Abstract
MRI at high magnetic fields (> 3.0 T ) is complicated by strong inhomogeneous radio-frequency fields, sometimes termed the “bias field”. These lead to nonuniformity of image intensity, greatly complicating further analysis such as registration and segmentation. Existing methods for bias field correction are effective for 1.5 T or 3.0 T MRI, but are not completely satisfactory for higher field data. This paper develops an effective bias field correction for high field MRI based on the assumption that the nonuniformity is smoothly varying in space. Also, nonuniformity is quantified and unmixed using high order neighborhood statistics of intensity cooccurrences. They are computed within spherical windows of limited size over the entire image. The restoration is iterative and makes use of a novel stable stopping criterion that depends on the scaled entropy of the cooccurrence statistics, which is a non monotonic function of the iterations; the Shannon entropy of the cooccurrence statistics normalized to the effective dynamic range of the image. The algorithm restores whole head data, is robust to intense nonuniformities present in high field acquisitions, and is robust to variations in anatomy. This algorithm significantly improves bias field correction in comparison to N3 on phantom 1.5 T head data and high field 4 T human head data.
Keywords: Restoration of MRI data, bias field compensation of MRI data, MRI brain data, image cooccurrence statistics, high order image statistics, deconvolution
1. INTRODUCTION
The accurate acquisition of MRI data requires a homogeneous radio-frequency field, which is not possible to achieve. The inhomogeneity, sometimes termed the “bias field”, is more pronounced at MRI of high fields (> 3.0 T ), where the radio-frequency wavelength gets shorter, approaching the dimension of the human head or body. The local dielectric properties of these regions lead to nonuniformity of image intensity, greatly complicating further analysis such as registration and segmentation. Existing methods for bias field correction have been reasonably effective for 1.5 T or 3.0 T MRI, but are not completely satisfactory for higher field data. This paper develops an effective bias field correction for high field MRI especially for quantitative brain imaging at 4 T .
There have been several attempts to correct for nonuniformity during acquisition based on its physical properties. The nonuniformity of the transmission coil has been estimated from its frequency response to parameterized acquisition sequences.1,2 In addition, the nonuniformity of transmission and receiver coil(s) combined has been approximated using phantoms.3 However, the physical correction methods are valid only for a particular MRI sequence, but do not account for the complicated interaction between the radio-frequency fields and the human body. It is not obvious how the combined nonuniformities can be accounted for during acquisition, especially for higher magnetic fields. Thus, existing methods for correction of the nonuniformities during acquisition are incomplete, time consuming, and clinically impractical.
As an alternative to physical corrections, several post-acquisition restoration methods have been proposed to account for the effect of nonuniformities that do not require additional acquisitions and are applicable to a range of MRI sequences. They make regularity assumptions about the field nonuniformity as well as anatomy and treat the nonuniformities in the same way irrespective of their sources. Typically, they operate on the logarithm of image intensities of the data4–6 and assume that the bias field can be approximated by basis functions such as Gaussians, splines, polynomials, or sinusoids. The simplest approach to address this problem has been homomorphic filtering.7,8 This is based on the assumption that the bias field corresponds to spatial frequencies lower than those of the anatomy. Thus, smoothing an image in the spatial or frequency domain gives the nonuniformity. Smoothing has also been applied to statistics of local histograms computed over image tiles.7,9 The basic assumption of homomorphic filtering, that the spatial frequencies of the bias field are lower than those of the anatomy can be problematic for high field MR images. In such images the nonuniformity can extend from the low frequency part of their spectrum until the intermediate frequency range.
Another class of approaches derives bias field estimations by registering the data to a tissue template or atlas of expected intensity distribution and by computing either directly10 the difference between actual and expected values or using the templates as a prior for subsequent processing.5,11 Bias field estimation has also been combined with tissue classification. Many of these approaches require initialization with manual presegmentation12–15 while others are fully automatic. Some of these approaches estimate tissue classification based on expectation maximization16 over the histogram,13–15,17 and fuzzy c-means classification.18,19 Intensity tissue classification has also been combined with spatial Markov random fields.4–6,13–15 Estimating the bias field with registration and segmentation depends extensively on the prior assumptions. The accuracy of registration is affected when the image has an intense bias field such as that present in > 3 T images. This poses a problem when registration is used to extract the brain region from a head image. Also the priors of an algorithm may be unable to represent the intensity variations of anatomy or pathology in the data. Moreover, there may not be adequate contrast between the assumed tissue classes.
