Abstract
Several genome-wide transcriptomics efforts have shown that a large percentage of the mammalian genome is transcribed into RNAs, however, only a small percentage (1–2%) of these RNAs is translated into proteins. Currently there is an intense interest in characterizing the function of the different classes of noncoding RNAs and their relevance to human disease. Using genomic approaches we discovered FMR4, a primate-specific noncoding RNA transcript (2.4 kb) that resides upstream and likely shares a bidirectional promoter with FMR1. FMR4 is a product of RNA polymerase II and has a similar half-life to FMR1. The CGG expansion in the 5′ UTR of FMR1 appears to affect transcription in both directions as we found FMR4, similar to FMR1, to be silenced in fragile X patients and up-regulated in premutation carriers. Knockdown of FMR4 by several siRNAs did not affect FMR1 expression, nor vice versa, suggesting that FMR4 is not a direct regulatory transcript for FMR1. However, FMR4 markedly affected human cell proliferation in vitro; siRNAs knockdown of FMR4 resulted in alterations in the cell cycle and increased apoptosis, while the overexpression of FMR4 caused an increase in cell proliferation. Collectively, our results demonstrate an antiapoptotic function of FMR4 and provide evidence that a well-studied genomic locus can show unexpected functional complexity. It cannot be excluded that altered FMR4 expression might contribute to aspects of the clinical presentation of fragile X syndrome and/or related disorders.
Introduction
While at least 40–50% of the human genome is transcribed into RNA, only 1.2% of the genome is translated into protein [1]–[3]. RNAs which do not code for proteins (noncoding RNAs) have been classified into different classes (tRNA, rRNA, snRNA, snoRNA, miRNA, siRNA, piRNA, natural antisense transcripts and long noncoding RNA) based on their size and function. Novel classes of noncoding RNAs have been shown to have a variety of functions which include translational inhibition (miRNA) [4], mRNA degradation (siRNA) [5], and repressing transposition (piRNA) [6], [7]. Also, thousands of protein coding genes have now been shown to have antisense transcripts [8]. Antisense transcript manipulation can in some cases lead to the repression of the sense transcript (discordant regulation) or enhance the stability of the sense transcript (concordant regulation) [8]. However, the exact mechanisms by which antisense transcripts regulate their sense partners are likely diverse and require further studies. Another class of noncoding RNAs is long noncoding RNAs (or “macroRNA”) [9], [10] which do not overlap with protein-coding genes and range from 300 nucleotides to over 10 kb in size with an average size of ∼2 kb [9], [11]. Notably, the sequence of noncoding RNAs, in contrast to other noncoding RNAs such as miRNAs and snoRNAs, is often not well conserved even between mammals [9] and can function both in cis (e.g., XIST) [12] and in trans (e.g., HOTAIR) [13]. In contrast to other noncoding RNAs, only a small number of long noncoding RNAs have been functionally characterized. Several studies have now shown that the expression pattern of noncoding RNAs can be altered in several human diseases such as cancers and heart disease [14], [15] suggesting that noncoding RNAs may have a functional relevance to human disease and that they may be considered as potential drug targets [11], [16].
Fragile X syndrome (FXS), the most common cause of inherited mental retardation, is caused by the expansion of CGG trinucleotide repeats in the 5′ UTR of the fragile X mental retardation 1 gene (FMR1) [17]–[19]. Normal individuals have a range of 5–50 CGG repeats in the 5′ UTR of FMR1 and express FMR1 in a wide range of adult and embryonic tissues [20]. The CGG repeats can expand in the female germ line or shortly after fertilization by an unknown mechanism. Individuals with 55–200 repeats are premutation carriers and generally express higher levels of FMR1 mRNA than normal individuals and may result in a clinical condition termed fragile X tremor and ataxia syndrome (FXTAS) [21], [22]. The expansion of CGG repeats above 200 leads to the repression or silencing of FMR1 and consequently to the absence of the fragile X mental retardation protein (FMRP). We have previously been part of effort to identify and characterize 2,113 bidirectional promoters from 42,887 transcriptional units in humans [23]. In the present manuscript, we report the discovery and functional characterization of a primate-specific noncoding RNA (FMR4) that becomes silenced as a result of the CGG expansion in the 5′ UTR of FMR1 in fragile X syndrome. In vitro manipulation of FMR4 indicates it has an antiapoptotic function in human cells.
Results
Identification and expression analysis of FMR4
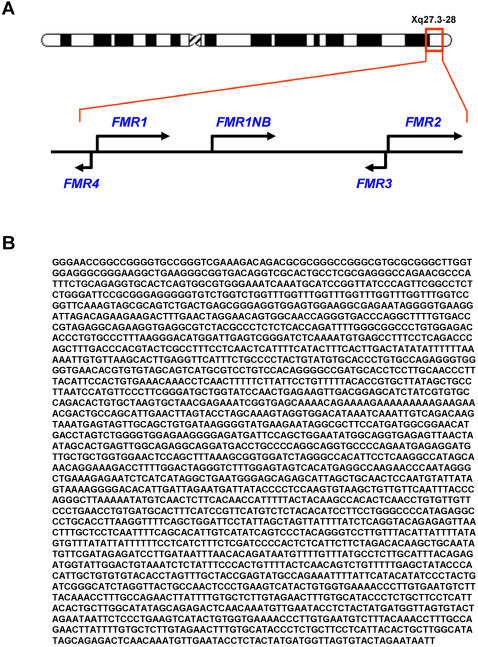
Previous work by us and others has shown that bidirectional promoters are relatively common in the mammalian genome [8], [23]. Therefore, using genomic approaches, including rapid amplification of cDNA ends (RACE), regular and real time PCR (RT-PCR), we searched for transcripts upstream of FMR1 that could also be affected by the CGG repeat expansion. Here, we report the identification of a novel 2.4 kb long noncoding RNA, which we named FMR4, that resides upstream and likely shares a bidirectional promoter with FMR1 (Figure 1A–B). Bioinformatic analysis suggests that FMR4 does not have a conventional open reading frame, to confirm that FMR4 is indeed a noncoding RNA we carried out in vitro transcription/translation followed by mass spectrometry analysis; however, no protein was detected suggesting that FMR4 is most likely a noncoding RNA (data not shown). Northern blot analysis shows that FMR1 is expressed in the majority of the human tissues examined consistent with previous reports [20]. FMR4 is expressed in several adult human tissues including brain, liver, placenta, small intestine, colon and spleen but not in the pancreas, testes, ovaries or prostate (Figure S1). Two bands corresponding to FMR4 were observed in several human adult tissues, one possibility is that there is alternative transcription start sites for FMR4, however, when we performed our RACE analysis we used RACE ready cDNA from SH-SY5Y cells and obtained only the longer 2.4 kb transcript.
