Abstract
Environmental polycyclic aromatic hydrocarbons (PAHs) are metabolically activated to diol epoxides that can react with DNA, resulting in covalent modifications to the bases. The (+)- and (–)-3,4-dihydroxy-1,2-epoxy-1,2,3,4-tetrahydro-benzo[c]phenanthrene (anti-BPhDE) isomers are diol epoxide metabolites of the PAH benzo[c]phenanthrene (BPh). These enantiomers readily react with DNA at the N6 position of adenine, forming bulky (+)-1R- or (–)-1S-trans-anti-[BPh]-N6-dA adducts. Transcription-coupled nucleotide excision repair clears such bulky adducts from cellular DNA, presumably in response to RNA polymerase transcription complexes that stall at the bulky lesions. Little is known about the effects of [BPh]-N6-dA lesions on RNA polymerase II, hence, the behavior of human RNA polymerase II was examined at these adducts. A site-specific, stereochemically pure [BPh]-N6-dA adduct was positioned on the transcribed or non-transcribed strand of a DNA template with a suitable promoter for RNA polymerase II located upstream from the lesion. Transcription reactions were then carried out with HeLa nuclear extract. Each [BPh]-dA isomer strongly impeded human RNA polymerase II progression when it was located on the transcribed strand; however, a small but significant degree of lesion bypass occurred, and the extent of polymerase blockage and bypass was dependent on the stereochemistry of the adduct. Molecular modeling of the lesions supports the idea that each adduct can exist in two orientations within the polymerase active site, one that permits nucleotide incorporation and another that blocks the RNA polymerase nucleotide entry channel, thus preventing base incorporation and causing the polymerase to stall or arrest.
INTRODUCTION
The very environment that provides oxygen, nutrients and water to organisms also exposes them to agents that damage their genetic material. Radiation, natural and industrial chemicals, pollutants, drugs and reactive oxygen species generate damaged bases and breaks in DNA that can potentially interfere with fundamental cellular processes like replication and transcription. Many of these agents are also carcinogenic by virtue of the fact that they damage DNA (1).
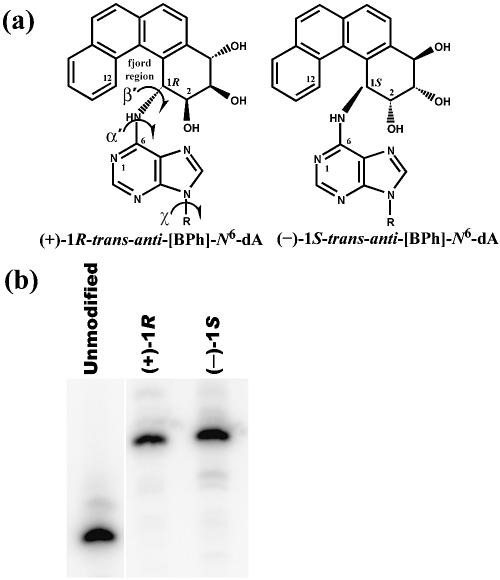
Polycyclic aromatic hydrocarbons (PAHs) are common environmental chemicals that are by-products of incomplete combustion. These substances are often procarcinogens that are metabolically activated to ultimate carcinogens in a stepwise process that occurs in several organs, particularly the liver (2–4). Benzo[c]phenanthrene (BPh), a four-ring, non-planar, fjord region aromatic compound, is metabolized to a number of intermediates, including the diol epoxide 3,4-dihydroxy-1,2-epoxy-1,2,3,4-tetrahydro-BPh (BPhDE), which exists as (+)- and (–)-anti and (+)- and (–)-syn enantiomeric pairs (3,4). While both anti-BPhDE enantiomers are mutagenic (5) and tumorigenic (6), (–)-anti-BPhDE is more mutagenic in mammalian systems (5) and is one of the most tumorigenic PAH diol epoxides (6). Both (+)- and (–)-anti-BPhDE readily react with purines in DNA, primarily attacking adenine at its exocyclic amino group to form the major (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA adducts, respectively (Fig. 1a) (7). High resolution NMR solution studies have yielded detailed structural data for (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA in the 11mer 5′-CTCT CA[BPh]CTTCC-3′, where the A is modified and its partner is T in a complementary oligomer (8,9). The phenanthrenyl group of the (+)-1R lesion intercalates on the 5′-side of the modified A, while that of the (–)-1S lesion intercalates on the 3′-side of the modified base, following the structural principle that adducts with opposite stereochemistry at the carcinogen linkage site are oriented in opposite directions relative to the damaged base (10). In both cases, Watson–Crick base pairing between the damaged base and its partner is somewhat distorted but not disrupted and the DNA helix is stretched and locally unwound. The aromatic rings of (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA twist about the fjord region to avoid steric clash between hydrogens at positions 1 and 12 of the BPh moiety (Fig. 1a). This twist is in opposite directions in each stereoisomer in order to maximize stacking with the adjacent DNA bases pairs. The structural data illustrate a general principle: the stereochemistry of a PAH–DNA adduct affects its three-dimensional orientation in DNA, and the orientation can profoundly affect the physiological consequences of the lesion (10).
