Abstract
The phytochrome family of informational photoreceptors has a central role in regulating light-responsive gene expression, but the mechanism of intracellular signal transduction has remained elusive. In a genetic screen for T DNA-tagged Arabidopsis mutants affected in early signaling intermediates, we identified poc1 (photocurrent 1), which exhibits enhanced responsiveness to red light. This phenotype is absent in a phyB (phytochrome B) null mutant background, indicating that the poc1 mutation enhances phyB signal transduction. The T DNA insertion in poc1 was found to be located in the promoter region of PIF3, a gene encoding a basic helix–loop–helix protein. The mutant phenotype seems to result from insertion-induced overexpression of this gene in red-light-grown seedlings, consistent with PIF3 functioning as a positively acting signaling intermediate. These findings, combined with data from a separate yeast two-hybrid screen that identified PIF3 as a phytochrome-interacting factor necessary for normal signaling, provide evidence that phytochrome signal transduction may include a direct pathway to photoresponsive nuclear genes via physical interaction of the photoreceptor molecules with the potential transcriptional regulator PIF3.
Keywords: photoreceptor, signal transduction
The phytochromes of higher plants are informational photoreceptors that influence many developmental processes in response to the red (R) and far-red (FR) regions of the light spectrum. Germination, growth strategies, and responses to seasonal changes of day length all respond to the light environment via these photoreceptors (1). In higher plants, the phytochromes comprise a family of photoreceptors, five of which, designated phyA through phyE, have been identified in Arabidopsis (2, 3). The availability of mutants with deficiencies in specific phytochromes has allowed detailed study of the functional roles of individual members of the phytochrome family (4–7). These studies have identified a complex array of phytochrome-controlled responses, some of which are specific to particular phytochromes and some of which are shared (6, 8).
The existence of both discrete and overlapping regulatory functions of the different phytochromes gives rise to the possibility that at least some components involved in the initial steps of signal transduction may be common to more than one phytochrome species. Indirect evidence for shared components has come from analysis of phytochrome-domain function and from photomorphogenic mutants. First, a series of deletion derivatives of both phyA and phyB expressed in Arabidopsis identified similar regions of the phyA (amino acids 617–687) and phyB (amino acids 633–652) COOH-terminal domains as functionally important in signal transduction (9, 10). Second, the COOH-terminal portions of both phytochromes have been shown to be functionally interchangeable by domain-swap experiments (11). Third, missense mutations shown to impair or inactivate signaling by photoactive phyA and phyB were found to cluster in the same “core” region of both photoreceptors (4). These data suggest that a similar region of the phyA and phyB molecules is important in regulatory function and that signal transduction could involve a common interaction partner. Potentially consistent with this view, mutants that are affected in the signal transduction of more than one phytochrome have been identified. The pef1 mutant (12) shows reduced R- and FR-mediated responses, whereas the psi1 mutant (13) shows enhanced R- and FR-mediated responses, suggesting disruption in both phyA and phyB signaling.
On the other hand, mutants have been isolated that seem to affect phyA or phyB signaling specifically. The mutants fhy1, fhy3, and spa1 affect only responses to FR and, hence, phyA signaling (14, 15). The fhy1 and fhy3 mutants show impaired phyA-regulated responses (16, 17), whereas the spa1 mutant shows amplified phyA signaling and is thought to be mutated at a locus encoding a negatively acting signaling component. By contrast, the pef2, pef3, and red1 mutants have reduced deetiolation only in R, indicating that these loci may be specific to phyB signal transduction and act as positive regulators in the signaling pathway (12, 18).
None of the genetically identified potential signaling-component-encoding loci described above have been characterized at the molecular level. Thus, with the aim of identifying and cloning early signaling intermediates, we screened Arabidopsis populations containing T DNA insertions and selected mutants displaying altered photomorphogenic responses under monochromatic light. Here, we describe the characterization of one such mutant, poc1, which had enhanced deetiolation under R, and the molecular identification of the tagged poc1 locus.
MATERIALS AND METHODS
Plant Material.
