Abstract
Somatic hypermutation (SHM) and class switch recombination (CSR) are initiated by activation-induced cytidine deaminase–mediated cytidine deamination of immunoglobulin genes. MutS homologue (Msh) 2−/− mice have reduced A-T mutations and CSR. This suggests that Msh2 may play a role in repairing activation-induced cytidine deaminase–generated G-U mismatches. However, because Msh2 not only initiates mismatch repair but also has other functions, such as signaling for apoptosis, it is not known which activity of Msh2 is responsible for the effects observed, and consequently, many models have been proposed. To further dissect the role of Msh2 in SHM and CSR, mice with a “knockin” mutation in the Msh2 gene that inactivates the adenosine triphosphatase domain were examined. This mutation (i.e., Msh2G674A), which does not affect apoptosis signaling, allows mismatches to be recognized but prevents Msh2 from initiating mismatch repair. Here, we show that, similar to Msh2−/− mice, SHM in Msh2G674A mice is biased toward G-C mutations. However, CSR is partially reduced, and switch junctions are more similar to those of postmeiotic segregation 2−/− mice than to Msh2−/− mice. These results indicate that Msh2 adenosine triphosphatase activity is required for A-T mutations, and suggest that Msh2 has more than one role in CSR.
Keywords: activation-induced cytidine deaminase, mismatch repair, affinity maturation, B cell, antibody
Introduction
High affinity antibodies of different classes are required to efficiently neutralize and clear pathogens and toxins from an animal's circulation. Affinity maturation is achieved by the somatic hypermutation (SHM) of immunoglobulin variable regions followed by selection in germinal centers for B cells producing high-affinity antigen-specific antibodies. Changes in antibody class are mediated by the class switch recombination (CSR) process, which recombines the V region to a downstream constant region in the immunoglobulin locus (1–3). Both processes require activation-induced cytidine deaminase (AID; references 4, 5). AID initiates SHM and CSR by deamination of cytidines in the V region and switch regions, respectively (6–10). Transcription of these sequences is expected to produce single-stranded DNA that would allow access to AID for deamination (6, 7, 11–13). This model for AID function is supported by findings that mice deficient for the uracil DNA glycosylase UNG have defects in both SHM and CSR (14). The uridine that is thus created by deamination is either replicated to produce T mutations or converted into an abasic site by the uracil DNA glycosylase, UNG. These abasic sites can be acted on by apurinic endonuclease and repaired by base excision repair.
Although this model might account for mutations at G-C basepairs, it does not explain how mutations occur at A-T basepairs. Papers investigating mismatch repair (MMR)–deficient mice suggest that this pathway is responsible for introducing mutations at A-T basepairs; mice deficient in MutS homologue (Msh) 2 or Msh6 have fewer A-T mutations than controls (15–18). These findings led to the hypothesis that SHM proceeds in two phases as follows: the first phase introduces mutations at G-C basepairs whereas the second phase, which is Msh2/6-dependent, is responsible for mutations at A-T basepairs (16). However, it remains unclear why the MMR system is mutagenic in B cells. One possibility is that Msh2/6 heterodimers participate directly in SHM by competing with UNG to bind to the AID-generated G-U mismatches, excising the DNA surrounding the mismatch and initiating error-prone repair with the translesional error-prone polymerases ι, ζ, and η (11). However, it has also been proposed that the defect in SHM and CSR in Msh2−/− mice is indirect and is due to reduced B cell viability resulting from a general loss of genomic stability (19, 20). This was supported by the finding in Msh2−/− mice that Peyer's patch B cells that had undergone many rounds of replication had increased microsatellite instability and the germinal centers in such mice were small and showed increased apoptosis (19, 20). Thus, these findings suggest that programmed cell death might influence the outcome of SHM. Indeed, SHM in Msh2−/− mice occurs at a reduced rate (16, 20), consistent with the notion that mutations do not accumulate in Msh2−/− mice because of reduced B cell viability. However, if MMR is directly involved in producing A-T mutations during SHM, a reduced mutation frequency would also be expected in Msh2−/− mice.
After Msh2/6 heterdimers bind to mismatches, the adenosine triphosphatase (ATPase) domain of Msh2 processes ATP (21). This is thought to induce a change in the Msh2/6 heterodimer that allows it to activate MMR and recruit downstream repair factors such as the MutL homologue (Mlh) 1, postmeiotic segregation (Pms) 2, and exonuclease 1 (21, 22). The ATPase domain of Msh2 contains a Walker “type A” motif (i.e., GNNNNGKS/T), which is found in other proteins involved in ATP hydrolysis and is known to be involved in binding to the phosphate group of ATP (see Fig. 1 A). Mutations in ATP processing domains occur frequently in hereditary nonpolyposis colorectal cancer (23) and result in MMR-deficient Saccharomyces cerevisiae (24, 25). Mice with a knockin mutation in the Msh2 gene that converts a glycine to an alanine at residue 674 (i.e., Msh2G674A mice) in the Walker type A motif (i.e., GNNNNAKS/T) express normal amounts of the mutant protein, are MMR deficient, and prone to B and T cell lymphomas and gastrointestinal adenocarcinomas (unpublished data). Although an S. cerevisiae Msh2 protein with a mutation in the equivalent residue to the mouse Msh2G674A functions as a dominant-negative (24, 25), this appears not to be the case for the murine Msh2G674A protein because mice heterozygous for the Msh2G674A mutation were essentially normal. In vitro studies showed that Msh2G674A/Msh6 heterodimers bound to G-G mismatches, but were MMR deficient (unpublished data). However, unlike Msh2−/− mouse embryo fibroblasts, wild-type and Msh2G674A mouse embryo fibroblasts cells were sensitive to cisplatin-induced apoptosis, indicating that Msh2G674A retains the ability to signal for this process (unpublished data). The apoptotic signaling function of Msh2 is likely related to its role in the S-phase checkpoint (26), where it acts as a “damage sensor” possibly through a large complex that contains checkpoint kinase CHK2 and ATM (23, 26).
