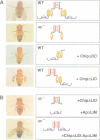
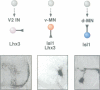
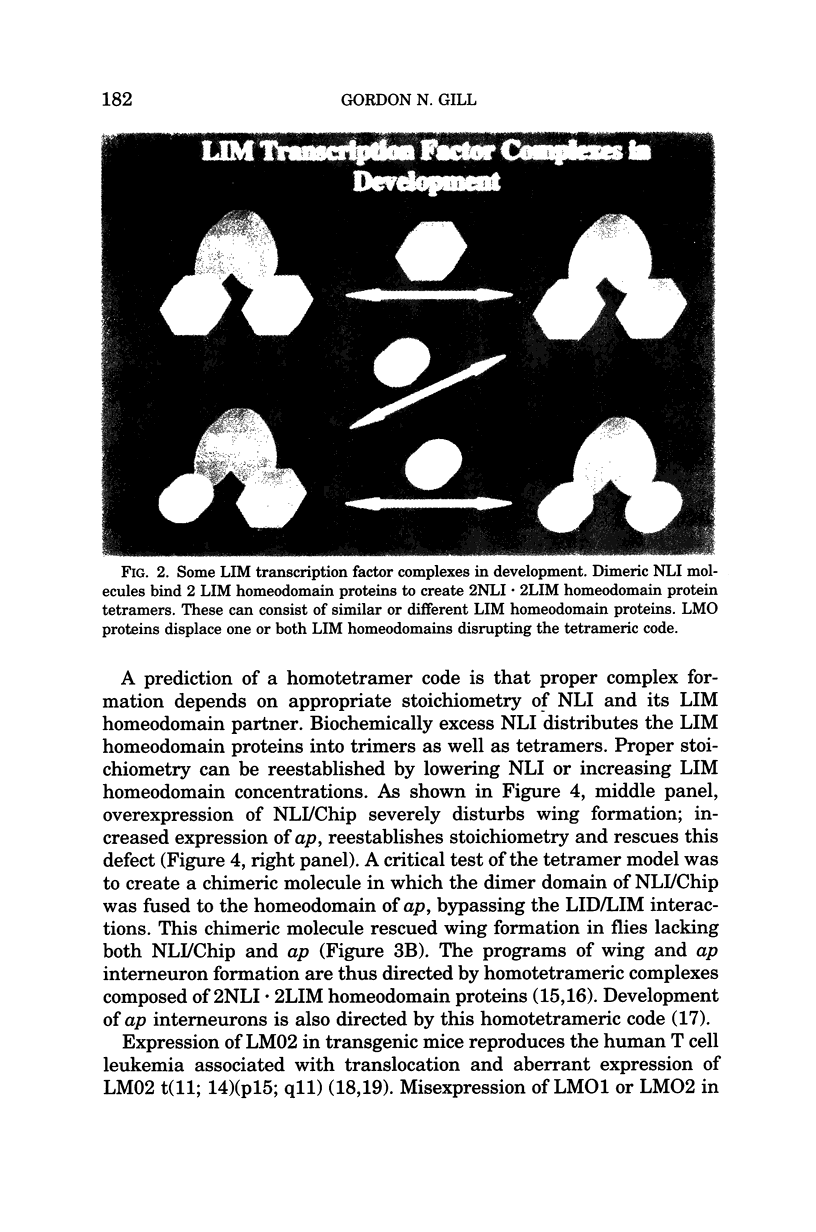
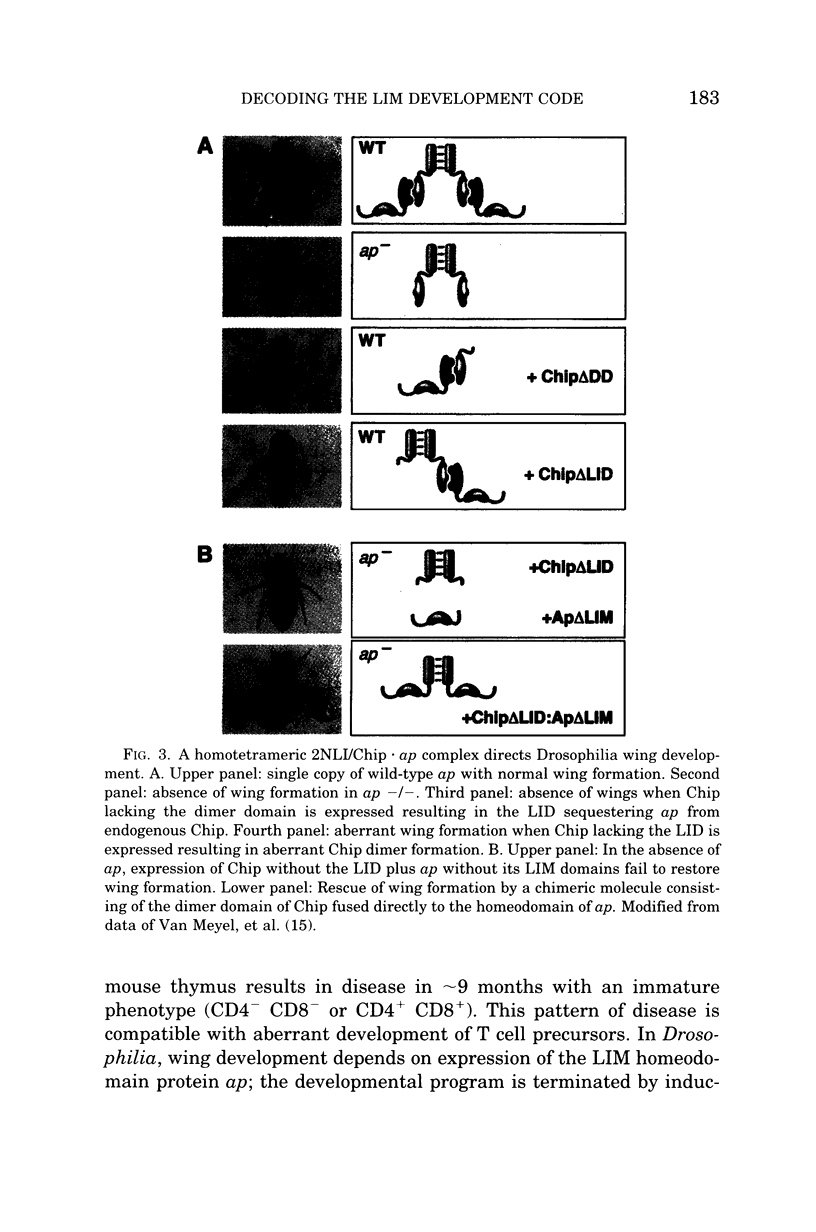
Abstract
During development a vast number of distinct cell types arise from dividing progenitor cells. Concentration gradients of ligands that act via cell surface receptors signal transcriptional regulators that repress and activate particular genes. LIM homeodomain proteins are an important class of transcriptional regulators that direct cell fate. Although in C. elegans only a single LIM homeodomain protein is expressed in a particular cell type, in vertebrates combinations of LIM homeodomain proteins are expressed in cells that determine cell fates. We have investigated the molecular basis of the LIM domain "code" that determines cell fates such as wing formation in Drosophilia and motor neuron formation in chicks. The basic code is a homotetramer of 2 LIM homeodomain proteins bridged by the adaptor protein, nuclear LIM interactor (NLI). A more complex molecular language consisting of a hexamer complex involving NLI and 2 LIM homeodomain proteins, Lhx3 and Isl1 determines ventral motor neuron formation. The same molecular "words" adopt different meanings depending on the context of expression of other molecular "words."