Figure 1. Identification and sequence analysis of FMR4.
(A) Schematic showing known genes in Xq27.3-28 including the newly identified FMR4. FMR4 is transcribed upstream of FMR1 and in the opposite direction. (B) Sequence of FMR4 obtained by rapid amplification of cDNA ends (RACE).
FMR4 is ubiquitously expressed during human development
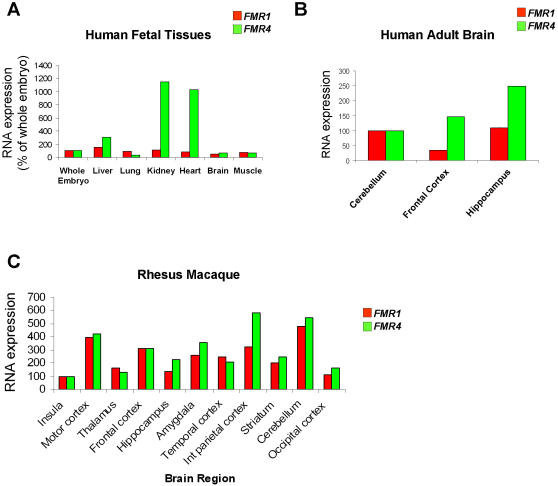
Since FMR4 is expressed in several human adult tissues, we next examined its expression levels in human fetal tissues. Using RT-PCR we measured RNA expression levels of FMR4 in seven different human fetal tissues (12 weeks); the RNA from each tissue was pooled from at least three different embryos (GBiosciences). We found FMR4 to be expressed in all the tissues examined including the brain. Notably, FMR4 is highly expressed in the kidney and heart at that stage of human development (Figure 2A). The high expression of FMR4 in the heart is consistent with a previous report that patients with fragile X syndrome have cardiac defects similar to those seen in other disorders of connective tissue such as Marfan's syndrome and Ehlers-Danlos syndrome [24].
Figure 2. Expression analysis of FMR4.
(A) RT-PCR analysis of FMR4 and FMR1 in seven different human fetal tissues (week 12), RNA from each tissue was pooled from at least three fetuses (GBiosciences). The RNA expression of FMR1 and FMR4 were normalized to whole embryo (set as 100%). Both transcripts are expressed in all the tissues tested with notably high expression of FMR4 in the kidney and heart. (B) RNA was extracted from six postmortem human adult brains from three different regions, thereafter; cDNA synthesis followed by RT-PCR was performed on all samples to measure the relative quantities of FMR1 and FMR4. Both FMR1 and FMR4 are highly expressed in all the human brain regions tested. (C) RT-PCR analysis of FMR4 and FMR1 in several regions of two monkeys brains. The RNA expression of FMR4 and FMR1 were normalized to the insula (set as 100%).
FMR4 is expressed in human and monkey brain
To determine whether FMR4 shows differential expression within different regions of the human brain, we examined the RNA concentrations of both FMR1 and FMR4 by Real-Time PCR (RT-PCR) using tissue from six postmortem human brains (from four males and two females aged 61–91 years) and studied three different regions (cerebellum, frontal cortex, and hippocampus). By this quantitative method, RT-PCR, both FMR1 and FMR4 were shown to display robust expression levels in all three brain regions tested (Figure 2B). To determine if FMR4 is also expressed in other primates, we examined the expression of FMR4 in rhesus monkey brain regions using RT-PCR. Total RNA from each brain region was isolated from two monkeys and DNAse treated prior to cDNA synthesis. We found FMR4 to be expressed in all the monkey brain regions tested with high expression in the cerebellum and interior parietal cortex (Figure 2C) confirming that FMR4 is expressed in other primates in addition to humans.
The CGG expansion affects transcription in both directions of a bidirectional promoter
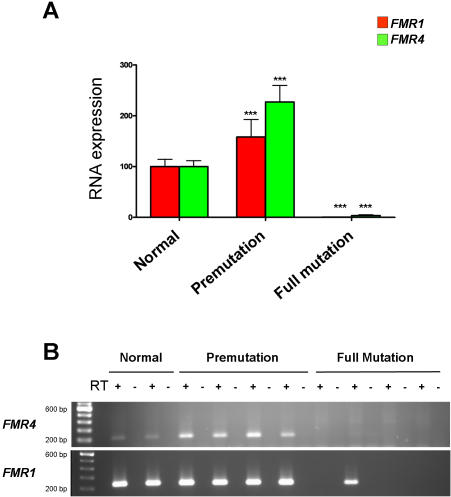
To determine if the expression of FMR4 is affected by the CGG expansion in the 5′ UTR of FMR1 that occurs in FXS and/or FXTAS, we investigated the relative expression of FMR4 and FMR1 by RT-PCR in untransformed leukocytes from four control, four premutation and four FXS patients. We found FMR4 expression, similar to FMR1, to be significantly up-regulated in premutation carriers, and shut down in full mutation (FXS) patients (P<0.0001) (Figure 3A). All the samples tested had a matching control without the reverse transcriptase to account for possible DNA contamination. We also utilized regular PCR (35 cycles), gel electrophoresis and ethidium bromide staining to examine the expression of FMR4 and FMR1 from the same samples. FMR4 was detectable in both the normal and premutation carriers but not in the full mutation fragile X patients. FMR1 was detectable in the normal, premutation carriers and one out of the four fragile X patients (Figure 3B); this is not surprising since variable levels of FMR1 mRNA have been previously shown to be present in some fragile X patients [25].