Figure 1.

The Msh2G674A mutation leads to a reduced anti-NP immunoglobulin response in Msh2G674A mice. (A) Schematic illustrating the domains of murine Msh2. Shown is the DNA binding domain, the domains involved in binding to Msh3 and Msh6, and the ATPase domain, which contains the Walker A motif. (B) Anti-NP titers in NP-immunized mice. Two Msh2G674A mice (M/M) and two heterozygous littermate controls (M/+) were immunized i.p. with NP23-CGG. ELISA plates coated with NP2-BSA or NP17-BSA were used to measure the serum titers of high affinity and total NP-specific IgG1, respectively. Serum titers were calculated using the serial dilution closest to 1/2 maximum. Standard errors are shown.
Because the Msh2G674A protein retains the apoptosis signaling capacity of Msh2, but is defective in initiating MMR, the G674A mutation represents a separation of function mutation for the Msh2 protein. Therefore, we examined Msh2G674A mice for defects in SHM and CSR because the repair activity of MMR is hypothesized to be responsible for introducing mutations at A-T basepairs (described in a previous paragraph) and might also be required for CSR.
Materials and Methods
Mice and Immunizations.
The Msh2G674A mice were created by a knockin targeting strategy and will be reported elsewhere (unpublished data). Msh2G674A were backcrossed to C57Bl/6 mice for four generations and were housed in a pathogen-free facility. Two Msh2G674A, two heterozygote littermates, and two wild-type littermates that were ∼70 d old were immunized by intraperitoneal injections of alum-precipitated nitrophenyl (NP) coupled to chicken γ globulin (NP)23-CGG and sera was monitored for antibodies specific to NP by ELISA, as described previously (27).
PCR Amplification and Sequence Analysis.
The JH3–JH4 region was sequenced as described previously (27). In brief, Peyer's patches were harvested from three Msh2G674A mice, three littermate controls, and one Msh2−/− mouse that were ∼8 mo old. PNAhi B cells were FACS® sorted. The JH3–JH4 region was amplified using Pfu turbo (Stratagene) and primers described previously (27). PCR products were cloned and sequenced. Statistics for data in Table I were calculated using the paired Student's t test comparing individual mice grouped into control and Msh2G674A mice.
Table I.
| Controls | Msh2G674A | Msh2−/− | |
|---|---|---|---|
| Mutation frequency (×10−3)c | 126/73,690 (1.71) | 115/158,580 (0.63) | 90/53,300 (1.69) |
| GC mutations/total | 53/126 (42%) | 98/115 (85%)e | 74/90 (82%) |
| Transition mutations/total | 69/126 (55%) | 84/115 (73%)e | 56/90 (62%) |
| Hotspot mutationsd/total | 20/126 (16%) | 38/115 (32%)e | 23/90 (26%) |
The JH2 to JH4 region is 1,460 bp long of which 1,300 bp was sequenced. Data obtained from Fig. 2.
PCR products amplified by PFU polymerase were cloned and plasmids sequenced. Identical mutations that were observed in more than one plasmid for each mouse were scored only once.
Frequency is defined as mutations per basepair sequenced.
Hotspot mutations occur in G-C basepairs within RGYW or WRCY motifs. 31/650 nucleotides (4.8%) of the JH3–JH4 region are hotspot nucleotides.
P < 0.05 compared to controls using the paired Student's t test.
In Vitro Switching Assay.
Switching was induced as described previously (27). In brief, unimmunized ∼6-mo-old mice were killed, spleens were harvested, and B cell splenocytes were purified. B cells were plated in RPMI 1640 media containing 10% FCS and 50 μg/ml LPS (Sigma-Aldrich) or with 50 μg/ml LPS and 50 ng/ml rIL-4 (R&D Systems) for 4 d, and cells were harvested and stained with anti-IgG1 or anti-IgG3 FITC (BD Biosciences) and anti-IgM CY5 (Jackson Immunochemicals). Cells were washed with PBS, fixed with 1% paraformaldehyde in PBS, and analyzed with a FACSCalibur™ (Becton Dickinson) apparatus. FACS® analysis was performed using the FlowJo software package (Treestar). Cells were gated for viability based on the forward- and side-scatter profiles. The IgG-positive population was scored according to the gating profile shown in Fig. 3. Statistics were calculated using the paired Student's t test comparing individual mice grouped into control and Msh2G674A mice.
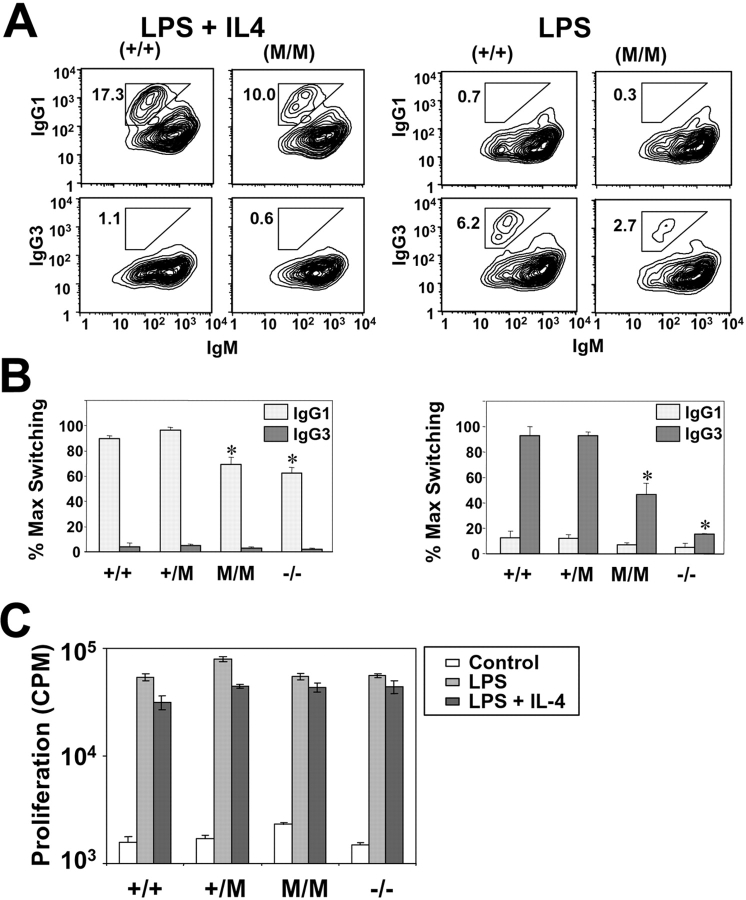
Figure 3.