Full text
PDF


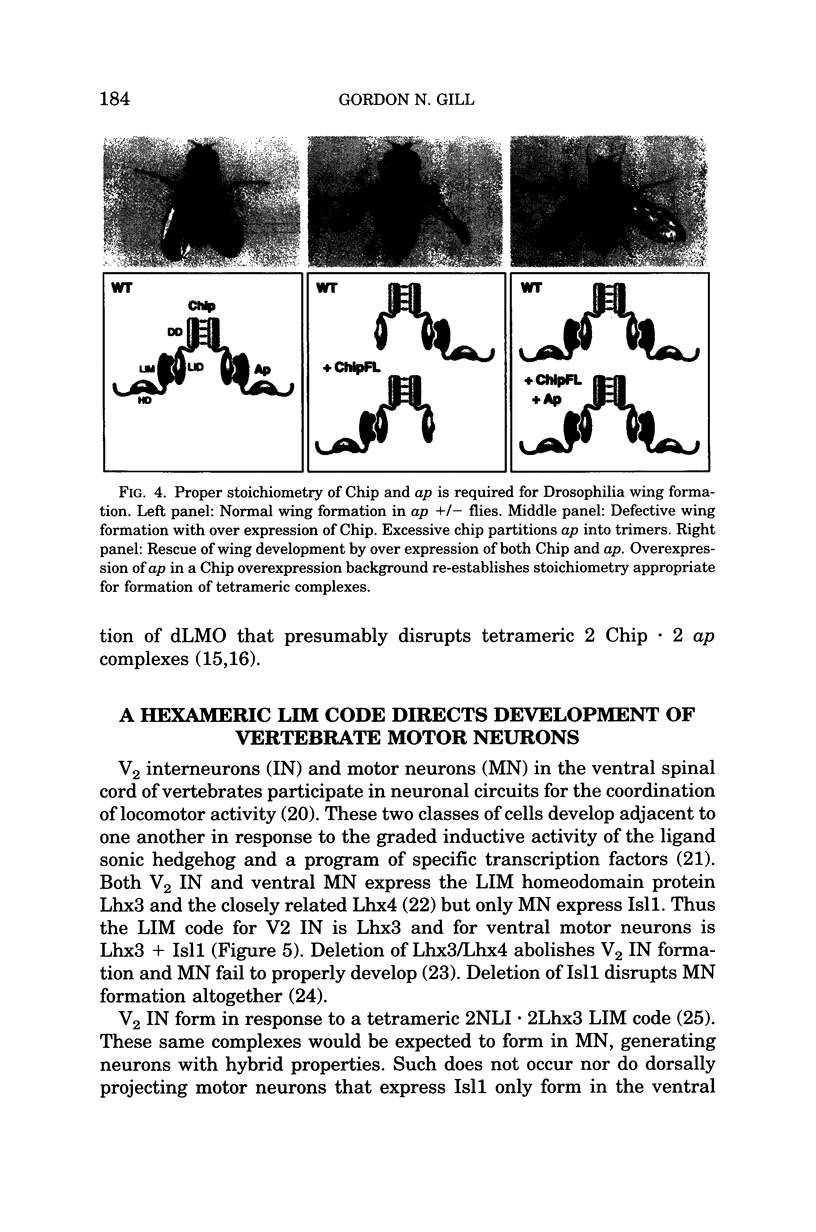
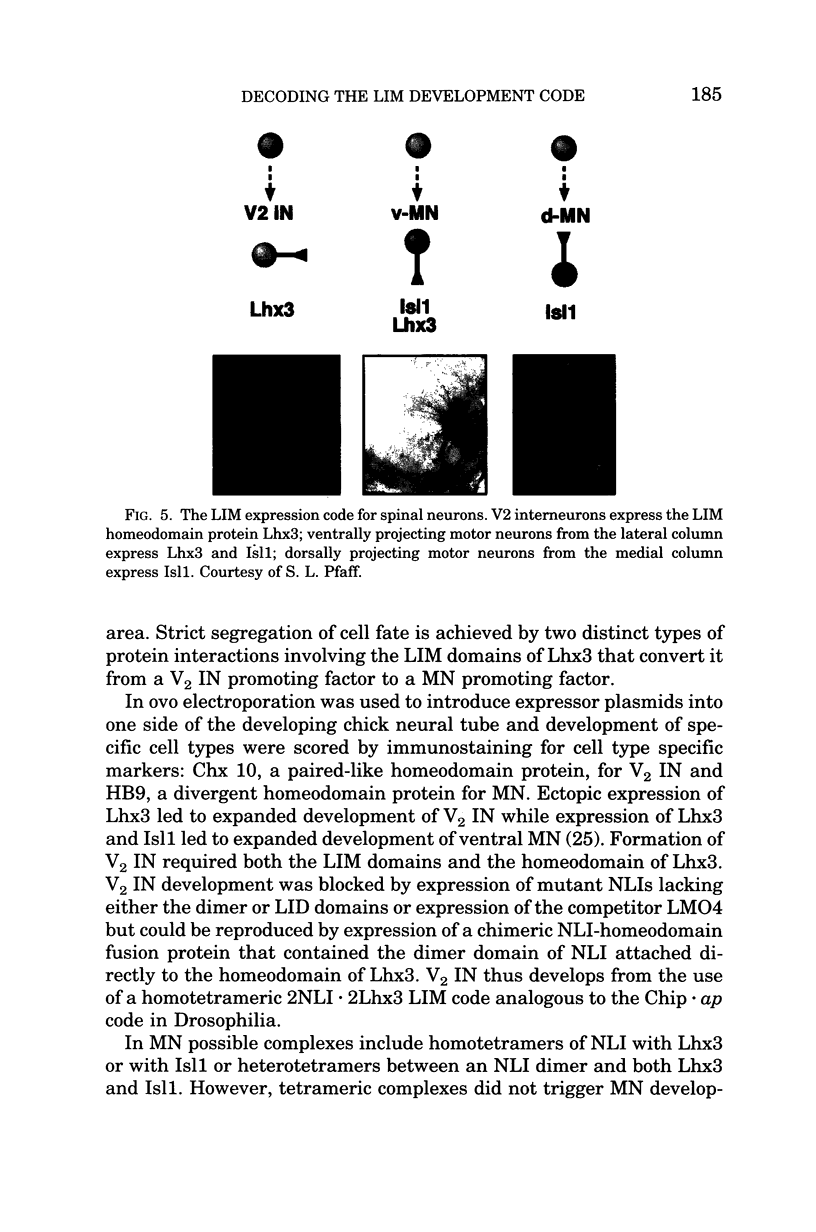




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agulnick A. D., Taira M., Breen J. J., Tanaka T., Dawid I. B., Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature. 1996 Nov 21;384(6606):270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- Bach I., Carrière C., Ostendorff H. P., Andersen B., Rosenfeld M. G. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 1997 Jun 1;11(11):1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- Boehm T., Foroni L., Kaneko Y., Perutz M. F., Rabbitts T. H. The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4367–4371. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B., McGuffin M. E., Pfeifle C., Segal D., Cohen S. M. apterous, a gene required for imaginal disc development in Drosophila encodes a member of the LIM family of developmental regulatory proteins. Genes Dev. 1992 May;6(5):715–729. doi: 10.1101/gad.6.5.715. [DOI] [PubMed] [Google Scholar]
- Edwards D. C., Sanders L. C., Bokoch G. M., Gill G. N. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol. 1999 Sep;1(5):253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- Fisch P., Boehm T., Lavenir I., Larson T., Arno J., Forster A., Rabbitts T. H. T-cell acute lymphoblastic lymphoma induced in transgenic mice by the RBTN1 and RBTN2 LIM-domain genes. Oncogene. 1992 Dec;7(12):2389–2397. [PubMed] [Google Scholar]
- Freyd G., Kim S. K., Horvitz H. R. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature. 1990 Apr 26;344(6269):876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- Isberg Ralph R., Barnes Penelope. Dancing with the host; flow-dependent bacterial adhesion. Cell. 2002 Jul 12;110(1):1–4. doi: 10.1016/s0092-8674(02)00821-8. [DOI] [PubMed] [Google Scholar]