The poc1 mutant was isolated by screening the publicly available T DNA insertion populations (Arabidopsis Biological Resources Center, Columbus, OH) developed by K. Feldmann (19) and colleagues in the Wassilewskija-ecotype background. For double-mutant analysis, we crossed poc1 with a T DNA-insertional phyB null allele in the Wassilewskija ecotype (hy3-464-19; ref. 20). Double mutants were derived from the F2 of the poc1 × hy3-464-19 cross as follows. F2 seedlings were selected for long hypocotyls in white light (phyB mutant phenotype), grown, and selfed. F3 seed from 10 such F2 plants then was screened for homozygosity by using the poc1 T DNA insertion as a marker. Screening was achieved by a three-primer PCR with primers F, R1, and R2 (sequence F, 5′-AGAAGCAATTTGGTCACCATGCTC-3′; R1, 5′-ATCCTGTATATCAGACATTAGGAAGC-3′; R2, 5′-TGCATACAAATAGTCGATCGTATG-3′). Primers F (to the PIF3 promoter region) and R1 (to the T DNA left border) amplified a 726-bp fragment from only the poc1 mutant DNA. Primers F and R2 amplified a 463-bp product from the PIF3 promoter from wild-type DNA but not from poc1 mutant DNA because of the T DNA insertion between the primer sites. DNA from a known heterozygote yielded both bands in this reaction (data not shown). This reaction was used to screen eight independently generated DNA preparations from individual F3 seedlings derived from each of the 10 selected F2 individuals. All eight progeny from 2 of the 10 F2 individuals showed only the band expected from plants homozygous for the poc1 T DNA insertion. These plants then were confirmed as phyB nulls by PCR from the eight progeny DNA preparations by using the primers BF and BR (BF, 5′-CCAACTTTCAAAGCAAATGGCTG-3′; BR, 5′-CCAAGTCGACTAAACCGAATAC-3′). These primers generate a product from the wild-type PHYB gene, which is not seen in the mutant because of the T DNA insertion between the primer sites. Both of the PCR-confirmed poc1 phyB double mutants provided data essentially identical to those shown below in Fig. 5.
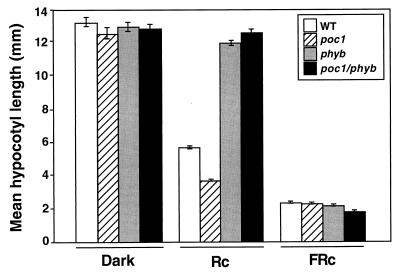
Figure 5.
Hypersensitivity of poc1 to R requires phyB. The mean hypocotyl lengths of the poc1 phyB double mutant, poc1 and phyB single mutants, and the wild type (WT) after Rc treatment (16 μmol⋅m−2⋅s−1, 3 days), FRc treatment (9 μmol⋅m−2⋅s−1, 3 days), and darkness (3 days).
Seedling Growth and Light Conditions.
For all experiments, seeds were surfaced sterilized in 20% bleach (1.05% sodium hypochlorite) and 0.03% Triton X-100 for 10 min, washed three times with sterile water, and plated on GM medium without sucrose (9). To synchronize germination, seeds were chilled for 5 days in darkness at 4°C before being given a 1 h white-light pulse at 21°C. The seeds were then returned to darkness at 21°C for a further 23 h before being placed in the appropriate light treatment. Seedling hypocotyl length was determined after a further 4 days of growth either in darkness or after transfer to continuous light, unless otherwise indicated. Hypocotyls were measured with a Pixera professional digital camera (Pixera, Cupertino, CA) and nih image software (version 1.61) from the National Institutes of Health. The white, R, and FR sources used have been described (18). Fluence rates were determined with a spectroradiometer (model LI-1800; Li-Cor, Lincoln, NE).
Cloning and Sequencing of the poc1 Locus.
A 4-kb fragment of genomic DNA flanking the T DNA-insert left border of the poc1 mutant was cloned by using plasmid rescue techniques (19). A 0.5-kb fragment from this cloned region was used as a probe in genomic Southern blot analysis to verify the origin of this DNA. Fragments flanking the T DNA right border and subsequent upstream fragments were cloned by using the PCR-based genome walker kit (CLONTECH). Sequencing was performed with an ABI automated sequencer by using a series of overlapping PCR products obtained from PCR amplification with sequential 5′, 21-bp primers (Advantage cDNA PCR kit, CLONTECH) by using the genomic sequence available from the database.
Analysis of PIF3 Transcript Levels.