Reduced in vitro switching of purified Msh2G674A B cells. (A) (left) B cell switching induced with LPS and IL-4. Representative FACS® profile of experiment showing the percent of IgG1 and IgG3 positive cells of the gated live population of B cells derived from wild-type (+/+) and Msh2G674A (M/M) mice. (right) Same, except that B cell switching was induced with LPS. (B) Means of switching experiment of B cells derived from three wild-type (+/+), four heterozygous Msh2G674A (+/M), three homozygous Msh2G674A (M/M), and two homozygous Msh2−/− (−/−) mice. Because of the high degree of variability between switching experiments, the data are normalized to percent maximum switching for each experiment. Standard errors are shown. *, P < 0.05 when compared with +/M controls. (C) B cell proliferation in control, Msh2G674A, and Msh2−/− B cells. Thymidine incorporation on day 3 of unstimulated B cells, or B cells stimulated with LPS or LPS and IL4. M/M are homozygous Msh2G674A B cells, whereas −/− are homozygous Msh2−/− B cells. Standard errors are shown.
Sμ-Sγ3 Switch-junction Analysis.
Three Msh2G674A and four littermate controls were used in this analysis. Genomic DNA was isolated using the DNAeasy kit (QIAGEN) from cells stimulated with 50 μg/ml LPS. Sμ-Sγ3 junctions were amplified by PCR using Expand Long Template Taq Polymerase (Roche) essentially as described previously (28). The primers used are as follows: μ3-H3, 5′-AACAAGCTTGGCTTAACCGAGATGAGCC-3′; and γ3–2, 5′-TACCCTGACCCAGGAGCTGCATAAC-3′. Amplified PCR products were cloned using the TOPO cloning kit (Invitrogen). Plasmid DNA was prepared using a 96-well miniprep kit (Millipore). T3 or T7 primers (Invitrogen) were used to sequence the PCR inserts. Sequence alignments were constructed by comparing the PCR sequence with Sμ references AF446347 and Sγ3 reference Musighana using Blast2 alignment (National Center for Biotechnology Information). Although sometimes there was more than one possible alignment due to the repetitive nature of the switch regions, the regions with the largest homology and highest identity were used. Statistics for data in Table II were calculated using the Student's t test, conditional on not having insertions. Based on homologies falling into each group and given weights due to the ordinal nature, this distribution was compared for each mouse between mutant and control mice.
Table II.
| Microhomologies
|
Insertions
|
||||||
|---|---|---|---|---|---|---|---|
| Blunt | 1–2 nt | 3–4 nt | ≥5 nt | 1–4 nt | ≥5 nt | Total | |
| Controls (%) | 16 (30) | 21 (39) | 6 (11) | 5 (9) | 5 (9) | 1 (2) | 54 (100) |
| Msh2G674A (%)c | 10 (17) | 12 (20) | 9 (15) | 14 (24) | 5 (8) | 9 (15) | 59 (100) |
Data presented in Fig. S1.
Unique switch junctions compiled from three Msh2G674/G674A mice and four littermate controls.
Microhomology lengths are statistically different when compared with controls (P = 0.04). See Materials and Methods.
nt, nucleotide.
Proliferation Assays.
5 × 105 total spleen cells were plated, as 5-well replicates in a 96-well culture plate, in 200 μl final volume of appropriate media. Three conditions were tested in these assays as follows: unstimulated, LPS stimulation, and LPS + IL-4 stimulation (same concentrations as described in the In Vitro Switching Assay section). Proliferation assays were performed on day 3 by pulsing with 1 μCi/well of [3H]Thymidine (NEN Life Science Products) for ∼16 h before harvesting onto 96-well printed filtermats (Wallac) and measuring radioactive incorporation on an LSC plate counter (model 1450; Microbeta; Wallac).
Online Supplemental Material.
In Fig. S1, μ-γ3 switch junctions from LPS-induced B cells are shown. In this figure, regions of homology between μ and γ3 switch regions and insertion sequences in μ-γ3 switch junctions are detailed. Fig S1 is available at http://www.jem.org/cgi/content/full/jem.20030880/DC1.
Results
Mice harboring a point mutation in the ATPase domain of the Msh2 gene were created by homologous recombination into the Msh2 loci. This mutation converts a glycine to an alanine at residue 674 of the Msh2 protein (i.e., Msh2G674A; Fig. 1 A). An equivalent mutation in the Msh2 gene of S. cerevisiae inhibits Msh2 ATP processing (24, 25). This defect was biochemically confirmed for murine Msh2G674A (unpublished data).