Figure 3. FMR4 is silenced in fragile X syndrome.
(A) RNA from four normal, four premutation and four full mutation FXS patients isolated from untransformed leucocytes (kindly provided by Flora Tassone and Paul Hagerman, UC Davis) was reverse transcribed using random hexamers. Quantitative RT-PCR analysis revealed that FMR4, similar to FMR1, is up-regulated in pre-mutation carriers and shut down in full mutation fragile X patients (P<0.0001). (B) RNA from untransformed leucocytes were reversed transcribed and the cDNA was used for PCR analysis. FMR4 is expressed in normal and premutation carriers but no bands were observed in the full mutation fragile X patients (35 cycles). FMR1 bands were observed in normal, premutation, and one of the full mutation patients (35 cycles). To account for any possible DNA contamination, no reverse transcriptase control for all samples were used in the PCR (lanes next to bands are all negative indicating no DNA contamination was present). Error bars: s.d.
FMR4 is a product of RNA polymerase II and has a similar half-life to FMR1
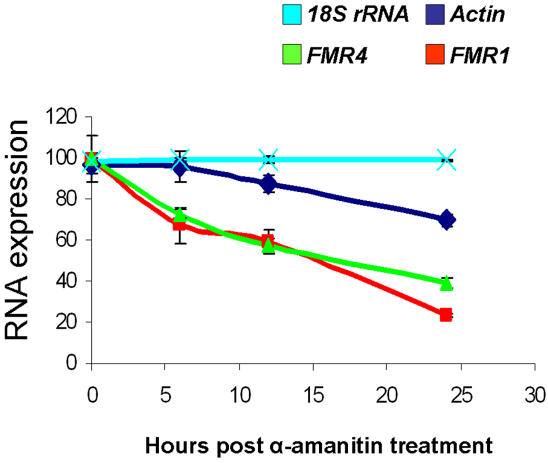
To measure the relative half-lives of FMR1 and FMR4 we treated HEK-293T cells with 50 µM of α-amanitin (an inhibitor of RNA polymerase II) [26] and RNA was isolated at 0, 6, 12 and 24 hours post treatment (six repeats each). All treated samples had a matching control which did not receive α-amanitin (untreated samples). Actin was used as a positive control and 18S rRNA was used as a negative control (a product of RNA polymerase I). By RT-PCR we measured the levels of Actin, 18S, FMR1, and FMR4. As expected 18S rRNA levels did not change at any of the time points tested since α-amanitin does not affect RNA polymerase I. By contrast Actin, FMR1 and FMR4 were all affected since they are all products of RNA polymerase II. The data indicates that FMR4 has a similar half-life to FMR1 (Figure 4).
Figure 4. FMR4 has a similar half-life to FMR1.
HEK-293T cells were treated with α-amanitin (blocks RNA polymerase II) and the levels of FMR4 and FMR1 were measured by RT-PCR at 0, 6, 12 and 24 hours post treatment. Both FMR4 and FMR1 have similar half-lives. These experiments also further confirm that FMR4 is a product of RNA polymerase II.
No evidence of direct cross-regulation between FMR1 and FMR4
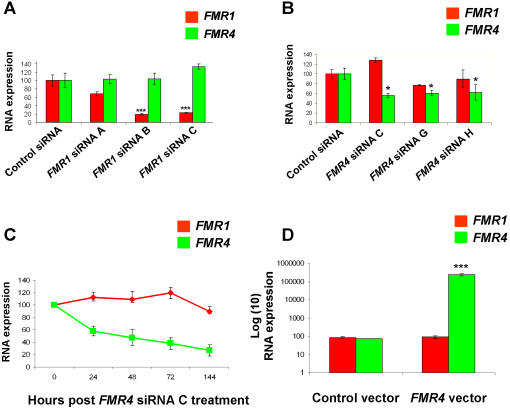
To determine whether FMR1 and FMR4 are functionally linked, we tested three different siRNAs against FMR1 and identified three different newly designed siRNAs against FMR4. It is important to utilize multiple efficacious siRNAs to any given target in order to avoid the possibility of off-target phenomena [11], [27]. Also, since FMR4 is highly expressed in human embryonic kidney (Figure 2A), we decided to use HEK-293T cells as an in vitro system to study FMR4 regulation and function. First, we tested three siRNAs against FMR1 by transfecting HEK-293T cells with 20 nM (final concentration) of FMR1 siRNAs (six repeats each). Two out of the three siRNAs tested were effective in knocking down FMR1 by 80% as early as 48 hours post transfection (siRNA B and C, Table 1). This level of knockdown was also observed at 72 hours post transfection and in repeated transfection experiments (144 hours total, two transfections at 0 hour and 72 hours post first transfection). However, in all cases the concentrations of FMR4 were not affected by FMR1 siRNAs (Figure 5A). We designed and tested nine different siRNAs for FMR4, three siRNAs (C, G, and H, see methods for sequences) caused a significant decrease in FMR4 at 48 and 72 hours post transfection, but did not affect the levels of FMR1 (Figure 5B). We used FMR4 siRNA C which caused the highest level of FMR4 knockdown among the siRNAs tested for a time course experiment. HEK-293T cells were transfected with FMR4 siRNA C and RNA was collected at 24, 48, 72 and 144 hours post transfection. This siRNA caused a significant knockdown of FMR4 but did not affect FMR1 levels at any of the time points tested (Figure 5C). We next cloned FMR4 into a pcDNA3.1 vector with a CMV promoter and overexpressed FMR4 in HEK-293T cells, a pcDNA3.1 vector without the FMR4 insert was used as a control. At 72 hours post transfection, RNA was isolated, reversed transcribed and used for RT-PCR analysis. As expected, FMR4 overexpression led to a substantial increase in FMR4 RNA levels, but did not have any effect on FMR1 RNA (Figure 5D). Collectively, these experiments suggest that the non-overlapping transcripts FMR1 and FMR4 do not regulate each other.