Several non-parametric approaches have been suggested that are more robust to inter and intra subject variability as well as pathology. One such class of non-parametric techniques is based on the retinex algorithm.20–22 This algorithm uses the logarithm of the intensities and normalizes them to the geometric mean of the intensities within a neighborhood. Thus it enhances the components of the nonuniformity whose wavelength is close to the size of the neighborhood considered and hence introduces overshooting contours at the borders between different tissue regions.21 This problem can be alleviated but not resolved by using the multiresolution of an image in terms of a Gaussian or a more general wavelet representation.22,23 Another class of non-parametric approaches assume that the image is a piecewise union of extensive regions of constant intensity. These methods compute image derivatives such as the gradient or Laplacian and threshold their low values assumed to be due to nonuniformities exclusively. The remaining derivatives are incorporated into a regularized cost functional whose minimization estimates the nonuniformity.24,25 The assumption of extensive homogeneous regions may not hold as in the brain cortex in head imaging. Both retinex and variational methods operate locally and may not restore global image statistics. They also assume a distinction between a low frequency nonuniformity and a high frequency anatomy. More generally, corrections that use differential image properties may lead to different restorations for different image regions.26
Another class of non-parametric techniques uses the global image intensity histogram.27,28 These methods can capture global image properties. They assume that the bias field widens the intensity distributions of the various tissues. Thus, the inverse is assumed to provide a deconvolution for sharpening the histogram and restoring image uniformity. This is achieved by estimating a smooth estimate of the nonuniformity. A commonly used technique, N3,28 was found to have a performance superior to those based on homomorphic filtering.29 The ability of the intensity histogram to discriminate between different distributions affected by high fields is limited due to the corruption of the distributions caused by the spatial nonuniformity. A common sharpness optimality criterion for histogram based methods is minimum entropy, which corresponds to single peak histograms resulting from a flat image.27,30,31 Thus, the entropy criterion can be problematic in terminating an iterative numerical algorithm.
To address the problem of uniformity restoration we use a nonparametric approach. In this study that is based on high order intensity coincidence or cooccurrence statistics. Since it is a nonparametric approach it is robust to subject anatomy, pathology, and intensity variations within a tissue. The cooccurrence statistics represent the intensities of different tissues as well as the joint intensities of adjacent tissues. The high order of coincidences favor dominant distributions and selectively decrease the contribution of noise to their variance. The unmixing of the bias field from these statistics is done by considering its physical effect on the original intensities, rather than their logarithm. The restoration is iterative with a robust stopping criterion based on the entropy of the distribution normalized with respect to the dynamic range, namely, the scaled entropy. Our algorithm was able to perform bias field restoration for human head data acquired under high field with an emphasis on the brain region that gives rise to the dominant distributions of the white matter and of the region around the interface between white matter and gray matter. It was found to have a better performance than N3 over the same data.28
2. INTENSITY NONUNIFORMITY MODEL
We assume that the measured image I and intrinsic anatomic image IA are related according to:
| (1) |
where matrix B contains the unknown nonuniform field and the noise N is additive. The statistics of IA are assumed to consist of distributions that can be discriminated.
The Taylor series expansion of the bias field around a voxel x0 gives:
| (2) |
Approximation up to the first order term leads to a bias field which is locally linear within a sphere of radius ρ = x − x0 around x0. Thus, quadratic and higher order terms are assumed to be negligible within distance ρ from x0. The nonuniformity is recovered within a scale factor which does not affect the discriminability between tissues. We also assume that the noise N is stationary, Gaussian, and white.32 In addition to the spatially smooth variations the nonuniformity also has abrupt variations at the borders between different tissues. However, we assume that these can be absorbed into the tissue statistics.
3. METHODS
The objective of this work is to decompose the product in equation (1) to estimate IA and a smooth B. We unmix the effect that nonuniformity B has on the statistics of I. This provides a rough estimate of the nonuniformity. We also assume that the nonuniformity B is spatially smooth, which is imposed directly by Gaussian smoothing. Thus, we do not consider noise in equation (1), which has high spatial frequencies. The restoration is iterative with a stable stopping criterion. An overview of the algorithm is sketched in figure 3.
Figure 3.
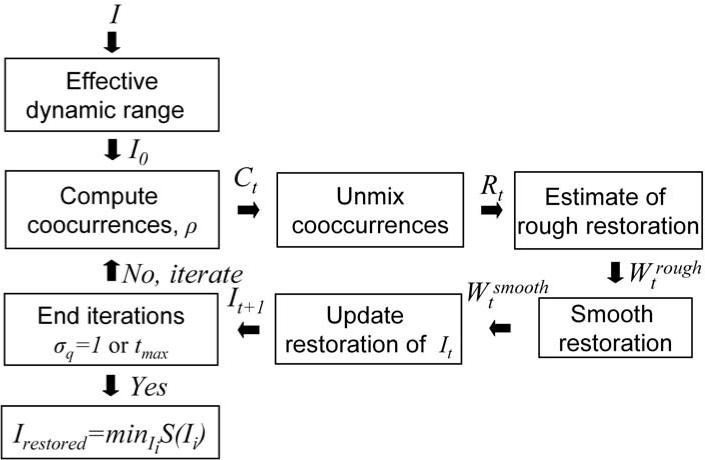
A block summary of the algorithm.