Msh2−/− mice show impaired antigen-specific humoral responses (15, 16, 29). This deficiency is probably due to defects in both the SHM and CSR processes. To determine whether Msh2G674A mice are defective in either of these processes, two heterozygous littermates and two Msh2G674A mice were immunized with the NP hapten coupled to chicken γ globulin. Sera were analyzed 10 d later by ELISA using NP2-BSA– and NP17-BSA–coated plates that bind to high affinity and total NP-specific IgG, respectively (27, 30). As shown in Fig. 1 B, NP-immunized Msh2G674A mice had approximately fivefold lower titers of high affinity and total NP-specific IgG1 than did the heterozygous littermate controls. The antibody titers to NP in wild-type and heterozygous littermates were similar (unpublished data). These results suggest that there is a defect in SHM and/or CSR in Msh2G674A mice.
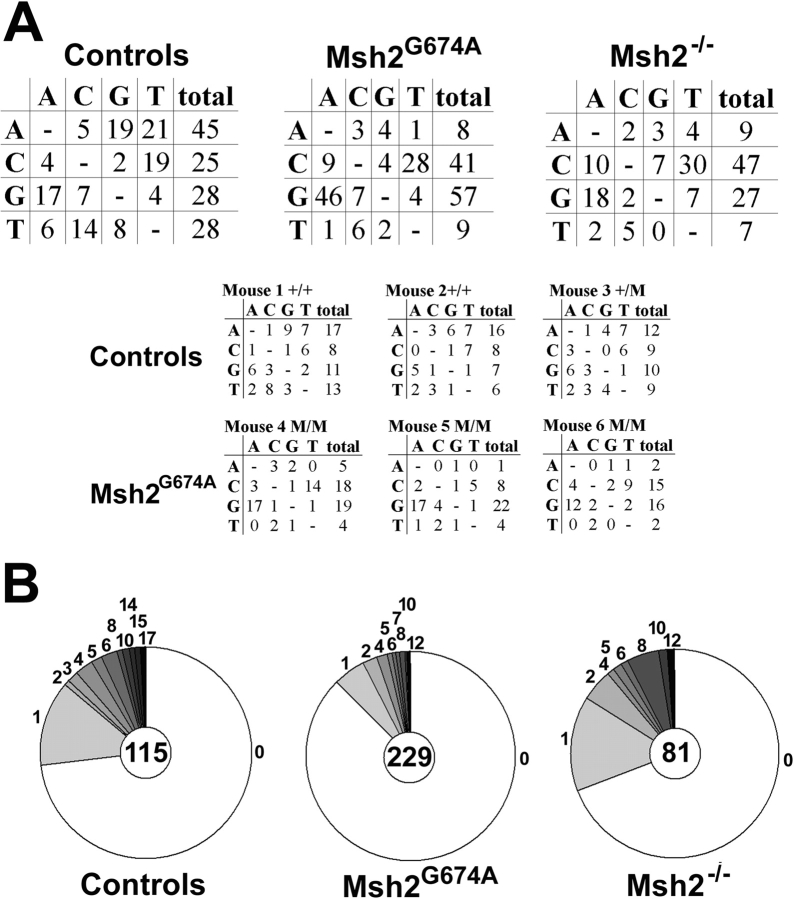
To determine whether SHM was compromised in Msh2G674A mice, the JH3 and JH4 exons and flanking introns in the heavy chain immunoglobulin locus were sequenced from FACS®-sorted PNAHi Peyer's patch B cells from three ∼8-mo-old Msh2G674A mice and three littermate controls. Numerous studies have analyzed this region to assess the effects of genetic deficiencies on the SHM process because mutations are unselected and occur at high rates (14, 17, 20, 27). One Msh2−/− mouse was also examined as a positive control for effects on the SHM spectrum. For each mouse, only unique mutations were tabulated. An analysis of the characteristics of mutations identified revealed that ∼42 and 82% of mutations occurred at G-C basepairs in control and Msh2−/− mice, respectively (Fig. 2 A and Table I). This finding is consistent to what has been reported previously for Msh2−/− mice (15, 16). Sequence analysis of the Msh2G674A mice showed a similar defect in SHM to Msh2−/− mice: ∼85% of mutations occurred at G-C basepairs (Fig. 2 A and Table I). This defect likely reflects a deficiency in A-T mutations rather than an increase in G-C mutations because the mutation frequency is lower in Msh2G674A mice than in littermate controls (Fig. 2 B and Table I; see Discussion). Mutation frequencies may not be comparable between normal controls of the Msh2G674A and Msh2−/− mice because they were not littermates, although they were age-matched and analyzed at the same time. Mutations at RGYW/WRCY hotspot motifs were also increased in Msh2G674A mice (Table I, 16 vs. 32% in control and Msh2G674A mice, respectively) and many of these base changes were transition mutations (Table I). Msh2−/− mice also showed a similar increase in hotspot and transition mutations to Msh2G674A mice (Table I). These findings show that SHM is similarly affected in Msh2−/− and Msh2G674A mice, indicating that the repair activity initiated by Msh2 is required for A-T mutations in SHM.
Figure 2.
Skewed SHM in Msh2G674A mice. (A) Sequencing analysis of the JH3-JH4 region in Peyer's patch B cells from Msh2−/− mice, Msh2G674A mice (M/M), and homozygous (+/+) or heterozygous (+/M) wild-type littermate controls. Unique mutations that occurred at the nucleotides depicted on the left column were mutated to the nucleotides shown in each row. The top three panels represent compiled data from one Msh2−/−, three Msh2G674A, and three littermate controls (Table I). The bottom six panels represent data derived from each individual Msh2G674A and littermate control mice. (B) Pie charts depict the distribution of mutation frequencies for plasmid clones that were sequenced (center of pie) from each mouse group and the proportion of sequences with 0, 1, 2 … mutations (numbers of mutations outside the pie slices).