Figure 1.
(a) Structure of (+)-1R-trans-anti-[BPh]-N6-dA and (–)-1S-trans-anti-[BPh]-N6-dA. The glycosidic torsion angle χ (O4′-C1′-N9-C5) is labeled, as well as α′ (N1-C6-N6-C1[BPh]) and β′ (C6-N6-C1[BPh]-C2[BPh]), the torsions governing the orientation of the BPh moiety. (b) PAGE analysis of the 11mers used in template construction. The contents are marked above each lane.
DNA polymerases behave in a variety of ways when they encounter damaged sites in the genome. Some lesions primarily block the progression of replicative DNA polymerases (10,11), a situation that often stimulates recombinagenic events (12). However, in some cases, translesion DNA polymerases may be employed to bypass the lesions, sometimes at the expense of fidelity, resulting in point and frameshift mutations (13). The mutagenicity of all four isomers of BPhDE, including (+)- and (–)-anti-BPhDE, was investigated using a shuttle vector randomly damaged by each agent and replicated in human cells, and most of the mutations observed were transversions, predominantly GC→TA and AT→TA. This mutational preference reflects the propensity of BPhDE to react primarily with G and A in DNA, as well as the tendency of these adducts to mispair with adenine during replication (14). A strand bias in mutations was also observed for racemic anti-BPhDE, with the non-transcribed strand giving rise to most of the mutations (15).
Cells have evolved a variety of genomic maintenance mechanisms that greatly reduce the deleterious effects of damaged DNA and help to avert mutagenic events during replication (1). Among these are nucleotide excision repair (NER), which is a pathway that facilitates the removal of bulky damaged bases from DNA, and base excision repair (BER), which tends to clear small, modified bases and base fragments from the genome. A third pathway, transcription-coupled DNA repair (TCR), removes damaged bases from the transcribed strand of actively expressed genes (16). The precise mechanism of TCR remains to be elucidated, but there appears to be overlap between it and other repair pathways, particularly NER, although the relationship of TCR to BER is less clear (17,18).
DNA damage recognition appears to occur differently for NER, BER and TCR (19,20). In humans, the XPC/HR23B heterodimer facilitates DNA damage recognition during NER (19). XPC is mutated in some patients with the disease xeroderma pigmentosum (XP) and NER is absent in these individuals, causing sun sensitivity and increased occurrence of skin cancer (1). Damage recognition during BER is facilitated by specific glycosylase enzymes that recognize the modified base. During TCR, RNA polymerase likely acts as the recognition factor of the damage, presumably by stalling at the damaged site on the template strand, an event that then acts as a signal to recruit the remaining repair factors (16,19).
Previous studies have shown that a wide variety of DNA lesions, such as thymine dimers, can impede transcription elongation by bacteriophage and mammalian RNA polymerases (21–23). Similarly, it has been shown that (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA impede T7 RNA polymerase elongation, resulting in a set of truncated transcripts in the vicinity of the adduct. Furthermore, the truncated transcripts often end with guanosine incorporated opposite the modified base rather than uridine, suggesting that translocation past a modified base in a DNA template might be a function of the base that is inserted (24).
Human RNA polymerase II is far more complex than bacteriophage RNA polymerases in that it relies on an array of transcription factors that influence promoter recognition and transcription initiation, elongation and termination (25). Little is known about the behavior of human RNA polymerase II at BPhDE lesions in DNA. Considering that there is a strand bias in mutagenesis for BPhDE adducts in DNA (15) that may well result from TCR operating on these lesions (26), it was of interest to assess the behavior of RNA polymerase II during transcription elongation past BPhDE lesions in DNA.
MATERIALS AND METHODS
Materials
Restriction endonucleases and enzymes were purchased from New England Biolabs Inc. (Beverly, MA) and Promega Corp. (Madison, WI). HeLaScribe® Nuclear Extract, Streptavidin Magnesphere Paramagnetic Particles (SA-PMPs), nucleotides, pCI-NeoI and DNA markers were also purchased from Promega. Radioactively labeled compounds were obtained from Perkin Elmer Inc. (Boston, MA). Materials for electrophoresis, chromatography supplies and plasmid preparation kits were obtained from Bio-Rad Laboratories (Hercules, CA) and Qiagen (Valencia, CA). Chemical reagents and supplies for bacterial growth medium were procured from Fisher Scientific (Suwanee, GA) and Sigma-Aldrich (St Louis, MO). All unmodified oligodeoxynucleotides were synthesized by Sigma-Genosys (The Woodlands, TX).