3.1. Determining the Valid Dynamic Range of the Image
The valid dynamic range of the MR contrast mechanism is detected and very bright artifacts due to instrumental imperfections or blood flow are removed. Such artifacts do not follow the nonuniformity assumption of equation (1). To this end we compute the histogram h of the original image I and the cumulative histogram . The latter is normalized to unit L1 norm . We compute the intensity value u0.9 that corresponds to the upper 0.9 percentile of . The intensity range up to 1.5 × u0.9 is preserved, whereas the intensity range beyond that value is linearly compressed to the range (1.5 × u0.9, 3.0 × u0.9] with maximum intensity umax = 3.0 × u0.9 to provide an initial estimate of image I0. The same intensity range [0, umax] is maintained during the iterations t. The image is a map to this intensity range from its domain D, It : D → [0, umax].
3.2. High Order Intensity Cooccurrence Statistics and their Properties
The statistics used in this work are the nth-order cooccurrences of pairs of intensities within a spherical neighborhood in the image It with radius ρ, assuming that the field in windows of this size is linear. The count of intensities in range u ∈ [0, umax] in a sphere of radius ρ around voxel x0 is given by:
| (3) |
In an nth-order cooccurrence of intensity range u1 and intensity range u2 the center of the spherical neighborhood has intensity within u1 and the remaining window contains at least n − 1 voxels of the same intensity and at least n voxels of intensity u2. Additional voxels of any of the two intensities within the spherical window ρ give the cooccurrence:
| (4) |
where ct(x0, u1, u2) is the cooccurrence at x0 for iteration t. The image cooccurrences are given by integrating the voxel cooccurrences over the entire image domain. The cooccurrence statistics are computed with a two dimensional Gaussian Parzen window G(σc) to give a 2D matrix:
| (5) |
This is the joint intensity distribution of nth-order of an image with itself computed within spheres of radius ρ. The resulting matrix Ct is not necessarily symmetric. The cooccurrences of a region with a contiguous tissue lie close to the diagonal of matrix Ct. The joint cooccurrences of tissues around an interface of different tissues lie farther from the diagonal. To examine the properties of the cooccurrence matrix we assume that the distributions of IA in the cooccurrence statistics CA are Gaussian. The noise variance of the dominant distributions in the cooccurrence matrix is decreased with increasing cooccurrence order. Thus, the discriminability between the dominant distributions in CA is increased.
The effect of the presence of additive zero mean Gaussian noise N in an image is an increase of the variance of the distributions in Ct. The effect of image intensity nonuniformity on an intensity cooccurrence between u1 and u2 where u1 is the intensity of the central voxel at x1 in a hypothetical anatomic image IA is to scale and rotate it about the origin of Ct. The zero order term Bt(x1) in equation (2) scales cooccurrences around the origin and the first order term ∇Bt(x1) in equation (2) rotates them around the origin. Thus, the effect of the nonuniformity on Ct is easier to examine in polar coordinates (r, ϕ), where and tan ϕ = u2/u1. We assume that the variation of the cooccurrence statistics of the nonuniformity Bt in the cooccurrence statistics CBt along the radial axis, r, is given by a Gaussian distribution for Δr, G(σr). It follows from33 that the effect of the nonuniformity on σr is given by:
| (6) |
The rotation angle Δϕ of the nonuniformity in its cooccurrence statistics CBt is given by:
| (7) |
where α = ∇B. This relation can be derived from figure 1 (a) and is plotted in figure 1 (b). We also assume that the variation of a distribution in the azimuthal axis Δϕ gives rise to a unimodal Gaussian distribution G(σϕ), where σϕ corresponds to a certain value of α.
Figure 1.
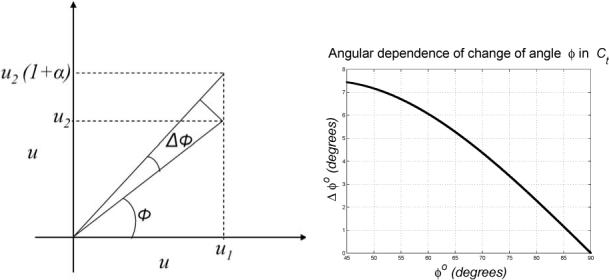
The effect of nonuniformity on the azimuthal size of the deconvolution filter. (a) An intensity pair (u1, u2) as a result of the nonuniformity becomes (u1, u2(1 + α)), α = ∇B. The angle ϕ of (u1, u2) with the horizontal axis becomes ϕ + Δϕ. (b) The effect of a constant nonuniformity α = 0.3 on the rotational angle Δϕ as a function of the angle ϕ. The effect on cooccurrences close to the diagonal is the highest.