The decreased titers to the NP-hapten could also be due to defects in the CSR process. To investigate this issue, B cell splenocytes from three Msh2G674A mice, seven littermate controls, and two age-matched Msh2−/− mice were isolated (Materials and Methods) and stimulated in culture with LPS or LPS and IL-4. LPS stimulates primary B cells to undergo CSR from IgM to IgG3, whereas LPS and IL-4 induces CSR to IgG1 (Fig. 3, A and B) . CSR to IgG1 was reduced ∼35% in Msh2−/− B cells and ∼25% in Msh2G674A B cells (Fig. 3, A and B, left). However, CSR to IgG3 was more severely affected in Msh2−/− and Msh2G674A B cells (Fig. 3, A and B, right) as follows: Msh2G674A B cells showed an ∼50% reduction in CSR to IgG3 whereas Msh2−/− B cells showed an ∼80% reduction when compared with wild-type B cells (Fig. 3, A and B, right). No significant differences were observed in the proliferative capacity of B cells from control, Msh2G674A, and Msh2−/− mice (Fig. 3 C), consistent with a previous paper (31). Thus, deficiencies in CSR activity in Msh2G674A and Msh2−/− B cells could not be ascribed to differences in proliferation. Collectively, these data indicate that the CSR process is affected in Msh2G674A mice, albeit not as severely as in Msh2−/− mice.
Previous works showed that the junctions of μ-γ3 switch regions were altered in Msh2−/− B cells (28, 32). Short regions of microhomology between μ and γ3 switch regions are typically observed in a large proportion of switch junctions from normal B cells. However, in Msh2−/− B cells, fewer switch junctions contain short regions of microhomologies (28). Although the significance of these findings to the CSR process is not clear, it suggests that microhomologies help resolve DNA breaks in an Msh2-dependent manner during normal CSR. To further investigate the CSR process in Msh2G674A mice, μ-γ3 switch junctions were analyzed in LPS-stimulated, control, and Msh2G674A B cells. Two other groups have performed switch-junction analysis in MMR-deficient mice (28, 32). Therefore, we have tabulated the switch junctions so that comparisons can be made between our data and those of the other two groups.
Three Msh2G674A mice and four littermate controls were used in this analysis and only unique switch junctions were tabulated (i.e., 58 and 54 sequences for Msh2G674A and controls, respectively). The complete dataset is shown in Fig. S1 available at http://www.jem.org/cgi/content/full/jem.20030880/DC1 and summarized in Table II. The switch junctions from Msh2G674A B cells differed in two ways from those of controls. First, more sites of recombination had longer microhomologies between the μ and γ3 in Msh2G674A B cells than in controls as follows: 24% of switch junctions from Msh2G674A B cells had microhomologies that were ≥5 nucleotides long compared with only 9% in littermate controls (Table II). Although Schrader et al. reported that only 3% of switch junctions from wild-type mice had ≥5 nucleotide-long microhomologies (28), they reported that ∼23% of switch junctions from Pms2−/− and Mlh1−/− B cells and 0% of switch junctions from Msh2−/− B cells had ≥5 nucleotide-long microhomologies (28). Thus, switch junctions from Msh2G674A B cells are more similar to those found in Pms2−/− and Mlh1−/− B cells. Ehrenstein et al. reported that the mean length of uninterrupted donor/acceptor sequence in Pms2−/− and control B cells is 3.5 and 0.8, respectively (32). Analyzing our data in that manner showed a mean length of uninterrupted donor/acceptor sequence from Msh2G674A and control B cells to be 2.6 and 1.8, respectively. Thus, the trend of longer microhomologies in Msh2G674A B cells was again observed, albeit not as pronounced as in Pms2−/− B cells. Therefore, switch junctions from Msh2G674A B cells are less like those in Msh2−/− B cells and more similar to those in Pms2−/− and Mlh1−/− B cells.
The second difference that was observed in this analysis was that 15% of the switch junctions in Msh2G674A B cells had large insertions compared with only 2% in control B cells (Table II). Most of these insertions were <24 basepairs long, but two were large and included sequences from a murine type C retrovirus (222 nucleotides) and a segment from chromosome 2 (114 nucleotides; Fig. S1). Although it is difficult to rule out the possibility that these two large insertions were not generated by a PCR artifact, large insertions, including some with viral sequences, have been observed in DNA rearrangements in immune and nonimmune cells (33). In addition, an increase in nucleotide insertions was also observed in Msh2−/− B cells (28). This indicates that switch junctions from Msh2G674A B cells have a mixed phenotype exhibiting similarities to switch junctions from Pms2−/−/Mlh1−/− mice and from Msh2−/− B cells. Collectively, these data show that CSR is perturbed in Msh2G674A mice, but in a manner that is distinct from Msh2−/− mice.
Discussion
Although AID initiates SHM and CSR by cytidine deamination of single-stranded DNA in the V region and switch regions, respectively, the subsequent events are likely to be quite different because proteins involved in nonhomologous end joining, such as Ku70 and Ku80, are required for CSR and not for SHM (34, 35). However, Msh2 has been found to be involved in both SHM and CSR, implying that these two processes might have other similarities. What complicates this interpretation is that Msh2 functions in at least three distinct physiological processes and, thus, might utilize a different function (or combination of functions) depending on the type of DNA lesion it encounters. Msh2 has been found to repair mismatches caused by point mutations or small insertions or deletions, to signal apoptosis in cells in response to DNA damage, and to inhibit recombination between nonidentical sequences (21). The goal of this paper was to dissect the activities of Msh2 and determine their requirement for SHM and CSR. The Msh2G674A mutation, which is located within the ATPase domain of Msh2, disrupts ATP binding and/or hydrolysis (24, 25; unpublished data). As a result, the Msh2G674A protein lacks the ability to initiate repair of mismatched DNA, but retains the apoptosis-inducing function of Msh2. It is not known whether Msh2G674A can function to inhibit recombination between nonidentical sequences. Thus, at least two functions for the Msh2 protein have been dissociated by the G674A mutation.