Table 1. Sequences of FMR1 and FMR4 siRNAs.
| siRNA | Sequence | Supplier |
| FMR1 A | GGUGUAUUCCAGAGCAAAUtt | Ambion (ID # 10824) |
| FMR1 B | GGGUGAGUUUUAUGUGAUAtt | Ambion (ID # 11010) |
| FMR1 C | GGAUGAUAAAGGGUGAGUUtt | Ambion (ID # 10919) |
| FMR4 A | GCCCUCUCUCACCAGAUUUtt | QIAGEN |
| FMR4 B | AGGGCCAGAACGCCCAUUUtt | QIAGEN |
| FMR4 C | GUGGCGUGGGAAAUCAAAUtt | QIAGEN |
| FMR4 D | GCAUCCGGUUAUCCCAGUUtt | Invitrogen |
| FMR4 E | UCGCCUUUCCUCAACUCAUtt | Invitrogen |
| FMR4 F | GCACUUGAGGUUCAUUUCUtt | Invitrogen |
| FMR4 G | AGAGAUCCUUGAUAAUUUAtt | QIAGEN |
| FMR4 H | CAUUGAUUAGAAUGAUUAUtt | QIAGEN |
| FMR4 I | GUAGGUGGACAUAAAUCAAtt | QIAGEN |
Figure 5. No direct cross-regulation between FMR1 and FMR4.
(A) We used three distinct siRNAs against FMR1 to transfect HEK-293T cells. Two out of the three siRNAs resulted in a significant knockdown of FMR1 (80%), but did not affect FMR4 RNA levels. (B) We used three distinct siRNAs against FMR4 to transfect HEK-293T cells. All three siRNAs resulted in a significant knockdown of FMR4 but did not affect FMR1 RNA levels. (C) Significant knockdown of FMR4 via siRNA C did not result in a change in FMR1 RNA levels at any of the time points tested (24, 48, 72, or 144 hours post transfection). (D) The entire sequence of FMR4 was cloned into a pcDNA3.1 vector with a CMV promoter. The pcDNA3.1 vector containing the FMR4 sequence and the original pcDNA3.1 (without the FMR4 insert) were transfected in HEK-293T cells. At 72 hours post transfection, RNA was isolated and reversed transcribed and used for RT-PCR analysis. There is a highly significant increase in the FMR4 RNA levels but no effect on FMR1 RNA. Error bars: s.d.
FMR4 affects cell proliferation in human cells
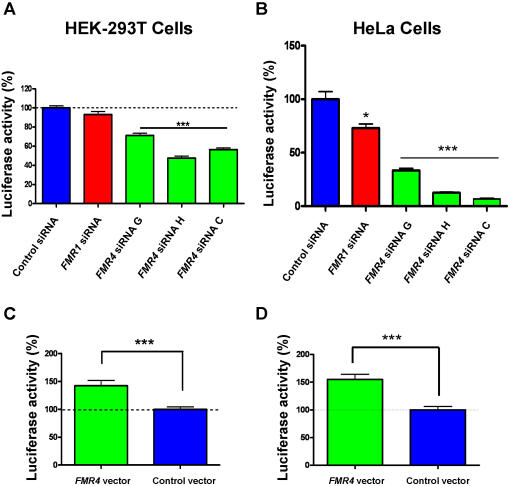
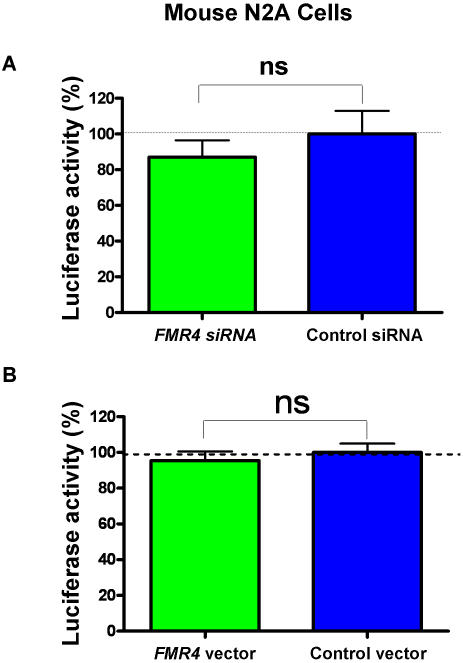
To uncover a possible function for FMR4, we examined the effects of three distinct and efficacious siRNAs against FMR4 on cell proliferation using a luciferase reporter system (see methods). HEK-293T cells were transfected with either siRNAs targeting FMR4 (C, G, and H), FMR1, or a control siRNA. All cells were simultaneously co-transfected with a pGL3 (luciferase) vector. At 72 hours post transfection, luciferase activity, which is a marker of cell proliferation, was measured using an Analyst GT Multimode Reader (Molecular Devices) and all data points were plotted as a percentage of control siRNA treated cells. All three siRNAs against FMR4, but not the siRNA against FMR1, resulted in significant decreases in cell proliferation in comparison to the control siRNA treated cells (Figure 6A). To determine if the effect of FMR4 on cell proliferation is reproducible in other human cell lines we carried out a similar experiment using HeLa cells and found that all three siRNAs against FMR4 also resulted in highly significant decreases in cell proliferation in comparison to control siRNA (P<0.0001) (Figure 6B). In contrast to HEK-293T cells, FMR1 siRNA resulted in a marginally significant decrease in cell proliferation in HeLa cells (Figure 6B). We also carried out a similar experiment using mouse N2A neuroblastoma cells as a negative control experiment and as expected siRNAs against FMR4 had no effect on cell proliferation in mouse N2A cells (Figure 7A). To determine if the overexpression of FMR4 have an opposite effect compared to FMR4 siRNAs knockdown on cell proliferation, we next transfected HEK-293T and HeLa cells with FMR4 overexpression vector. We found that the overexpression of FMR4 in both cell lines resulted in an increase in cell proliferation compared to the control vector treated cells (P<0.0001) (Figure 6C–D). The overexpression of FMR4 in mouse N2A cells had no effect on cell proliferation (Figure 7B).