- Jessell T. M. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000 Oct;1(1):20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Jurata L. W., Gill G. N. Functional analysis of the nuclear LIM domain interactor NLI. Mol Cell Biol. 1997 Oct;17(10):5688–5698. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata L. W., Kenny D. A., Gill G. N. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci U S A. 1996 Oct 15;93(21):11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata L. W., Pfaff S. L., Gill G. N. The nuclear LIM domain interactor NLI mediates homo- and heterodimerization of LIM domain transcription factors. J Biol Chem. 1998 Feb 6;273(6):3152–3157. doi: 10.1074/jbc.273.6.3152. [DOI] [PubMed] [Google Scholar]
- Lee S. K., Pfaff S. L. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001 Nov;4 (Suppl):1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- Lundgren S. E., Callahan C. A., Thor S., Thomas J. B. Control of neuronal pathway selection by the Drosophila LIM homeodomain gene apterous. Development. 1995 Jun;121(6):1769–1773. doi: 10.1242/dev.121.6.1769. [DOI] [PubMed] [Google Scholar]
- Marquardt T., Pfaff S. L. Cracking the transcriptional code for cell specification in the neural tube. Cell. 2001 Sep 21;106(6):651–654. doi: 10.1016/s0092-8674(01)00499-8. [DOI] [PubMed] [Google Scholar]
- McGuire E. A., Rintoul C. E., Sclar G. M., Korsmeyer S. J. Thymic overexpression of Ttg-1 in transgenic mice results in T-cell acute lymphoblastic leukemia/lymphoma. Mol Cell Biol. 1992 Sep;12(9):4186–4196. doi: 10.1128/mcb.12.9.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milán M., Cohen S. M. Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol Cell. 1999 Aug;4(2):267–273. doi: 10.1016/s1097-2765(00)80374-3. [DOI] [PubMed] [Google Scholar]
- Morcillo P., Rosen C., Baylies M. K., Dorsett D. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev. 1997 Oct 15;11(20):2729–2740. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostendorff Heather P., Peirano Reto I., Peters Marvin A., Schlüter Anne, Bossenz Michael, Scheffner Martin, Bach Ingolf. Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature. 2002 Mar 7;416(6876):99–103. doi: 10.1038/416099a. [DOI] [PubMed] [Google Scholar]