3.3. Unmixing the effect of the Nonuniformity from the Cooccurrence Statistics
To decompose the product in equation (1) and estimate It and a smooth Bt we use the cooccurrence statistics to unmix the effect that the nonuniformity Bt has on Ct,where Bt is the estimate of B at iteration t. To unmix the effect of the nonuniformity we compute the inverse of the Gaussian nonuniformity distributions for Δr and Δϕ. The inverses of these distributions give the restoration filters of the cooccurrence matrix. The radial restoration filter is and the angular restoration filter is , where ∊ is very small nonzero constant. In equation (6) the radial distortion is proportional to r. Thus, the size of the radial deconvolution filter has the same linear dependence, σr ∝ r. The size of the radial restoration filter in the algorithm is its standard deviation normalized with respect to the dynamic range to give its normalized standard deviation:
| (8) |
The azimuthal restoration filter is represented in terms of angle in radians and hence it is inherently normalized to the radius r in Ct, which is the dynamic range. The standard deviation of the angular restoration filter, σϕ, can be determined from equation (7). The 2D restoration filter fq = fr * fϕ is applied to the cooccurrence matrix Ct. This will map every cooccurrence (u1, u2) to the expected one to give the expected restored cooccurrence matrix . The restoration of the cooccurrence matrix gives a restoration matrix Rt with a gain factor for each intensity cooccurrence. The gain is the ratio of the new position of the intensity of the central pixel over the initial position u1 of the same intensity. Thus, the restoration factor or gain is .
3.4. Estimation of the Spatial Nonuniformity
The restoration matrix Rt is backprojected to the image. We consider a sphere of radius ρ around a voxel x1. The size of this neighborhood is the same as the size of the neighborhood that was used to compute the cooccurrence statistics Ct. This sphere gives intensity cooccurrences with the intensity of voxel x1. These intensity cooccurrences index the restoration matrix Rt to get an equal number of gain factors. Their expected value gives a rough estimate of the restoration image at x1:
| (9) |
This is the inverse of the rough estimate of the nonuniformity . The nonuniformity field is assumed to be smooth. Thus, the restoration image is filtered with a spatial Gaussian filter G(σw) to give .
In practice the Gaussian distributions of the nonuniformity statistics can only be approximately inverted to give the deconvolution filters. As a result they contain a low frequency component that tends to contract the dynamic range of the image with iterations t. It is necessary to normalize the effective dynamic range since the standard deviation of the radial deconvolution filter is a function of umax. The normalization is done with the cumulative histogram of the image It by rescaling the intensity range to ensure that the upper 0.9 percentile, u0.9,t, of the cumulative histogram remains constant with iterations t. That is:
| (10) |
The restoration from equation (10) is applied to the restored image pixelwise to improve its estimate.
3.5. Condition for End of Iterations
The Shannon entropy of the cooccurrence statistics of an image monotonically increases as it is corrupted with a smooth nonuniformity from which it follows that the Shannon entropy of the cooccurrence statistics of an image is larger than the one of the assumed intrinsic anatomic image, S(C0) > S(CA). However, the Shannon entropy of the cooccurrence statistics of an image with the normalized effective dynamic range, namely, the scaled entropy of the cooccurrence matrix is a non-monotonic function of the iterations t. The scaled entropy is used as a stopping condition of the iterations. When the scaled entropy increases S(Ct+1) > S(Ct) the size of the deconvolution filter is halved σq,t+1 = σq,t/2. The iterations end when the size of the deconvolution filter becomes unity, σq,tend = 1. The restored image is the one whose cooccurrence matrix has minimum scaled entropy tq = mint∈[0,tmax] S(Ct), where tmax is the maximum allowed number of iterations.
3.6. Implementation Issues of the Algorithm
The computational cost of the algorithm can be accelerated with minimal loss of accuracy. The deconvolution of Ct to get Rt can be accelerated by considering that the deconvolution filter fq is separable. The size of the deconvolution filter is bounded in the implementation. This improves the robustness of the deconvolution with respect to noise and flow artifacts that can produce very high intensities. A bound on the size also avoids an unnecessary increase of the computational cost. Its maximum size corresponds to radial distance in Ct equal to u0.9,0 of the histogram of the original image I0. That is, the maximum radial size of the filter is . In effect determines the radial size of the deconvolution filter. We also set a maximum value for the size of the angular deconvolution filter σϕ,max for similar reasons. It is obtained for u0.9,0 as well as ϕ = 45° in Ct and a certain value of α in equation (7). The small value of the spatial window of radius ρ for the computation of the cooccurrence statistics makes the significance of the radial standard deviation σr greater than that of the angular standard deviation σϕ.
The computation of the cooccurrence matrix Ct with the discretized version of equation (5) can be accelerated by spatially subsampling the voxels in a sphere of radius ρ with a factor of Δρ. The same subsampling Δρ can be used to compute the rough nonuniformity correction in the discretized version of equation (9). The Gaussian filter applied to is separable. The algorithm can be further accelerated by using a multiplicative factor for the smooth nonuniformity correction prior to applying it to the image. The computations can be limited by bounding the maximum allowed number of iterations tmax. The speed of the algorithm can also be increased without loss of accuracy by taking advantage of the fact that all the steps of the algorithm are fully parallelizable.
