7.Banchereau, J., B. Schuler-Thurner, A.K. Palucka, and G. Schuler. 2001. Dendritic cells as vectors for therapy. Cell. 106:271–274. [DOI] [PubMed] [Google Scholar]
- 8.Samson, M., A.L. Edinger, P. Stordeur, J. Rucker, V. Verhasselt, M. Sharron, C. Govaerts, C. Mollereau, G. Vassart, R.W. Doms, et al. 1998. ChemR23, a putative chemoattractant receptor, is expressed in dendritic cells and is a coreceptor for SIV and some HIV-1 strains. Eur. J. Immunol. 28:1689–1700. [DOI] [PubMed] [Google Scholar]
- 9.Methner, A., G. Hermey, B. Schinke, and I. Hermans-Borgmeyer. 1997. A novel G protein-coupled receptor with homology to neuropeptide and chemoattractant receptors expressed during bone development. Biochem. Biophys. Res. Commun. 233:336–342. [DOI] [PubMed] [Google Scholar]
- 10.Mognetti, B., M. Moussa, J. Croitoru, E. Menu, D. Dormont, P. Roques, and G. Chaouat. 2000. HIV-1 co-receptor expression on trophoblastic cells from early placentas and permissivity to infection by several HIV-1 primary isolates. Clin. Exp. Immunol. 119:486–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy, P.M., M. Baggiolini, I.F. Charo, C.A. Hebert, R. Horuk, K. Matsushima, L.H. Miller, J.J. Oppenheim, and C.A. Power. 2000. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol. Rev. 52:145–176. [PubMed] [Google Scholar]
- 12.Stables, J., A. Green, F. Marshall, N. Fraser, E. Knight, M. Sautel, G. Milligan, M. Lee, and S. Rees. 1997. A bioluminescent assay for agonist activity at potentially any G-protein-coupled receptor. Anal. Biochem. 252:115–126. [DOI] [PubMed] [Google Scholar]
- 13.Kotani, M., M. Detheux, A. Vandenbogaerde, D. Communi, J.M. Vanderwinden, E. Le Poul, S. Brezillon, R. Tyldesley, N. Suarez-Huerta, F. Vandeput, et al. 2001. The metastasis-suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J. Biol. Chem. 276:34631–34636. [DOI] [PubMed] [Google Scholar]
- 14.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858. [DOI] [PubMed] [Google Scholar]
- 15.Costagliola, S., P. Rodien, M.C. Many, M. Ludgate, and G. Vassart. 1998. Genetic immunization against the human thyrotropin receptor causes thyroiditis and allows production of monoclonal antibodies recognizing the native receptor. J. Immunol. 160:1458–1465. [PubMed] [Google Scholar]
- 16.Kotani, M., C. Mollereau, M. Detheux, E. Le Poul, S. Brezillon, J. Vakili, H. Mazarguil, G. Vassart, J.M. Zajac, and M. Parmentier. 2001. Functional characterization of a human receptor for neuropeptide FF and related peptides. Br. J. Pharmacol. 133:138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vulcano, M., C. Albanesi, A. Stoppacciaro, R. Bagnati, G. D'Amico, S. Struyf, P. Transidico, R. Bonecchi, A. Del Prete, P. Allavena, et al. 2001. Dendritic cells as a major source of macrophage-derived chemokine/CCL22 in vitro and in vivo. Eur. J. Immunol. 31:812–822. [DOI] [PubMed] [Google Scholar]
- 18.Sozzani, S., P. Allavena, G. D'Amico, W. Luini, G. Bianchi, M. Kataura, T. Imai, O. Yoshie, R. Bonecchi, and A. Mantovani. 1998. Differential regulation of chemokine receptors during dendritic cell maturation: a model for their trafficking properties. J. Immunol. 161:1083–1086. [PubMed] [Google Scholar]
- 19.Nagpal, S., S. Patel, H. Jacobe, D. DiSepio, C. Ghosn, M. Malhotra, M. Teng, M. Duvic, and R.A. Chandraratna. 1997. Tazarotene-induced gene 2 (TIG2), a novel retinoid-responsive gene in skin. J. Invest. Dermatol. 109:91–95. [DOI] [PubMed] [Google Scholar]
- 20.Joost, P., and A. Methner. 2002. Phylogenetic analysis of 277 human G-protein-coupled receptors as a tool for the prediction of orphan receptor ligands. Genome Biol. 3:0063.10–0063.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meunier, J.C., C. Mollereau, L. Toll, C. Suaudeau, C. Moisand, P. Alvinerie, J.L. Butour, J.C. Guillemot, P. Ferrara, B. Monsarrat, et al. 1995. Isolation and structure of the endogenous agonist of opioid receptor-like ORL1 receptor. Nature. 377:532–535. [DOI] [PubMed] [Google Scholar]
- 22.Lee, D.K., S.R. George, J.F. Evans, K.R. Lynch, and B.F. O'Dowd. 2001. Orphan G protein-coupled receptors in the CNS. Curr. Opin. Pharmacol. 1:31–39. [DOI] [PubMed] [Google Scholar]
- 23.Im, D.S. 2002. Orphan G protein-coupled receptors and beyond. Jpn. J. Pharmacol. 90:101–106. [DOI] [PubMed] [Google Scholar]
- 24.Adams, A.E., Y. Abu-Amer, J. Chappel, S. Stueckle, F.P. Ross, S.L. Teitelbaum, and L.J. Suva. 1999. 1,25 dihydroxyvitamin D3 and dexamethasone induce the cyclooxygenase 1 gene in osteoclast-supporting stromal cells. J. Cell. Biochem. 74:587–595. [PubMed] [Google Scholar]
- 25.Yang, D., O. Chertov, and J.J. Oppenheim. 2001. The role of mammalian antimicrobial peptides and proteins in awakening of innate host defenses and adaptive immunity. Cell. Mol. Life Sci. 58:978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, D., Q. Chen, A.P. Schmidt, G.M. Anderson, J.M. Wang, J. Wooters, J.J. Oppenheim, and O. Chertov. 2000. LL-37, the neutrophil granule- and epithelial cell–derived cathelicidin, utilizes formyl peptide receptor-like 1 (FPRL1) as a receptor to chemoattract human peripheral blood neutrophils, monocytes, and T cells. J. Exp. Med. 192:1069–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verbanac, D., M. Zanetti, and D. Romeo. 1993. Chemotactic and protease-inhibiting activities of antibiotic peptide precursors. FEBS Lett. 317:255–258. [DOI] [PubMed] [Google Scholar]
- 28.Salvesen, G., C. Parkes, M. Abrahamson, A. Grubb, and A.J. Barrett. 1986. Human low-Mr kininogen contains three copies of a cystatin sequence that are divergent in structure and in inhibitory activity for cysteine proteinases. Biochem. J. 234:429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schutyser, E., S. Struyf, P. Proost, G. Opdenakker, G. Laureys, B. Verhasselt, L. Peperstraete, I. Van de Putte, A. Saccani, P. Allavena, et al. 2002. Identification of biologically active chemokine isoforms from ascitic fluid and elevated levels of CCL18/pulmonary and activation-regulated chemokine in ovarian carcinoma. J. Biol. Chem. 277:24584–24593. [DOI] [PubMed] [Google Scholar]