Figure 6. FMR4 affects proliferation in human cells.
(A) Cell proliferation assay showing that the knockdown of FMR4 via three distinct siRNAs in HEK-293T cells, but not knockdown of FMR1, resulted in decrease in cell proliferation in comparison to cells which are treated with a negative control siRNA. Cell proliferation was measured based on luciferase activity in these cells at 72 hours post siRNA transfection. (B) Cell proliferation assay showing that the knockdown of FMR4 via three distinct siRNAs in HeLa cells resulted in decrease in cell proliferation in comparison to cells which are treated with a negative control siRNA (P<0.0001). (C–D) In both HEK-293T and HeLa cells, overexpression of FMR4 resulted in an increase in cell proliferation in comparison to cells treated with a control vector. Error bars: s.d.
Figure 7. The effect of FMR4 on cell proliferation is not observed in non-primates.
Since FMR4 is a primate-specific transcript we examined its effect on cell proliferation in non-primates using mouse N2A cells. We examined both the siRNA knockdown of FMR4 (as a negative control experiment) and the over-expression of FMR4 on cell proliferation in N2A cells. (A) Mouse N2A cells were transfected with FMR4 siRNA C and a control siRNA. Simultaneously, cells were transfected with pGL3 (luciferase) vector. At 72 hours post transfection luciferase activity was measured and data are graphed as a percentage of control siRNA. (B) Mouse N2A cells were transfected with FMR4 over-expression vector and a control vector (no FMR4 insert). Simultaneously, cells were transfected with pGL3 (luciferase) vector. At 72 hours post transfection luciferase activity was measured and data are graphed as a percentage of control vector. Unlike human cells which show an increase in cell proliferation when transfected with the FMR4 vector, mouse N2A cells did not show any change in proliferation.
FMR4 has antiapoptotic properties
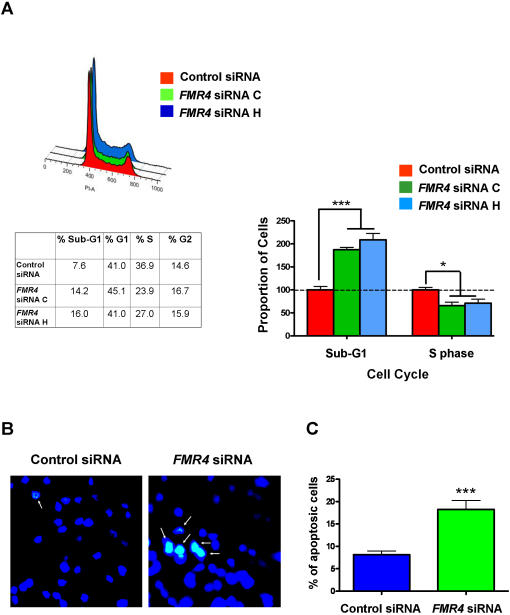
To further characterize the manner in which FMR4 affects cell proliferation, we knocked down FMR4 using two different siRNAs (four repeats each), and we used a control negative siRNA (four repeats also) to examine the effects of FMR4 on the cell cycle in HEK-293T cells. At 72 hours post transfection, we performed propidium iodide FACS analysis (see methods) and found that the siRNAs knockdown of FMR4 resulted in an increase in the number of cells in the Sub-G1 phase and a modest but significant decrease in the number of cells in S phase of the cell cycle (Figure 8A). This suggested that FMR4 may have an antiapoptotic function in human cells; therefore, we next performed a fluorescent TUNEL assay and found a significant increase in apoptosis in cells treated with FMR4 siRNA (72 hours post transfection) compared to cells treated with a control siRNA, further indicating that FMR4 has an antiapoptotic function in human cells (Figure 8B–C).
Figure 8. FMR4 has an antiapoptotic function in human cells.
(A) Cell cycle analysis of control cells (red) and cells treated with two different siRNAs against FMR4 (green and blue) shows that knockdown of FMR4 resulted in a highly significant increase in the number of cells in Sub-G1 and a modest but significant decrease in the number of cells in the S phase suggesting a possible role in apoptosis. (B) Microscope images of cells (DAPI stained) treated with a control siRNA and cells treated with FMR4 siRNA for 72 hours prior to a TUNEL assay. A significant number of cells are undergoing apoptosis (FITC) in the FMR4 siRNA treated cells in comparison to the control siRNA treated cells. (C) Quantification of cells following a TUNEL assay indicated that there is at least a two-fold change in the number of cells undergoing apoptosis in the FMR4 siRNA treated cells in comparison to the control siRNA treated cells. Error bars: s.d.