PIF3, cinnamyl alcohol dehydrogenase (CAD), and PHYE transcript levels were determined by reverse transcription (RT)-PCR analysis (21) of total RNA, which was extracted with an RNeasy plant mini kit (Qiagen, Valencia, CA), and treated with RQI RNase-free DNase (Promega). The three transcripts were assayed simultaneously in parallel in separate tubes by using identical RNA aliquots and the respective gene-specific primers. RNA (8 ng) was used in each RT-PCR, which was performed with the access RT-PCR system kit (Promega) with minor modifications of the manufacturer’s recommended procedures. Briefly, RT was performed at 48°C for 45 min by first adding the downstream primer to the reaction mix, followed by a denaturation step at 94°C for 2 min, and a chilling step at 4°C for 10 min. During the chilling period, the upstream primer was added to the reaction mix, and the PCR amplification was done for a total of 18 cycles as follows: denaturation at 94°C for 30 s, annealing at 60°C for 1 min, and extension at 68°C for 2 min. The final extension was carried out at 68°C for 7 min, and the reaction was then stopped at 4°C. In preliminary experiments, we determined that 18 cycles lay within the linear range of PCR product amplification (data not shown) and therefore terminated all RT-PCR reactions reported here after 18 cycles. Primers used for amplification of PIF3 were the 5′ primer 15F (5′-CGCAGGAACCACTAATTACTA-3′) and the 3′ primer 15R (5′-CAGGCAAGCCCATTGCATAAG-3′). For the amplification of CAD, the 5′ primer 1F (5′-TAAATCCAGGACTTGTGCTC-3′) and the 3′ primer 1R (5′-ATAGGATGATCACACAAGAC-3′) were used. The PCR products were separated by gel electrophoresis on 1.2% agarose gels, blotted, and hybridized with a probe (see Fig. 3) for detection of PIF3 and with a probe that was PCR-amplified from genomic DNA by using 1F and 1R for detection of CAD. As an unrelated, low-abundance control for this experiment, PHYE transcript levels also were measured by RT-PCR. The PHYE-specific primers used were E5 (5′-CAGCTGCAAGCAACATGAAACCTC-3′) and E3 (5′-TCCTCCGGGAAGTGACTGCAGCCTAGA-3′), and blots were hybridized with a E5–E3 probe PCR-amplified from genomic DNA.
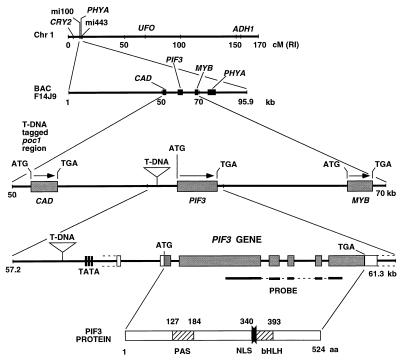
Figure 3.
The T DNA insert is located in the promoter region of PIF3. The diagram depicts (from top to bottom) the position of the poc1 locus on chromosome 1; the sequenced BAC F14J9 containing this locus; the T DNA insertion site in the promoter region of the PIF3 gene of poc1 and its location with respect to the predicted neighboring genes CAD and a MYB-related gene (MYB); the structure of the PIF3 gene and the location of the T DNA insert; and the structure of the PIF3 protein. The protein coding regions within the PIF3 gene are shown as filled boxes, the untranslated regions as open boxes, the introns and flanking DNA as a line, and the putative TATA box as vertical lines. The identity and location of motifs within the PIF3 protein are shown: PAS, Per–Arnt–Sim-like domain; bHLH, basic helix–loop–helix domain; NLS, nuclear localization signal. The position of the probe used in quantitative PCR analysis of PIF3 transcript levels also is shown (Probe).
RESULTS
Identification of the poc1 Mutant.
A preliminary screen of pooled T DNA-tagged lines identified poc1, a mutant that has a short hypocotyl phenotype compared with that of the wild type when seedlings were grown under white light photoperiods with end-of-day FR pulses. The nptII kanamycin resistance marker within the T DNA sequence was used to assess the presence and number of tagged insertion sites. Analysis of the F2 progeny from a back-cross of poc1 to the Wassilewskija wild-type parent indicated that the T DNA insert was at a single locus and that the poc1 mutation was monogenic with partial dominance (data not shown). The F2 progeny of poc1 crosses with both phyA and phyB mutants had the wild-type hypocotyl-length segregation ratios expected for independent segregation for each of the loci tested (data not shown). These tests indicated that the poc1 mutation was not located in either the PHYA or the PHYB gene.