The findings reported here show that SHM is compromised in Msh2G674A mice in two ways. First, the majority of mutations occur at G-C basepairs (Table I). Second, the mutation frequencies are lower in Msh2G674A mice than in littermate controls. The preferential mutation of G-C basepairs is also seen in Msh2−/− mice (15, 16) but is not as dramatic in Pms2−/− and Mlh1−/− mice (15, 32, 36–38), suggesting that Pms2 or Mlh1 is partially redundant. The findings reported here also suggest that the repair of mismatches, which is deficient in Msh2G674A mice, is required for A-T mutations. Thus, Msh2/6 heterodimers might bind to AID-generated G-U mismatches, a mismatch that can be recognized by Msh2/6 heterodimers (39), and initiate error-prone repair. Although it is unclear how Msh2-directed repair can lead to mutations at A-T basepairs, one possibility is that the DNA surrounding the mismatch is excised and the resulting gap is filled in by the translesional error-prone polymerases ι, ζ, and η that have been implicated in SHM (40–46). In fact, polymerase η has been implicated as being the A-T mutator in SHM (42, 43). Such a proposition would suggest that secondary mutations introduced by these error-prone polymerases might trigger a second round of MMR because their activities would be expected to produce additional novel substrates for Msh2/6 heterodimers.
The lower mutation frequencies observed in Msh2G674A mice could simply reflect a loss of mutations at A-T basepairs. Indeed, adding back the A-T mutations that were calculated to have been lost in Msh2G674A mice would give a mutation frequency of 1.47 × 10−3 (i.e., 98 G-C mutations in Msh2G674A mice divided by 42% G-C mutations in littermates would give 233 total mutations in 15,8580 base pairs sequenced; Table I), which is similar to that of the littermate controls (Table I, 1.71 × 10−3). Conversely, this lower mutation frequency is consistent with the hypothesis that fewer mutations accumulate in Msh2G674A B cells because of reduced B cell viability (19, 20). In this scenario, the effects of Msh2 deficiency on SHM would be an indirect consequence of a general defect in DNA repair. This issue was partially addressed with findings that SHM in Msh2−/− mice transgenic for the bcl-2 oncogene was similar to Msh2−/− mice (47), showing that programmed cell death does not contribute significantly to SHM effects. However, in contrast to the work presented here and those of others (15, 16), that analysis did not find a significant skewing of mutations toward G-C basepairs in Msh2−/− and bcl 2-transgenic Msh2−/− mice when compared with controls (47). Nevertheless, it is possible that SHM in Msh2G674A mice is skewed due to a reduced overall mutation frequency coupled with an increase in mutations at G-C basepairs. The increase in G-C mutations might be the result of preferential repair of G-U mispairs by Msh2/6 heterodimers over other mispairs (15, 48). Thus, the reduced A-T mutations in Msh2−/− and Msh6−/− mice would be a consequence of MMR deficiency, and would possibly indicate that A-T mutations occur independent of Msh2. However, the results presented here add further support to the notion that Msh2/6 dimers directly participate in the SHM process because Msh2G674A mice still show a bias toward mutating G-C basepairs, even though the Msh2G674A protein retains a normal capacity to signal apoptosis, a process that was deficient in Msh2−/− mice.
CSR in Msh2G674A mice was also perturbed in two ways. First, CSR activity was reduced in Msh2G674A B cells compared with controls, albeit, not to the same degree as in Msh2−/− B cells (Fig. 3). This indicates that the Msh2G674A protein retains some activities that allow CSR to proceed at a reduced capacity. Second, μ-γ3 switch junctions in Msh2G674A B cells were altered when compared with controls; a larger proportion of switch junctions from Msh2G674A B cells contained long microhomologies and large insertions when compared with normal controls (Table II). Although large insertions were also observed in Msh2−/− B cells (28), long microhomologies were observed in Mlh1−/− and Pms2−/− B cells (28, 32) and not in Msh2−/− B cells (28). Thus, switch junctions in Msh2G674A mice display a mixed phenotype between Msh2−/− and Pms2−/− or Mlh1−/− mice. And because Msh2G674A B cells have more CSR activity than Msh2−/− B cells (Fig. 3), these data suggest that Msh2 has more than one role in CSR, one (or more) that is ATPase dependent and one (or more) that is ATPase independent. However, we cannot exclude the possibility that one of these roles is not directly involved in the CSR process.
The data presented here suggest that DNA lesions produced during CSR are repaired by a number of independent mechanisms. At least two pathways appear to be Msh2 dependent, whereas one or more are Msh2 independent. The latter is inferred because CSR still proceeds at significant levels in MMR-deficient mice. The similarity in CSR activity between Msh2G674A and Pms2−/− or Mlh1−/− mice might be due to the uncoupling of Pms2/Mlh1 function from Msh2 by the Msh2G674A mutation. Therefore, the difference between Msh2−/− and MshG674A might reveal a pathway that is Pms2/Mlh1 independent. This notion is supported by findings in S. cerevisiae that Msh2 functions with Msh3 and Rad1 in an Mlh1–Pms2-independent pathway to remove 3′ overlaps during gene conversion (49). Indeed, Msh2 was suggested recently to have more than one role in CSR because switch junctions from mice doubly deficient in Mlh1 and Msh2 are intermediate to switch junctions from the singly deficient mice (50), similar to that of Msh2G674A mice. But because the phenotype of these Mlh1/Msh2 doubly deficient mice was not similar to Msh2−/− mice, which would be expected if Msh2 acted upstream of Mlh1, the authors concluded that Mlh1 functions either upstream of Msh2 or functions independently of Msh2 (50). However, it is interesting to note that the Msh2G674A mutation produced a CSR phenotype that was similar to Mlh1/Msh2 doubly deficient mice, suggesting that Msh2 is dominant or functions independently of Mlh1. Although the work reported here does not fully resolve the questions about whether Msh2 and other MMR proteins are directly or indirectly involved in either SHM or CSR, the use of such mutated proteins should ultimately lead to the dissection of the different roles of MMR in each of these processes.