4. EVALUATION OF THE ALGORITHM
Our algorithm has been implemented in the C++ programming language. The algorithm was applied to whole head imaging data. A preprocessing step was used to remove the low signal regions with background or noise. To this end a Rayleigh distribution was fit to the diagonal self-cooccurrence statistics. The nonuniformity was computed over the foreground region and subsequently extrapolated over the low signal regions. The cooccurrence algorithm has been evaluated on phantom as well as real data. It was compared with N3,28 a commonly used algorithm for bias field correction.29 The parameters of N3 are the FWHM of the deconvolution kernel and the distance between the knots of the spline bases. The values used for these parameters are listed in table 1 and were combined with the least spatial decimation factor of two. The algorithm was evaluated for phantom images with the noise free and bias free MNI head phantom. The algorithm was also evaluated with human head images using the sharpness of the histograms of the restored images that was measured with the Shannon entropy. The nonuniformity was initialized to unity everywhere B0(x) = 1, ∀x. The standard deviation of the Parzen window in equation (5) was σc = 1.5. The cooccurrence statistics in equation (4) were computed in a sphere of ρ = 9 mm with a subsampling factor of Δρ = 3 mm and the order of the statistics in the same equation was n = 3. The size of the deconvolution kernel was and α = 0.3 in equation (7). The maximum number of iterations was set to tmax = 36.
Table 1.
Parameters used for N3 to correct phantom and human brain imaging data.
| Suggested phantom28 | Tested for brain 4T | Used for brain 4T | |
|---|---|---|---|
| FWHM of deconvolution | 0.15 | 0.06,0.13,0,20 | 0.20 |
| Distance of knots in space (mm) | 200 | 20,110,200 | 20 |
| Decimation in space | 2−4 | 2 | 2 |
4.1. Phantom Data
We used the 1.5 T phantom brain and the phantom nonuniformities provided by MNI.34,35 The spatial smoothing of the nonuniformity in the cooccurrence algorithm was σw = 77 mm. N3 was run with the parameters suggested for this dataset28 given in the first column of table 1. We also computed the error maps for that data compared to the uncorrupted original phantom. The original as well as the restored images were normalized to zero mean and unit L1 norm. They were subsequently subtracted to give the error image. The L1 norm of the error image was used as a measure of the error.
The error value was lower in all cases with our algorithm compared to N3. This is summarized in table 2. The lower error demonstrates the improved performance of the algorithm as well as the improved stability of its stopping criterion. This is an indication that subsequent applications of the algorithm to an image will have a limited or no effect. Two example corrections with the highest level of simulated nonuniformity used, 40%, and noise levels of 3% and 5% are shown in figures 4(a) and (b), respectively. In each example the first row shows the original corrupted phantom with bias field and noise, the second row shows the restoration with N3, and the last row shows the restoration with the cooccurrence algorithm. The images are shown with the dynamic range windowed to that of the white matter. The N3 restoration contains a considerable residual of the bias. The white matter intensity in the image restored with the cooccurrence algorithm is more uniform. The average time duration of the cooccurrence algorithm is 37 min whereas that of N3 is 31 min.
Table 2.
The error is the absolute value of the difference between a corrupted phantom image and the noise free and bias free MNI phantom. A variety of synthetic bias fields B and noise levels N were used. In all cases the error of the cooccurrence algorithm is lower. The greater difference is that for the bias and noise free image.
| N = 0% | N = 3% | N = 5% | N = 5% | N = 5% | |
|---|---|---|---|---|---|
| B = 0% | B = 40% | B = 0% | B = 20% | B = 40% | |
| N3 (10−2) | 1.40 | 8.17 | 12.76 | 12.56 | 12.77 |
| Cooccurrence (10−2) | 0 (−100%) | 7.50 (−8.21%) | 12.43 (−2.54%) | 12.20 (−2.92%) | 12.27 (−3.92%) |
Figure 4.
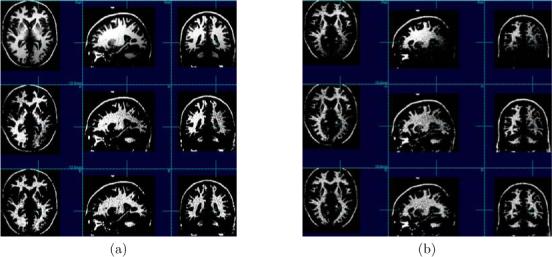
Restorations of two phantom images with nonuniformity of 40% and noise levels 3% and 5% in (a) and (b), respectively. In the first row is the original corrupted phantom, in the second row is the restoration with N3, and in the third row is the restoration with the cooccurrence algorithm. The N3 restoration contains a considerable residual of the nonuniformity. The improved performance of the cooccurrence algorithm is demonstrated by the higher uniformity of the white matter intensity in the image.
4.2. Human Head Data
Human brain MRI data were obtained at 4 T using a T 1-weighted magnetization prepared rapid gradient echo (MPRAGE) sequence and an 8-channel array head coil. The size of the images is 256 × 256 × 176 voxels with uniform voxel resolution of 1.0 mm × 1.0 mm × 1.0 mm. The parameters of the sequence are TR/TE = 2300/3.37 ms, TI = 950 ms, and a flip angle of 7°. The algorithm was applied to geriatric head data images, which were selected randomly from a large database. The 8-channel receiver coil had larger field nonuniformity than a conventional birdcage coil but provided better signal to noise, especially at the outer brain regions close to the coils. The spatial smoothing of the nonuniformity in the cooccurrence algorithm was σw = 25 mm. N3 was run for a variety of parameters in the ranges recommended by the authors and given in the third column of table 1. The objective of N3 is to sharpen the histogram distributions. We observed the histograms and selected the parameters that gave the image that had the histogram with the most compact distributions. We also verified our selection by measuring the sharpness with the decrease in the Shannon entropy of a histogram and selected the parameter set that minimized it. The best performing parameters were FWHM = 0.2 mm and distance of 20 mm between the knots, in the fourth column of table 1. The low distance between the knots is consistent with the fact that the 4 T magnetic field gives rise to nonuniformities with a small wavelength. The minimal entropy values are in table 3. In all test cases the entropy of the histogram of the image restored with our algorithm was lower than that of the image restored with N3 indicating that the new algorithm provides an improvement over N3.