Discussion
In the present manuscript we report the discovery of a 2.4 kb noncoding RNA (FMR4) which is transcribed upstream of FMR1. There is no overlap between the FMR1 and FMR4 transcripts, and therefore, FMR4 is not a natural antisense transcript to FMR1. FMR4 is expressed in human adult and fetal tissues, and in several regions of human and rhesus monkey adult brain but at varying concentrations. Despite the likelihood that FMR4 shares a bidirectional promoter with FMR1, FMR4 is not expressed in the adult testes, ovary and prostate where FMR1 is highly expressed. It is possible however that FMR4 is expressed in these tissues during embryonic and/or fetal development as the RNAs used in our experiments from these tissues (testes, ovary, and prostate) were obtained from human adults. Notably, we found FMR4 to be highly expressed in fetal heart and kidney. The cardiac expression of FMR4 may possibly be of functional relevance considering the fact that many patients with fragile X syndrome exhibit heart defects such as dilation of the aortic root and mitral valve prolapse [24], [28]. Moreover the high expression of FMR4 in the kidney (Figure 2A) appears consistent with our observations that the human embryonic kidney cell line, HEK-293, also expresses FMR4.
Bioinformatics analysis shows that the genomic sequence encompassing FMR4 is conserved in other primates (chimp and rhesus monkey) with only partial homology to the mouse (Figure S2). Interestingly, however, there is an apparent transcript in the mouse X chromosome that is on the minus strand that starts approximately 100 bp upstream of the mouse Fmr1 gene. This transcript (NCBI accession number AK148387) does not have significant homology with the human FMR4 transcript (∼3% nucleotide identity by ClustalW using default analysis parameters for nucleotide sequences). Furthermore, this mouse transcript appears to be highly spliced and contains 4 exons, which is an additional distinction from the human FMR4 transcript. However, we can not rule out that FMR4 and AK148387, despite their genomic differences, still perform a similar function. The majority of noncoding RNAs identified to date seem to be poorly conserved even among mammals; this is in contrast to other noncoding RNAs (e.g., microRNAs and snoRNAs) which show a high level of conservation among diverse species [9]. It is worth noting that while the human and mouse Xist show 66% homology [29]; however, there seems to be rapid evolution of unique sequences [30]. Also, several attempts to find an orthologue for XIST in marsupials have failed [31], [32] despite the fact that imprinted X inactivation still occurs in marsupials, a process that requires XIST in eutharians [33] suggesting the possibility that a non-conserved noncoding RNA in marsupials may perform a similar function to XIST.
We found that FMR4 regulates human cell proliferation in vitro; knockdown of FMR4 resulted in alterations in the cell cycle and apoptosis while the overexpression of FMR4 leads to an increase in cell proliferation. While the involvement of poorly studied protein coding gene(s) within the FMR1 genomic locus cannot be excluded at this time, our results suggest that the FMR4 function involves a noncoding mechanism. Recently, a long noncoding RNA, similar in size to FMR4, was identified in the HOXC locus (HOTAIR) [13]. The HOTAIR noncoding RNA represses transcription in trans across 40 kb of the HOXD locus by altering the chromatin modifications through enhancement of the PCR2 activity at the HOXD locus [13]. It is therefore possible that FMR4 may also target a set of genes in trans resulting in its antiapoptotic properties. In conclusion, our findings should add further evidence that novel nonconserved noncoding RNAs may be functional and that they do not simply represent “transcriptional noise”.
While our manuscript was under review, Ladd et al. published on an antisense transcript that spans the CGG repeats in the 5′ UTR of FMR1 [34]. This transcript, ASFMR1, is up-regulated in premutation carriers and shut down in fragile X patients similar to FMR1 and FMR4. Also, one splice variant of ASFMR1 overlaps with FMR4, therefore it is possible that FMR4 is nested in the 3′ UTR of ASFMR1, a phenomenon seems to be prevalent in many genes throughout mammalian genomes [1] or another possibility is that these two transcripts may also have contiguous isoforms which is another intriguing possibility. ASFMR1 appears to be highly spliced and has multiple transcription start sites. One transcription start site is in intron 2 of FMR1 at position +10243. This transcription start site was identified by utilizing a 5′ RACE primer in exon 1 of FMR1. Also, multiple transcription start sites for ASFMR1 were also identified in −99 to −208 upstream of FMR1 by designing 5′ RACE primers in the −1000 position relative to FMR1. From these experiments it is not clear if the transcript which starts in the +10243 is the same transcript as the one that initiates in the −99 to −208. If these two transcripts were both ASFMR1, the authors should have been able to clone the longer transcript which initiates at +10243 using the 5′ RACE primers which they used to identify the multiple transcription start sites in the −99 to −208 positions. Therefore, another possibility is that one of the transcripts that Ladd et al. cloned and identified as an alternative transcript to ASFMR1 is the FMR4 transcript. In essence, our findings along with Ladd et al. suggest a complex transcription within the FMR1 locus than previously thought and should provide an incentive for further studies to determine the exact nature of all the transcripts, and their function, throughout the FMR1 region and their relevance to FMR1-associated human disorders (fragile X syndrome and FXTAS).