The poc1 Mutant Phenotype Is Light-Dependent and R Specific.
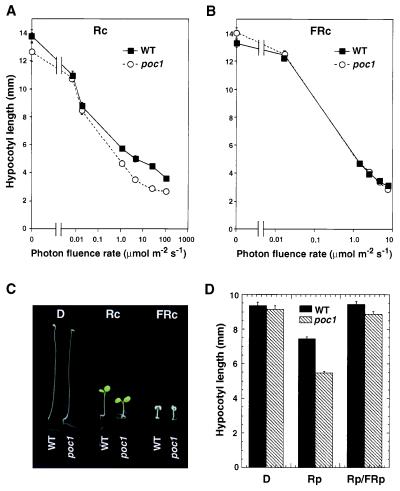
To test the specificity of the poc1 mutant phenotype, hypocotyl length was measured in seedlings grown in darkness, or under continuous R (Rc) or FR (FRc). Fig. 1 A and B shows that, when grown in darkness and under all FRc fluence rates tested, poc1 mutant seedlings were indistinguishable from wild-type seedlings. By contrast, poc1 seedlings displayed enhanced responsiveness to Rc compared with the wild type (Fig. 1A). Fig. 1C shows the visible phenotypes shown by poc1 seedlings compared with the wild type under these growth conditions. In addition to greater hypocotyl inhibition, cotyledon expansion is enhanced specifically by Rc in poc1 relative to the wild type (Fig. 1C and data not shown). Together, these data indicate that the enhanced deetiolation phenotype of poc1 is strictly light-dependent and that Rc selectively induces this phenotype.
Figure 1.
The poc1 mutant shows enhanced deetiolation under R. Photon fluence-rate response curves for hypocotyl length in wild-type (WT) and poc1 seedlings grown for 4 days in (A) continuous R (Rc) or (B) continuous FR (FRc). (C) Seedling phenotype for WT and poc1 seedlings grown in constant darkness (D), Rc, or FRc for 4 days. (D) Hypocotyl length of WT and poc1 seedlings grown for 3 days in darkness, in darkness but irradiated every 4 h with 5-min R pulses (Rp), or in the same conditions but with an FR pulse (FRp) given immediately after each Rp (Rp/FRp). Error bars represent SEM.
To establish whether the Rc hypersensitivity of the poc1 mutant results from altered phytochrome-induced responsiveness, the R/FR reversibility of the poc1 phenotype was tested. Fig. 1D shows that R pulses delivered at 4-h intervals caused partial hypocotyl inhibition in wild-type seedlings and that this effect was enhanced in the poc1 mutant. Pulses of FR given immediately after each R pulse negated the effects of the R pulse for both poc1 and wild-type seedlings. This result suggests that the poc1 phenotype is specific to phytochrome action.
To determine whether the poc1 phenotype simply could be attributed to an elevation in the level of phyA or phyB, poc1 and wild-type seedlings were assayed for immunochemically detectable phyA and phyB. Both the absolute levels and degradation kinetics for phyA were identical in poc1 and wild-type seedlings. Likewise, phyB levels were unchanged in poc1 seedlings relative to the wild type (data not shown).
Identification of the poc1 Locus.