Table 3.
In all cases the entropy of the image processed with the cooccurrence algorithm is lower than that of the entropy of the histogram of the image corrected with N3. This demonstrates the higher performance of our algorithm.
| Image | N3 | Cooccurrence |
|---|---|---|
| 1 | 4.85 | 4.63 |
| 2 | 4.68 | 4.50 |
| 3 | 4.46 | 4.30 |
| 4 | 4.84 | 4.60 |
| 5 | 4.80 | 4.53 |
| 6 | 4.62 | 4.45 |
| 7 | 4.85 | 4.53 |
| 8 | 5.44 | 5.33 |
| 9 | 4.92 | 4.90 |
| 10 | 5.15 | 4.76 |
Representative corrections of four sets of data are shown in figure 5. In each example the first row shows the acquired image, the second row shows the restoration with N3, and in the last row is the restoration with the cooccurrence algorithm. The images are shown with the dynamic range windowed to that of the white matter. The N3 restoration contains a considerable residual of the nonuniformity and sometimes even accentuates it. Also N3 decreases image contrast by darkening the bright white matter and introducing overshooting contours at the borders between different regions such as between gray matter and white matter at the cortex. The white matter intensity in the image restored with the cooccurrence algorithm is more uniform. It took on average 1 : 23 hrs with the new algorithm to correct the volumetric MPRAGE datasets with 256 × 256 × 176 dimensions given in table 1. The duration of the restoration depends on the amount of the nonuniformity in an image. It took on average 2 : 21 hrs with N3. The cooccurrence algorithm is significantly more efficient.
Figure 5.
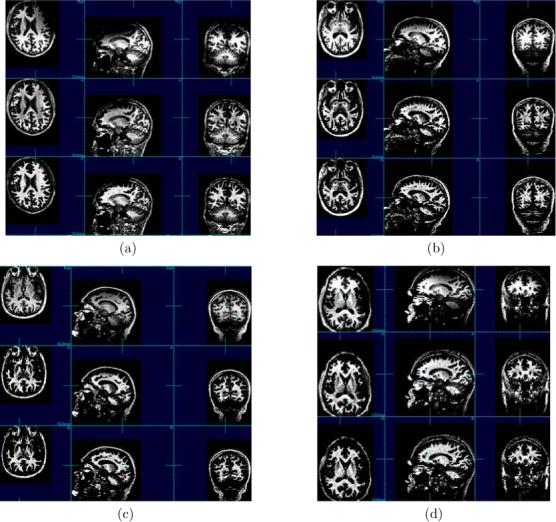
Examples of restorations of four human head images. In each example the first row shows the acquired image, the second row shows the restoration with N3, and in the third row is the restoration with the cooccurrence algorithm. The N3 restoration contains a considerable residual of the nonuniformity and sometimes even accentuates it. Also, it darkens the white matter and introduces overshooting contours at the borders between different regions such as between gray matter and white matter at the cortex. The improved performance of the cooccurrence algorithm is demonstrated by the higher uniformity of the white matter intensity in these images.
5. DISCUSSION AND CONCLUSION
Most intensity uniformity restoration algorithms operate on the logarithm of the image intensities. Even though this intensity transformation can make the problem additive it warps the dynamic range in a non physical way. As a result it can increase noise and decrease the contrast to noise ratio. In this work we operate on the original dynamic range with a physically motivated deconvolution. It is assumed that the corruption of the cooccurrence statistics is caused by a nonuniformity which has a unimodal Gaussian distribution in its cooccurrence statistics. This provides the deconvolution filter fq. The assumption for the statistics of the nonuniformity corruption may not hold, particularly for high field data. However, the deconvolution is robust with respect to the shape of the deconvolution filter and is mainly dependent on its overall variance. The deconvolution based on cooccurrence statistics takes advantage of the fact that the nonuniformity B is smooth across the borders of different tissue regions.