Materials and Methods
Rapid amplification of cDNA ends (RACE)
The genomic sequence for FMR1 locus was obtained from the UCSC website (http://genome.ucsc.edu/). RACE-ready cDNA (0.5 ng/µl) from SH-SY5Y cells for 5′ and 3′ RACE was custom made by Ambion (Austin, TX). RACE primers were designed using Primer3 software and the name and sequence of the RACE primers for FMR4 is listed here: FMR4 5′ outer 1: TGAGTTGAGGAAAGGCGAGT; FMR4 5′ inner 1: TTGAGATCCCGACTCAATCC. We carried out two rounds of PCR using the outer and inner primers sequentially. First PCR was carried out using 10 µl of RACE-ready cDNA and the second PCR was carried out using 2 µl of the first PCR product. PCR conditions were as follow: 94 C for 5 minutes, (94 C for 30 seconds, 59 C for 30 seconds, 72 C for 2 minutes) for 35 cycles, 72 C for 10 minutes. Both first and second PCR products were ran on a 2% agarose gel and bands of interest were cut, purified, cloned into pGEM T-easy vector (Promega, Madison, WI) before sequencing (UC Davis Sequencing Core Facility).
RNA Extraction and cDNA synthesis
Total RNA was extracted using Quiagen RNeasy mini kit (catalogue # 74106). RNA concentrations were measured using The NanoDrop® ND-1000 UV-Vis Spectrophotometer. Equal amounts of RNA were reversed transcribed using TaqMan reverse transcription reagents (part # N808-0234) according to the manufacturer's protocol.
Real-Time PCR
FMR4 forward primer: ACACCCTGTGCCCTTTAAGG, FMR4 reverse primer: TCAAAGCTGGGTCTGAGGAAAG, Reporter (probe): TCGGGATCTCAAAATGT. Real-Time PCR (RT-PCR) was carried out with the GeneAmp 7900 (Applied Biosystems, Foster City, CA). The PCR reactions contained 20–40 ng cDNA, Universal Mastermix (Applied Biosystems, Foster City, CA), 300 nM of forward and reverse primers, and 200 nM of probe in a final reaction volume of 15 µl. The primers and probe were designed using File-Builder software (Applied Biosystems, Foster City, CA). The PCR conditions were as follows: 50 C for 2 min then 95 C for 10 min then 40 cycles of 95 C for 15 s and 60 C for 1 min. The results are based on cycle threshold (Ct) values. Differences between the Ct values for experimental and reference genes (18srRNA) were calculated as ΔΔCt.
In vitro transcription and translation
1 ug of pcDNA3.1 containing the FMR4 sequence (or without the sequence as a control) was utilized for in vitro transcription using a MAXIscript kit (Cat # AM1200, Ambion, Austin, TX) according to manufacturer's instructions. Subsequently, in vitro translation was carried out using the Retic Lysate IVT kit (Ambion, Austin, TX) according to manufacturer's instructions, except that Fluorotect Green-Lys (Promega, Madison, WI) was included in the translation mixture according to manufacturer's suggestions. Fluorotect Green-Lys, is a BIODPY labeled lysine that allows the addition of a fluorescent amino acid to any newly synthesized peptide (unlabeled lysine is also included in the reaction mixture) allowing for fluorescent detection of such peptides.
Northern blot analysis
We purchased a human ready-to-hybridize northern blot membrane from Ambion (cat# 3141) which has 2 µg of poly(A) RNA per lane isolated from human brain, liver, placenta, small intestine, colon, pancreas, spleen, prostate, testes, and ovary. The membrane was initially incubated with 15 ml of pre-warmed hybridization solution (Ambion, cat#8670) for 1 hour at 65 C. Northern blot probes (32P) were generated using Amersham rediprime II random prime labeling system (RPN1633) according to the manufacturer's protocol (GE Healthcare, Piscataway, NJ). We added 14 µl of the probe per 5 ml of hybridization buffer (42 µl total) to the membrane for overnight incubation at 42 C. Two low-stringency and two high-stringency washes were performed for 15 minutes each prior to exposing the membrane for phosphor imager.
Cell culture, siRNA transfection and RNA isolation
HEK-293T and N2A cells were cultured in MEM plus 10% FBS. Cells in the logarithmic growth phase were transfected with 20 nM of siRNA using 0.2% Lipofectamine 2000 according to manufacturer's instructions (Invitrogen, Carlsbad, CA). Cells were incubated for 72 h or transfected a second time for an additional 72 hours prior to RNA isolation. RNA was isolated using QIAGEN RNeasy mini-kit (QIAGEN, Valencia, CA). All samples were treated with RNAse-free DNase (QIAGEN, Valencia, CA, #79254) for 20 minutes as described in the manufacturer's protocol. The sequence of FMR1 and FMR4 siRNAs are listed in table 1.
FMR4 over-expression
The entire cDNA sequence of FMR4 was cloned into pcDNA3.1 vector. The vector was sequenced to verify the insertion of the FMR4 sequence. The vector was transfected into several human and mouse cell lines using standard procedures.
Stability and α-amanitin treatment
HEK-293T cells were cultured in 6 well plates. Twenty-four h later, cells were treated with 50 µg/ml of α-amanitin (Sigma, St. Louis, MO). Cells were harvested for RNA isolation and RT-PCR at 0, 6, 12, and 24 hours post treatment. Three independent samples were taken for each data point and all samples had untreated matching samples for RNA purification and data analysis.
Cell proliferation assay
Using Multidrop 384Titan, 50 ng of PGL3 vector (luciferase vector with SV40 promoter), 20 nM of siRNA and transfection reagents (Lipofectamine 2000 0.2% and OptiMEM, Invitrogen, CA) were plated in 96 well plates. Equal number of cells (20,000 per well) were added to each well and incubated at 37°C for 72 hours. Bright-Glo luciferase reagent (Promega, Madison, WI) was added to each well and incubated at room temperature for 5 minutes. Luciferase activity, as a marker of cell proliferation, was measured by Analyst GT Multimode Reader (Molecular Devices, Sunnyvale, CA) and plotted against control siRNA.