- Pfaff S. L., Mendelsohn M., Stewart C. L., Edlund T., Jessell T. M. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996 Jan 26;84(2):309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Pérez-Alvarado G. C., Miles C., Michelsen J. W., Louis H. A., Winge D. R., Beckerle M. C., Summers M. F. Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP. Nat Struct Biol. 1994 Jun;1(6):388–398. doi: 10.1038/nsb0694-388. [DOI] [PubMed] [Google Scholar]
- Sharma K., Sheng H. Z., Lettieri K., Li H., Karavanov A., Potter S., Westphal H., Pfaff S. L. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998 Dec 11;95(6):817–828. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- Tanabe Y., Jessell T. M. Diversity and pattern in the developing spinal cord. Science. 1996 Nov 15;274(5290):1115–1123. doi: 10.1126/science.274.5290.1115. [DOI] [PubMed] [Google Scholar]
- Thaler J., Harrison K., Sharma K., Lettieri K., Kehrl J., Pfaff S. L. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999 Aug;23(4):675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- Thaler J., Harrison K., Sharma K., Lettieri K., Kehrl J., Pfaff S. L. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999 Aug;23(4):675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- Wadman I., Li J., Bash R. O., Forster A., Osada H., Rabbitts T. H., Baer R. Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. EMBO J. 1994 Oct 17;13(20):4831–4839. doi: 10.1002/j.1460-2075.1994.tb06809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle Hynek, Lieberam Ivo, Porter Jeffery A., Jessell Thomas M. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002 Aug 9;110(3):385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- van Meyel D. J., O'Keefe D. D., Jurata L. W., Thor S., Gill G. N., Thomas J. B. Chip and apterous physically interact to form a functional complex during Drosophila development. Mol Cell. 1999 Aug;4(2):259–265. doi: 10.1016/s1097-2765(00)80373-1. [DOI] [PubMed] [Google Scholar]
- van Meyel D. J., O'Keefe D. D., Thor S., Jurata L. W., Gill G. N., Thomas J. B. Chip is an essential cofactor for apterous in the regulation of axon guidance in Drosophila. Development. 2000 May;127(9):1823–1831. doi: 10.1242/dev.127.9.1823. [DOI] [PubMed] [Google Scholar]