One of the main assumptions of the deconvolution process is that in the intrinsic anatomic image IA the variation of intensities within a tissue class is low compared to the distance between the means of the distributions of different tissue classes. The deconvolution is problematic if this discriminability assumption between the distributions in IA is low or does not exist. In these cases the deconvolution introduces an error that tends to decrease the contrast to noise ratio. If the spatial frequencies of the error are higher than those of the anatomy, the smoothing step can remove them. Otherwise, the restoration will have a lower contrast. In effect the algorithm is unable to unmix nonuniformity between two regions with very similar statistics that are spatially close. The smoothing of is done with a Gaussian which has a local effect. Smoothing with more global bases that are polynomials or splines may increase the nonuniformity in . In conclusion, we developed a new non-parametric algorithm based on neighborhood intensity cooccurrences for image restoration from nonuniform field distortion. The new algorithm performed significantly better than the well-known N3 approach on high field, 4 T , phantom, and human brain MRI data.
Figure 2.
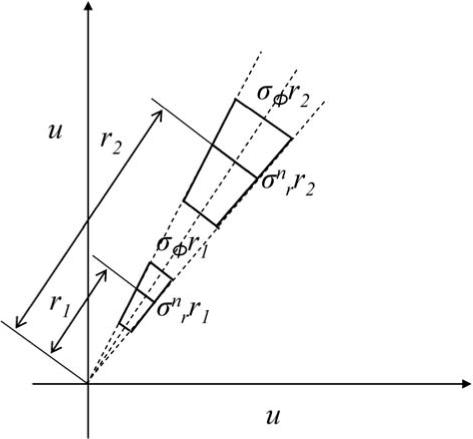
The radial and azimuthal size of the deconvolution filter change with its position on the cooccurrence matrix.
REFERENCES
- 1.Stollberger R, Wach P. “Imaging of the active B1 field in vivo,”. Magnetic Resonance in Medicine. 1996;35:246–251. doi: 10.1002/mrm.1910350217. [DOI] [PubMed] [Google Scholar]
- 2.Vita E, Thomas D, Deichmann R, Turner R, Oldidge R. “Fast B1 mapping with EPI,”. Proc. of ISMRM. 2004;11 [Google Scholar]
- 3.Axel L, Costantini J, Listerud J. “Intensity correction in surface-coil MR imaging,”. Am.J.Roentgenology. 1987;148:418–420. doi: 10.2214/ajr.148.2.418. [DOI] [PubMed] [Google Scholar]
- 4.Guillemaud R, Brady M. “Estimating the bias field of MR images,”. IEEE Transactions on Medical images. 1997;16:238–251. doi: 10.1109/42.585758. [DOI] [PubMed] [Google Scholar]
- 5.Leemput K, Maes F, Vandermeulen D, Suetens P. “Automated model based bias field correction of MR images of the brain,”. IEEE Transactions on Medical Imaging. 1999;18(10):885–896. doi: 10.1109/42.811268. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Brady M, Smith S. “Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm,”. IEEE Transactions on Medical Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 7.Brinkmann B, Manduca A, Robb R. “Optimized homomorphic unsharp masking for MR grayscale intensity correction,”. IEEE Transactions on Medical Imaging. 1998;17(2):161–171. doi: 10.1109/42.700729. [DOI] [PubMed] [Google Scholar]
- 8.Cohen M, DuBois R, Zeineh M. “Rapid and effective correction of RF inhomogeneity for high field magnetic resonance imaging,”. Human Brain Mappring. 2000;10:204–211. doi: 10.1002/1097-0193(200008)10:4<204::AID-HBM60>3.0.CO;2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeCarli C, Murphy D, Teichberg D, Campbell G, Sobering G. “Local histogram correction of MRI spatially dependent image pixel intensity nonuniformity,”. Journal of Magnetic Resonance Imaging. 1996;6:519–528. doi: 10.1002/jmri.1880060316. [DOI] [PubMed] [Google Scholar]
- 10.Studholme C, Cardenas V, Song E, Ezekiel F, Maudsley A, Weiner M. “Accurate template based correction of brain MRI intensity distortion with application to dementia and aging,”. IEEE Transactions on Medical Imaging. 2004;23(1):99–110. doi: 10.1109/TMI.2003.820029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pohl K, Wells W, Guimond A, Kasai K, Shenton M, Kikinis R, Grimson W, Warfield S. “Incorporating nonrigid registration into expectation maximization algorithm to segment MR images,”. Proc. of MICCAI. 2002 doi: 10.1007/3-540-45786-0_70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawant B, Zijdenbos A, Margolin R. “Correction of intensity variations in MR images for computer aided tissue classification,”. IEEE Transactions on Medical Imaging. 1993;12(4):770–781. doi: 10.1109/42.251128. [DOI] [PubMed] [Google Scholar]
- 13.Wells W, Grimson W, Kikinis R, Jolesz F. “Adaptive segmentation of MRI data,”. IEEE Transactions on Medical Imaging. 1996;15:429–442. doi: 10.1109/42.511747. [DOI] [PubMed] [Google Scholar]
- 14.Held K, Korps E, Krause B, WellsIII W, Kikinis R, Gartner H-WM. “Markov random field segmentation of brain MR images,”. IEEE Transactions on Medical Imaging. 1997;16:878–886. doi: 10.1109/42.650883. [DOI] [PubMed] [Google Scholar]