Acknowledgments
We would like to thank Dr. D. Ronai for critical review of the manuscript and M. Fan for technical help.
This work was supported by grants from the National Institutes of Health to W. Edelmann (CA76329 and CA93484), to P.D. Bardwell (grant 5T32 CA09173), and to M.D. Scharff (CA 72649, CA102705, and AI 43937), who is also supported by the Harry Eagle Chair provided by the National Women's Division of the Albert Einstein College of Medicine. Z. Li is a Cancer Research Institute Fellow. M.D. Iglesias-Ussel is a fellow of the Ministerio de Educacion, Cultura y Deporte. A. Martin is a special fellow of the Leukemia and Lymphoma Society.
The online version of this article includes supplemental material.
Abbreviations used in this paper: AID, activation-induced cytidine deaminase; ATPase, adenosine triphosphatase; CSR, class switch recombination; MMR, mismatch repair; Mlh, MutL homologue; Msh, MutS homologue; NP, nitrophenyl; Pms, postmeiotic segregation; SHM, somatic hypermutation.
References
- 1.Kenter, A.L. 2003. Class-switch recombination: after the dawn of AID. Curr. Opin. Immunol. 15:190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manis, J.P., M. Tian, and F.W. Alt. 2002. Mechanism and control of class-switch recombination. Trends Immunol. 23:31–39. [DOI] [PubMed] [Google Scholar]
- 3.Stavnezer, J. 2000. Molecular processes that regulate class switching. Curr. Top. Microbiol. Immunol. 245:127–168. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 5.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. [DOI] [PubMed] [Google Scholar]
- 6.Bransteitter, R., P. Pham, M.D. Scharff, and M.F. Goodman. 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA. 100:4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudhuri, J., M. Tian, C. Khuong, K. Chua, E. Pinaud, and F.W. Alt. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 422:726–730. [DOI] [PubMed] [Google Scholar]
- 8.Dickerson, S.K., E. Market, E. Besmer, and F.N. Papavasiliou. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197:1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen-Mahrt, S.K., R.S. Harris, and M.S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 418:99–104. [DOI] [PubMed] [Google Scholar]
- 10.Martin, A., P.D. Bardwell, C.J. Woo, M. Fan, M.J. Shulman, and M.D. Scharff. 2002. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature. 415:802–806. [DOI] [PubMed] [Google Scholar]
- 11.Martin, A., and M.D. Scharff. 2002. AID and mismatch repair in antibody diversification. Nat. Rev. Immunol. 2:605–614. [DOI] [PubMed] [Google Scholar]
- 12.Ramiro, A.R., P. Stavropoulos, M. Jankovic, and M.C. Nussenzweig. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452–456. [DOI] [PubMed] [Google Scholar]
- 13.Yu, K., F. Chedin, C.L. Hsieh, T.E. Wilson, and M.R. Lieber. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4:442–451. [DOI] [PubMed] [Google Scholar]
- 14.Rada, C., G.T. Williams, H. Nilsen, D.E. Barnes, T. Lindahl, and M.S. Neuberger. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748–1755. [DOI] [PubMed] [Google Scholar]
- 15.Phung, Q.H., D.B. Winter, A. Cranston, R.E. Tarone, V.A. Bohr, R. Fishel, and P.J. Gearhart. 1998. Increased hypermutation at G and C nucleotides in immunoglobulin variable genes from mice deficient in the MSH2 mismatch repair protein. J. Exp. Med. 187:1745–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rada, C., M.R. Ehrenstein, M.S. Neuberger, and C. Milstein. 1998. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 9:135–141. [DOI] [PubMed] [Google Scholar]
- 17.Wiesendanger, M., B. Kneitz, W. Edelmann, and M.D. Scharff. 2000. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J. Exp. Med. 191:579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertocci, B., L. Quint, F. Delbos, C. Garcia, C.-A. Reynaud, and J.-C. Weill. 1998. Probing immunoglobulin gene hypermutation with microsatellites suggests a nonreplicative short patch DNA synthesis process. Immunity. 9:257–265. [DOI] [PubMed] [Google Scholar]
- 19.Vora, K.A., K.M. Tumas-Brundage, V.M. Lentz, A. Cranston, R. Fishel, and T. Manser. 1999. Severe attenuation of the B cell immune response in Msh2-deficient mice. J. Exp. Med. 189:471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frey, S., B. Bertocci, F. Delbos, L. Quint, J.C. Weill, and C.A. Reynaud. 1998. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity. 9:127–134. [DOI] [PubMed] [Google Scholar]
- 21.Harfe, B.D., and S. Jinks-Robertson. 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34:359–399. [DOI] [PubMed] [Google Scholar]
- 22.Tishkoff, D.X., A.L. Boerger, P. Bertrand, N. Filosi, G.M. Gaida, M.F. Kane, and R.D. Kolodner. 1997. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. USA. 94:7487–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinen, C.D., C. Schmutte, and R. Fishel. 2002. DNA repair and tumorigenesis: lessons from hereditary cancer syndromes. Cancer Biol. Ther. 1:477–485. [DOI] [PubMed] [Google Scholar]
- 24.Drotschmann, K., A.B. Clark, H.T. Tran, M.A. Resnick, D.A. Gordenin, and T.A. Kunkel. 1999. Mutator phenotypes of yeast strains heterozygous for mutations in the MSH2 gene. Proc. Natl. Acad. Sci. USA. 96:2970–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Studamire, B., T. Quach, and E. Alani. 1998. Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch repair. Mol. Cell. Biol. 18:7590–7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown, K.D., A. Rathi, R. Kamath, D.I. Beardsley, Q. Zhan, J.L. Mannino, and R. Baskaran. 2003. The mismatch repair system is required for S-phase checkpoint activation. Nat. Genet. 33:80–84. [DOI] [PubMed] [Google Scholar]