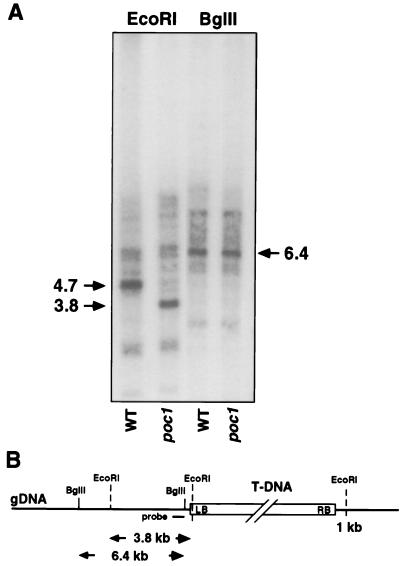
Preliminary evidence that the poc1 mutation was caused by a single T DNA insertion at the poc1 locus was obtained from analyses showing cosegregation of the poc1 phenotype with a T DNA-specific PCR fragment (data not shown). Molecular cloning of the genomic DNA flanking the T DNA insert was then achieved by left-border plasmid rescue. Southern blot analysis confirmed that the genomic fragment cloned in this way corresponded to the T DNA insertion site (Fig. 2). An EcoRI digest of genomic DNA probed with a subfragment from the sequence adjoining the tag yielded a polymorphism between wild type and poc1, caused by the insertion and the presence of an EcoRI site in the terminal region of the T DNA left border. This additional EcoRI site in poc1 was effective in reducing the fragment size by 0.9 kb to 3.8 kb (Fig. 2). A control digest with BglII, which has a site adjacent and distal to the probe region, yielded identical fragments of the expected size (6.4 kb) in both wild-type and poc1 extracts.
Figure 2.
Identification of the genomic region flanking the T DNA insert in poc1. (A) Southern blot analysis of total genomic DNA from wild type (WT) and poc1 digested with EcoRI or BglII and hybridized with a 0.5-kb probe from the cloned region flanking the T DNA left border (LB). (B) Diagram of the T DNA insert within the poc1 genomic DNA (gDNA) illustrating the locations of the restriction sites, the size of the restriction fragments generated, and the fragment used as the probe.
Genomic DNA flanking the right border was cloned by using the PCR-based Genome Walker kit (CLONTECH) and sequenced. Database searches located this sequence to a region within a sequenced bacterial artificial chromosome (BAC), F14J9, at the top of chromosome I (Fig. 3). The T DNA tag was found to be located at a position 1,040 bp upstream of the predicted ATG translation start site of a gene with homology to the bHLH protein family. Remarkably, in a separate project, by using a yeast two-hybrid screen, we recently identified this gene as encoding a factor designated PIF3 (phytochrome interacting factor 3), that interacts molecularly with both phyA and phyB (22). The PIF3 gene encodes regions with sequence homology to PAS and bHLH motifs and contains a nuclear localization signal motif at the 5′ end of the bHLH domain. Because the BAC, F14J9, had not been annotated, we identified potential ORFs in the sequence flanking the PIF3 locus. A CAD homolog was found to be situated 6 kb upstream of the T DNA tag, and a gene with MYB sequence homology was identified some 8 kb downstream of PIF3, interestingly, just upstream of the PHYA gene (Fig. 3).
poc1 Expresses Enhanced Levels of PIF3 mRNA in Rc.
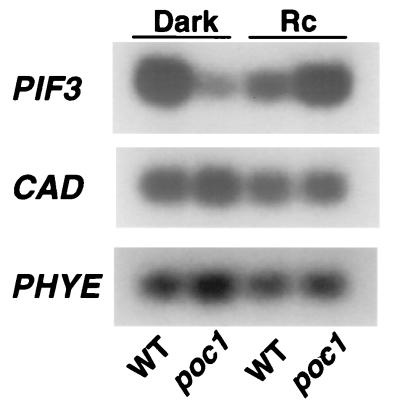
Potential effects of the transgene on both PIF3 and CAD transcript levels were assayed by using quantitative RT-PCR. Southern blot analysis with probes generated from the primers used in the RT-PCR procedure identified both PIF3 and CAD as apparently single-copy genes (data not shown). For the RT-PCR analysis, seedlings were grown in either darkness or Rc. Whereas CAD mRNA levels were indistinguishable in wild-type and poc1 mutants, PIF3 transcript levels were clearly altered (Fig. 4). PIF3 mRNA was lower in dark-grown poc1 seedlings than in dark-grown wild-type seedlings. However, the converse was true for the Rc-grown seedlings; under these conditions, poc1 had higher PIF3 mRNA levels than the wild type and also had higher PIF3 mRNA levels than the dark-grown poc1 seedlings (Fig. 4). These results indicate that the T DNA insert influences the expression of PIF3 but not that of the putative CAD gene situated upstream of the insert. Furthermore, the data indicate that the effect of the T DNA insert on PIF3 expression is influenced by Rc. In the absence of light, PIF3 expression is low in poc1, but it is enhanced in Rc. Time-course experiments indicated that Rc given from germination onwards suppresses the accumulation of PIF3 mRNA in wild-type seedlings, thereby maintaining low transcript levels throughout the growth period (data not shown). Conversely, Rc enhances PIF3 mRNA accumulation in poc1 over the growth period, resulting in higher net levels in the mutant than the wild type in light-grown seedlings (Fig. 4). Thus, the hypersensitive poc1 mutant phenotype seems to be correlated with elevated levels of the PIF3 transcript under Rc conditions.