- 15.Styner M, Brechbuhler C, Szekely G, Gerig G. “Parametric estimate of intensity inhomogeneities applied to MRI,”. IEEE Transactions on Medical Imaging. 2000;19(3):153–165. doi: 10.1109/42.845174. [DOI] [PubMed] [Google Scholar]
- 16.Dempster A, Laird N, Rubin D. “Maximum likelihood from incomplete data via the EM algorithm (with discussion),”. Journal of the Royal Statistical Society B. 1977;39:1–38. [Google Scholar]
- 17.Ashburner A, Friston K. “Voxel-based morphometry–the methods,”. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 18.Pham D, Prince J. “An adaptive fuzzy c-means algorithm for image segmentation in the presence of intensity inhomogeneities,”. Pattern Recognition Letters. 1999;20:57–68. [Google Scholar]
- 19.Ahmed M, Yamany S, Mohamed N, Farag A, Moriarty T. “A modified fuzzy c-means algorithm for bias field estimation and segmentation of MRI data,”. IEEE Transactions on Medical Imaging. 2002;21(3):193–199. doi: 10.1109/42.996338. [DOI] [PubMed] [Google Scholar]
- 20.Kimmel R, Elad M, Shaked D, Keshet R, Sobel I. “A variational framework for retinex,”. International Journal of Computer Vision. 2003;52(1):7–23. [Google Scholar]
- 21.Brainard D, Wandell B. “Analysis of the retinex theory of color vision,”. Journal of the Optical Society of America A. 1986;3(10):1651–1661. doi: 10.1364/josaa.3.001651. [DOI] [PubMed] [Google Scholar]
- 22.Rising H., III “Analysis and generalization of retinex by recasting the algorithm in wavelets,”. Journal of Electronic Imaging. 2004;13(1):93–99. [Google Scholar]
- 23.Han C, Hatsukami T, Yuan C. “A multi-scale method for automatic correction of intensity nonuniformity in MR images,”. Journal of Magnetic Resonance Imaging. 2001;13:428–436. doi: 10.1002/jmri.1062. [DOI] [PubMed] [Google Scholar]
- 24.Vokurka E, Thacker N, Jackson A. “A fast model independent method for automatic correction of intensity nonuniformity in MRI data,”. Journal of Magnetic Resonance Imaging. 1999;10:550–562. doi: 10.1002/(sici)1522-2586(199910)10:4<550::aid-jmri8>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 25.Lai S, Fang M. “A new variational shape-from-orientation approach to correcting intensity inhomogeneities in magnetic resonance images,”. Medical Image Analysis. 1999;3(4):409–424. doi: 10.1016/s1361-8415(99)80033-4. [DOI] [PubMed] [Google Scholar]
- 26.Vovk U, Pernus F, Likar B. “MRI intensity inhomogeneity correction by combining intensity and spatial information,”. Institute of Physics Publishing. 2004;49:4119–4133. doi: 10.1088/0031-9155/49/17/020. [DOI] [PubMed] [Google Scholar]
- 27.Likar B, Viergever M, Pernus F. “Retrospective correction of MR intensity inhomogeneity by information minimization,”. IEEE Transactions on Medical Imaging. 2001;20(12):1398–1410. doi: 10.1109/42.974934. [DOI] [PubMed] [Google Scholar]
- 28.Sled J, Zijdenbos A, Evans A. “A nonparametric method for automatic correction of intensity nonuniformity in MRI data,”. IEEE Transactions on Medical Imaging. 1998;17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 29.Arnold J, Liow J, Schaper K, Stern J, Sled J, Shattuck D, Worth A, Cohen M, Leahy R, Mazziota J, Rottenberg D. Qualitative and quantitative evaluation of six algorithms for correcting intensity nonuniformity effects,”. Neuroimage. 2001;13:931–943. doi: 10.1006/nimg.2001.0756. [DOI] [PubMed] [Google Scholar]
- 30.Mangin J. “Entropy minimization for automatic correction of intensity nonuniformity,”. Proc. of IEEE workshop on MMBIA. 2000:162–169. [Google Scholar]
- 31.Bansal R, Staib L, Peterson B. “Correcting nonuniformities in MRI intensities using entropy minimization based on an elastic model,”. Proc. of MICCAI. 2004 [Google Scholar]
- 32.Gudbjartsson H, Patz S. “The Rician distribution of noisy MRI data,”. Magnetic Resonance in Medicine. 1995;34:910–914. doi: 10.1002/mrm.1910340618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ware R, Lad F. “Approximating the distribution for sums of products of normal variables,”. Research report, University of Caterbury. UCDMS 2003/15. [Google Scholar]
- 34.Collins D, Zijdenbos A, Kollokian V, Sled J, Kabani N, Holmes C, Evans A. “Design and construction of a realistic digital brain phantom,”. IEEE Transactions on Medical Imaging. 1998;17(3):463–468. doi: 10.1109/42.712135. [DOI] [PubMed] [Google Scholar]
- 35.Cocosco C, Kollokian V, Kwan R-S, Evans A. “Brainweb: Online interface to a 3D MRI simulated brain database,”. NeuroImage. 1997;5(4−2/4):S425. [Google Scholar]