- 27.Bardwell, P.D., A. Martin, E. Wong, Z. Li, W. Edelmann, and M.D. Scharff. 2003. Cutting edge: the G-U mismatch glycosylase methyl-CpG binding domain 4 is dispensable for somatic hypermutation and class switch recombination. J. Immunol. 170:1620–1624. [DOI] [PubMed] [Google Scholar]
- 28.Schrader, C.E., J. Vardo, and J. Stavnezer. 2002. Role for mismatch repair proteins Msh2, Mlh1, and Pms2 in immunoglobulin class switching shown by sequence analysis of recombination junctions. J. Exp. Med. 195:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ehrenstein, M.R., and M.S. Neuberger. 1999. Deficiency in msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation. EMBO J. 18:3484–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi, Y., P.R. Dutta, D.M. Cerasoli, and G. Kelsoe. 1998. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. V. affinity maturation develops in two stages of clonal selection. J. Exp. Med. 187:885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schrader, C.E., W. Edelmann, R. Kucherlapati, and J. Stavnezer. 1999. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med. 190:323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehrenstein, M.R., C. Rada, A.M. Jones, C. Milstein, and M.S. Neuberger. 2001. Switch junction sequences in PMS2-deficient mice reveal a microhomology-mediated mechanism of Ig class switch recombination. Proc. Natl. Acad. Sci. USA. 98:14553–14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth, D.B., X.B. Chang, and J.H. Wilson. 1989. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol. Cell. Biol. 9:3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casellas, R., A. Nussenzweig, R. Wuerffel, R. Pelanda, A. Reichlin, H. Suh, X.F. Qin, E. Besmer, A. Kenter, K. Rajewsky, and M.C. Nussenzweig. 1998. Ku80 is required for immunoglobulin isotype switching. EMBO J. 17:2404–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manis, J.P., Y. Gu, R. Lansford, E. Sonoda, R. Ferrini, L. Davidson, K. Rajewsky, and F.W. Alt. 1998. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J. Exp. Med. 187:2081–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winter, D.B., Q.H. Phung, A. Umar, S.M. Baker, R.E. Tarone, K. Tanaka, R.M. Liskay, T.A. Kunkel, V.A. Bohr, and P.J. Gearhart. 1998. Altered spectra of hypermutation in antibodies from mice deficient for the DNA mismatch repair protein PMS2. Proc. Natl. Acad. Sci. USA. 95:6953–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, N., G. Bozek, J.C. Lo, and U. Storb. 1999. Different mismatch repair deficiencies all have the same effects on somatic hypermutation: intact primary mechanism accompanied by secondary modifications. J. Exp. Med. 190:21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phung, Q.H., D.B. Winter, R. Alrefai, and P.J. Gearhart. 1999. Hypermutation in Ig V genes from mice deficient in the MLH1 mismatch repair protein. J. Immunol. 162:3121–3124. [PubMed] [Google Scholar]
- 39.Gu, L., J. Wu, L. Qiu, C.D. Jennings, and G.M. Li. 2002. Involvement of DNA mismatch repair in folate deficiency-induced apoptosis. J. Nutr. Biochem. 13:355–363. [DOI] [PubMed] [Google Scholar]
- 40.Poltoratsky, V., M.F. Goodman, and M.D. Scharff. 2000. Error-prone candidates vie for somatic mutation. J. Exp. Med. 192:F27–F30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poltoratsky, V., C.J. Woo, B. Tippin, A. Martin, M.F. Goodman, and M.D. Scharff. 2001. Expression of error-prone polymerases in BL2 cells activated for Ig somatic hypermutation. Proc. Natl. Acad. Sci. USA. 98:7976–7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogozin, I.B., Y.I. Pavlov, K. Bebenek, T. Matsuda, and T.A. Kunkel. 2001. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat. Immunol. 2:530–536. [DOI] [PubMed] [Google Scholar]
- 43.Zeng, X., D.B. Winter, C. Kasmer, K.H. Kraemer, A.R. Lehmann, and P.J. Gearhart. 2001. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2:537–541. [DOI] [PubMed] [Google Scholar]
- 44.Zan, H., A. Komori, Z. Li, A. Cerutti, A. Schaffer, M.F. Flajnik, M. Diaz, and P. Casali. 2001. The translesion DNA polymerase zeta plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 14:643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diaz, M., L.K. Verkoczy, M.F. Flajnik, and N.R. Klinman. 2001. Decreased frequency of somatic hypermutation and impaired affinity maturation but intact germinal center formation in mice expressing antisense RNA to DNA polymerase zeta. J. Immunol. 167:327–335. [DOI] [PubMed] [Google Scholar]
- 46.Faili, A., S. Aoufouchi, E. Flatter, Q. Gueranger, C.A. Reynaud, and J.C. Weill. 2002. Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature. 419:944–947. [DOI] [PubMed] [Google Scholar]
- 47.Alabyev, B., and T. Manser. 2002. Bcl-2 rescues the germinal center response but does not alter the V gene somatic hypermutation spectrum in MSH2-deficient mice. J. Immunol. 169:3819–3824. [DOI] [PubMed] [Google Scholar]
- 48.Reynaud, C.A., B. Bertocci, S. Frey, F. Delbos, L. Quint, and J.C. Weill. 1999. Mismatch repair and immunoglobulin gene hypermutation: did we learn something? Immunol. Today. 20:522–527. [DOI] [PubMed] [Google Scholar]
- 49.Paques, F., and J.E. Haber. 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6765–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrader, C.E., J. Vardo, and J. Stavnezer. 2003. Mlh1 can function in antibody class switch recombination independently of Msh2. J. Exp. Med. 197: 1377–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]