Figure 4.
PIF3 mRNA levels are elevated in R-grown poc1 seedlings. Quantitative RT-PCR analysis of PIF3, CAD, and PHYE mRNA levels in wild-type (WT) and poc1 seedlings grown for 4 days in either constant darkness (D) or Rc. Southern blotting was used to detect the PCR products as described in Materials and Methods.
phyB Is Necessary for Expression of the poc1 Phenotype.
The Rc specificity of the poc1 phenotype (Fig. 1) suggested that phyB might mediate the effects of the photosignal. To test this possibility, we constructed poc1 phyB double mutants by using a T DNA-induced phyB null allele (20) and examined seedling photoresponsiveness to Rc and FRc. Because both the poc1 and phyB single mutants were derived by T DNA insertion, we used PCR primers specific to each insert to verify homozygosity at both mutant loci in the lines selected as double mutants (data not shown). Fig. 5 shows that poc1 phyB double mutants have strongly reduced responsiveness to Rc, equivalent to that of the phyB single mutant. This result indicates that phyB is necessary for the enhanced Rc responsiveness of the poc1 mutant.
DISCUSSION
The genetic and photobiological evidence presented here indicates that the poc1 mutation represents aberrant activity of a component involved in phyB signal transduction. The molecular data indicate further that this mutation results from a T DNA insertion in the promoter region of the PIF3 gene that leads to overexpression of this gene in Rc-grown seedlings. Recently, PIF3 was identified by yeast two-hybrid screening in a separate investigation in our laboratory as a phytochrome interacting factor that binds to both phyA and phyB (22). Subsequent reverse genetic experiments showed that PIF3 is a protein necessary for normal phytochrome signal transduction (22). Thus, our elucidation of the molecular basis of the poc1 mutant phenotype constitutes independent evidence for the functional importance of the PIF3 locus for phytochrome activity in vivo.
poc1 Is Defective in phyB Signaling.
The absence of an observable mutant phenotype in dark-grown poc1 seedlings indicates that this phenotype is light-conditional. The induction of the phenotype by Rc and, in a FR-reversible fashion, by R pulses, provides photobiological evidence that the phytochrome system mediates these light signals. The absence of a detectable difference in photoresponsiveness between wild type and poc1 in FRc indicates that the phyA-mediated FR-high irradiance response is unaffected by the mutation. On the other hand, the elimination of the differential in Rc photoresponsiveness between poc1 +/+ and poc1 −/− seedlings in a phyB null mutant background (Fig. 5) establishes that phyB is necessary for expression of the poc1 phenotype and that none of the other phytochromes in the Wassilewskija ecotype, alone or in combination, can substitute for phyB in this capacity. The data suggest, therefore, that poc1 is enhanced specifically in phyB signaling. The absence of changes in phyB levels in the poc1 mutant relative to the wild type (data not shown) indicates that this effect is not caused by an indirect effect that results in phyB overexpression.
The poc1 Mutant Phenotype Is the Consequence of T DNA Induced Overexpression of PIF3.
The cosegregation of the single T DNA locus and its flanking genomic DNA with the poc1 phenotype provides evidence that the mutation is the result of this T DNA insertion. The location of this insertion in the promoter of the PIF3 gene and the selectively enhanced PIF3 transcript levels in Rc-grown poc1 seedlings support the conclusion that the mutant phenotype is generated by PIF3 overexpression induced by the insert. The mechanism by which this overexpression is induced is unknown but could involve either disruption of a negatively acting, Rc-responsive PIF3 promoter element or the introduction of an element present in the neighboring T DNA sequence that fortuitously responds positively to Rc. The insert also suppresses the accumulation of PIF3 mRNA that normally occurs in dark-grown seedlings, suggesting disruption of an additional positively acting promoter element. The biological role of the Rc-imposed suppression of PIF3 mRNA accumulation in wild-type seedlings is undetermined but could represent a negative-feedback desensitization or a reduction in capacity of the phyB signaling pathway that occurs once the deetiolation process has been initiated.