Cell cycle analysis
We knocked down FMR4 using two different siRNAs (4 repeats each), and we used a control negative siRNA (4 repeats also) to examine the effects of FMR4 on the cell cycle. At 72 hours post transfection, we prepared the cells for flow cytometry as follow: cells were washed with PBS, trypsynized and centrifuged at 1,000 rpm for 10 minutes. Then the cells were washed again with PBS before being fixed with 70% ethanol at −20C overnight. The next day the cells were centrifuged, washed with PBS and re-suspended in 38 mM sodium citrate, 69 µM propidium iodide and 19 µg/ml RNAse A for flow cytometry analysis. Results were analyzed using FlowJo analysis software.
TUNEL assay
HEK-293T cells were plated on cover slips prior to treatment with a control siRNA or siRNAs against FMR4. At 72 hours post transfection, the cells were fixed with 4% paraformaldehyde (pH 7.4) for 15 min at room temperature, followed by permeabilization with 0.1% triton-X in PBS. TdT- mediated dUTP nick end labeling (TUNEL) reaction mixture was added to the cells and incubated at 37C for 1 hour (Roche, Indianapolis, IN). The cells were then stained with DAPI and images were captured using a confocal microscope.
Supporting Information
Northern blot analysis of FMR1 and FMR4 in human adult tissues. FMR1 and FMR4 are co-expressed in some but not all of the human adult tissues examined.
(0.38 MB TIF)
Bioinformatic analysis of the genomic DNA sequence encompassing FMR4.
(0.17 MB DOC)
Acknowledgments
We are grateful to Drs. Flora Tassone and Paul Hagerman (UC Davis) for helpful discussions and their kind gift of total RNA from untransformed leukocytes from normal, premutation and full mutation individuals. We thank Drs. Nagi Ayad and Charles Weissmann (Scripps) for critically reading the manuscript. Diane Stephenson (Pfizer) kindly provided tissues from rhesus monkey brain regions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Conquer Fragile X Foundation and the Scripps Research Institute have provided funding.
References
- 1.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 3.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 8.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 9.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Migeon BR. X chromosome inactivation: theme and variations. Cytogenet Genome Res. 2002;99:8–16. doi: 10.1159/000071568. [DOI] [PubMed] [Google Scholar]
- 13.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, et al. Altered microRNA expression in human heart disease. Physiol Genomics. 2007 doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 16.Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- 17.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 18.Garber K, Smith KT, Reines D, Warren ST. Transcription, translation and fragile X syndrome. Curr Opin Genet Dev. 2006;16:270–275. doi: 10.1016/j.gde.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Oostra BA, Chiurazzi P. The fragile X gene and its function. Clin Genet. 2001;60:399–408. doi: 10.1034/j.1399-0004.2001.600601.x. [DOI] [PubMed] [Google Scholar]
- 20.Hinds HL, Ashley CT, Sutcliffe JS, Nelson DL, Warren ST, et al. Tissue specific expression of FMR-1 provides evidence for a functional role in fragile X syndrome. Nat Genet. 1993;3:36–43. doi: 10.1038/ng0193-36. [DOI] [PubMed] [Google Scholar]
- 21.Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, et al. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oostra BA, Willemsen R. A fragile balance: FMR1 expression levels. Hum Mol Genet. 2003;12 Spec No 2:R249–257. doi: 10.1093/hmg/ddg298. [DOI] [PubMed] [Google Scholar]
- 23.Engstrom PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, et al. Complex Loci in human and mouse genomes. PLoS Genet. 2006;2:e47. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sreeram N, Wren C, Bhate M, Robertson P, Hunter S. Cardiac abnormalities in the fragile X syndrome. Br Heart J. 1989;61:289–291. doi: 10.1136/hrt.61.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tassone F, Hagerman RJ, Taylor AK, Hagerman PJ. A majority of fragile X males with methylated, full mutation alleles have significant levels of FMR1 messenger RNA. J Med Genet. 2001;38:453–456. doi: 10.1136/jmg.38.7.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 27.Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldstein G, Hagerman R. Aortic hypoplasia and cardiac valvular abnormalities in a boy with fragile X syndrome. Am J Med Genet. 1988;30:83–98. doi: 10.1002/ajmg.1320300107. [DOI] [PubMed] [Google Scholar]
- 29.Chureau C, Prissette M, Bourdet A, Barbe V, Cattolico L, et al. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 2002;12:894–908. doi: 10.1101/gr.152902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nesterova TB, Slobodyanyuk SY, Elisaphenko EA, Shevchenko AI, Johnston C, et al. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res. 2001;11:833–849. doi: 10.1101/gr.174901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davidow LS, Breen M, Duke SE, Samollow PB, McCarrey JR, et al. The search for a marsupial XIC reveals a break with vertebrate synteny. Chromosome Res. 2007;15:137–146. doi: 10.1007/s10577-007-1121-6. [DOI] [PubMed] [Google Scholar]
- 32.Hore TA, Koina E, Wakefield MJ, Marshall Graves JA. The region homologous to the X-chromosome inactivation centre has been disrupted in marsupial and monotreme mammals. Chromosome Res. 2007;15:147–161. doi: 10.1007/s10577-007-1119-0. [DOI] [PubMed] [Google Scholar]
- 33.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 34.Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, et al. An Antisense Transcript Spanning the CGG Repeat Region of FMR1 is Upregulated in Premutation Carriers but Silenced in Full Mutation Individuals. Hum Mol Genet. 2007 doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Northern blot analysis of FMR1 and FMR4 in human adult tissues. FMR1 and FMR4 are co-expressed in some but not all of the human adult tissues examined.
(0.38 MB TIF)
Bioinformatic analysis of the genomic DNA sequence encompassing FMR4.
(0.17 MB DOC)