The conclusion that the Rc hypersensitivity of the poc1 mutant is caused by enhanced PIF3 levels is consistent with the separate observation that transgenic Arabidopsis seedlings engineered to overexpress the PIF3-sense sequence under control of the 35S cauliflower-mosaic-virus promoter also display marginally increased Rc sensitivity (22). Together with the converse effects of PIF3-antisense expression in strongly reducing the Rc photoresponsiveness of Arabidopsis seedlings (22), these data indicate that PIF3 acts positively in phyB signal transduction, as suggested by the genetically defined, partially dominant behavior of the poc1 locus. By contrast, although PIF3-antisense expression reduces FRc responsiveness (22), PIF3 overexpression has either no (Fig. 1) or a barely detectable (22) effect on the FRc responsiveness of poc1 and transgenic, PIF3-sense expressers, respectively. These data suggest that, whereas PIF3 is necessary for normal phyA signaling, endogenous PIF3 levels are essentially saturating for the transduction pathway. Thus, although the Rc-specific phenotype of poc1 initially suggested that the mutant phenotype could represent a phyB-specific signaling component, the observed reduction in responsiveness of PIF3-antisense seedlings to both Rc and FRc (22) indicates PIF3 involvement in both phyA and phyB signal transduction. This conclusion is consistent with the observed physical interaction of PIF3 with both phyA and phyB molecules (22).
Signaling-Pathway Configuration.
Although the data presented here and elsewhere (22) provide evidence that PIF3 is a primary reaction partner in a putative shared transduction pathway between phyA and phyB, there is considerable genetic evidence for the existence of signaling components specific to phyA or phyB. For example, because the mutants fhy1, fhy3, and spa1 (14, 15) have specific effects on only FRc-induced responses, these signaling components seem to act specifically in phyA signaling. Genetically, therefore, they would seem to act upstream of or in parallel with PIF3. Similarly, the mutants red1, pef2, and pef3 are affected specifically only in Rc-induced responses (12, 18), suggesting activity upstream of or in parallel with PIF3 in phyB signaling. It seems unlikely that such components could lie upstream between PIF3 and phyA or phyB in a conventional linear pathway, because PIF3 has a direct molecular interaction with both photoreceptors presumably essential for signal transmission (22).
Therefore, one alternative that seems able to reconcile the genetic and molecular data is that parallel signaling pathways originate from each photoreceptor molecule: one pathway via the shared factor PIF3 that would result in immediate convergence of phyA and phyB signaling and a second pathway via separate factors providing independent signaling from each photoreceptor. A second alternative, also consistent with the data, is that a single signaling pathway emanates from each photoreceptor but is subject to modification or “tuning” by factors specific to each photoreceptor type. Architecturally, this pathway configuration could be achieved with twin complexes involving signaling interactions centered around both phyA and phyB, with some factors acting specifically on one complex (e.g., SPA1, FHY1, and FHY3 on the phyA complex) and others acting on a second complex (e.g., RED1, PEF2, and PEF3 on the phyB complex). PIF3 and possibly other factors would form essential component(s) of both complexes. PIF3 and possibly other factors would interact directly with the phytochrome molecule, whereas others may act as modifiers or transduce secondary signals without direct molecular contact with the phytochromes. Such components could still behave in a manner genetically upstream of PIF3 and could also act temporally upstream, if they fulfilled a molecular function on the complex before PIF3 was able to transduce a signal. Precedence for signal-pathway channeling via separate multimolecular complexes, composed of both shared and distinct components assembled on scaffolding proteins, is found in networks of mitogen-activated protein kinases (23). We suggest, therefore, that a configuration similar to the scaffold complexes of mitogen-activated protein kinases, in which PIF3 is shared, and other components (such as SPA1, FHY1, FHY3, RED1, PEF2, and PEF3) are either phyA- or phyB-specific, could account for the currently available data.
CONCLUSIONS
In two separate contemporaneous studies aimed at defining phytochrome signaling intermediates by different strategies, we independently identified PIF3 as a candidate. We suggest that this unanticipated convergence of the two distinct strategies on the same molecular component provides compelling mutual reinforcement of the proposition that PIF3 does indeed function as a phytochrome transduction-pathway component in vivo. Moreover, because PIF3 interacts physically with both phyA and phyB, the evidence indicates that this factor acts immediately downstream of both photoreceptors as a direct recipient of signaling information (22). Together with the identification of PIF3 as a member of the bHLH class of transcriptional regulators, these data suggest a direct pathway for signal transfer from the phytochrome molecule to photoresponsive genes.
Acknowledgments
We thank Yurah Kang for technical assistance, all members of the laboratory for invaluable advice and lively discussion, Ron Wells for preparation and editing of the manuscript, and Jim Tepperman for figure preparation. The research was supported by National Institutes of Health Grant GM47475; Department of Energy, Basic Energy Sciences Grant DE-FG03-87ER13742; and U.S. Department of Agriculture, Current Research Information Service Grant 5335-21000-0010-00D.
ABBREVIATIONS
- R
red light
- FR
far red light
- RT
reverse transcription
- Rc
continuous R
- FRc
continuous FR
- BAC
bacterial artificial chromosome
References
- 1.Kendrick R E, Kronenberg G H M. Photomorphogenesis in Plants. 2nd Ed. Dordrecht, the Netherlands: Kluwer; 1994. [Google Scholar]
- 2.Clack T, Mathews S, Sharrock R A. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- 3.Sharrock R A, Quail P H. Genes Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- 4.Quail P H, Boylan M T, Parks B M, Short T W, Xu Y, Wagner D. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 5.Aukerman M J, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino R M, Sharrock R A. Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitelam G C, Devlin P F. Plant Cell Environ. 1997;20:752–758. [Google Scholar]
- 7.Devlin P F, Patel S R, Whitelam G C. Plant Cell. 1998;10:1479–1487. doi: 10.1105/tpc.10.9.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quail P H. Philos Trans R Soc London B. 1998;353:1399–1403. doi: 10.1098/rstb.1998.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boylan M, Douglas N, Quail P H. Plant Cell. 1994;6:449–460. doi: 10.1105/tpc.6.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner D, Quail P H. Proc Natl Acad Sci USA. 1995;92:8596–8600. doi: 10.1073/pnas.92.19.8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner D, Koloszvari M, Quail P H. Plant Cell. 1996;8:859–871. doi: 10.1105/tpc.8.5.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad M, Cashmore A R. Plant J. 1996;10:1103–1110. doi: 10.1046/j.1365-313x.1996.10061103.x. [DOI] [PubMed] [Google Scholar]
- 13.Genoud T, Millar A J, Nishizawa N, Kay S A, Schäfer E, Nagatani A, Chua N-H. Plant Cell. 1998;10:889–904. doi: 10.1105/tpc.10.6.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoecker U, Xu Y, Quail P H. Plant Cell. 1998;10:19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitelam G C, Johnson E, Peng J, Carol P, Anderson M L, Cowl J S, Harberd N P. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnes S A, Quaggio R B, Whitelam G C, Chua N-H. Plant J. 1996;10:1155–1161. doi: 10.1046/j.1365-313x.1996.10061155.x. [DOI] [PubMed] [Google Scholar]
- 17.Johnson E, Bradley M, Harberd N P, Whitelam G C. Plant Physiol. 1994;105:141–149. doi: 10.1104/pp.105.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner D, Hoecker E, Quail P H. Plant Cell. 1997;9:731–743. doi: 10.1105/tpc.9.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldmann K A. In: Methods in Arabidopsis Research. Kocz C, Chua N-H, Schell J, editors. London: World Scientific; 1992. pp. 274–288. [Google Scholar]
- 20.Reed J W, Nagpal P, Poole D S, Furuya M, Chory J. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rappollee D A, Mark D, Banda M J, Werb Z. Science. 1988;241:708–712. doi: 10.1126/science.3041594. [DOI] [PubMed] [Google Scholar]
- 22.Ni M, Tepperman J M, Quail P H. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- 23.Elion E A. Science. 1998;281:1625–1626. doi: 10.1126/science.281.5383.1625. [DOI] [PubMed] [Google Scholar]