Preparation and purification of transcription templates
A plasmid designated pCI-Neo-G-less was utilized to prepare template DNA for the transcription reactions. pCI-Neo-G-less contained the cytomegalovirus (CMV) immediate-early enhancer/promoter element and a G-less cassette; a detailed description of its preparation has been published, as has the preparation of site-specifically modified DNA templates for transcription by human RNA polymerase II (27). pCI-neo-G-less was digested with BbsI for 2 h at 37°C in 10 mM Tris–HCl (pH 7.4), 50 mM NaCl, 10 mM MgCl2 and 1 mM dithiothreitol (DTT), resulting in a non-palindromic 5′-overhang located 256 bases downstream from the +1 transcription start site. BbsI was heat-inactivated at 65°C for 20 min.
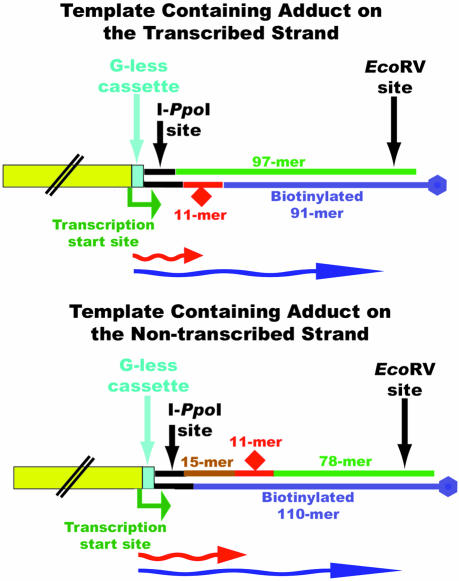
The method for preparing a DNA template containing the site-specifically modified base on the transcribed strand is illustrated in Figure 2, and the protocol has been published (27). The 11mer 5′-CTCTCACTTCC-3′ with a BPhDE attached to the N6 position of the sole adenine served as the source of a site-specific, stereochemically pure adduct (28). The oligomers used to prepare the template DNA were phosphorylated at the 5′-end with 10 U of T7 polynucleotide kinase and 1 mM ATP in 10 µl of 70 mM Tris–HCl (pH 7.6), 10 mM MgCl2 and 5 mM DTT. Equimolar amounts of phosphorylated 97mer, BPhDE-modified or unmodified 11mer and a 91mer that contained a biotin tag on the 5′-end were mixed. The oligomers were annealed by heating the mixture to 90°C and then cooling slowly to 4°C. The annealed oligomers were ligated to BbsI-digested pCI-neoG-less using 2000 U of T4 DNA ligase in 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2 10 mM DTT, 1 mM ATP and 25 µg/ml BSA for 16 h at 16°C.
Figure 2.
Schematic diagram of the DNA templates used in transcription assays. The CMV immediate-early enhancer/promoter is shown in yellow and the EcoRV site is used to cut the templates off the streptavidin paramagnetic particles during purification. The red and blue wavy arrows represent the truncated and full-length transcripts, respectively.
To construct the templates containing the DNA damage on the non-transcribed strand, the modified and unmodified 11mers, a 15mer and a 78mer were phosphorylated, subsequently annealed to a biotinylated 110mer and ligated to linear pCI-neo-G-less as shown in Figure 2 (27).
To purify the template, the ligation reaction products were incubated with SA-PMPs. The beads were washed three times with 500 µl of 0.5× SSC (75 mM NaCl, 7.5 mM sodium citrate, pH 7.0) to remove any impurities in the storage buffer. Non-specific DNA-binding sites on the particles were blocked by incubation with 1 mg/ml salmon sperm DNA for 15 min at room temperature in a final volume of 500 µl. Excess salmon sperm DNA was removed by washing three times with 500 µl of 0.5× SSC. The template reaction mix was then incubated with the SA-PMPs for 1 h at room temperature with agitation. The SA-PMPs were pelleted, the supernatant was removed and the pellet was washed three times with 500 µl of wash buffer (100 mM NaCl, 50 mM Tris–HCl pH 7.9, 10 mM MgCl2, 1 mM DTT). This was followed by two successive digestions with 100 U BglII for 2 h each at 37°C. BglII cuts immediately upstream from the CMV enhancer/promoter, thus removing ∼4500 bp of the vector that are irrelevant for the transcription reaction. Finally, the template was removed from the beads by digesting with 100 U EcoRV. Template integrity was confirmed by 7% PAGE analysis in the presence of 8 M urea.
In vitro transcription
Multiple round transcription reactions were carried out using HeLa nuclear extract. The reaction contained 5 mM Tris–HCl (pH 7.8), 6.4 mM HEPES, 44 mM KCl, 3 mM MgCl2, 88 µM EDTA, 9.4% glycerol, 60 µM phenylmethylsulfonyl chloride, 220 µM DTT, 400 µM each ATP, GTP and UTP, 16 µM [α-32P]CTP (25 Ci/mmol), 200 ng (0.1 pmol) DNA template and 8 U HeLa nuclear extract. The samples were incubated at 30°C and the reactions were stopped by the addition of 175 µl of HeLa extract stop solution (0.3 M Tris–HCl (pH 7.4), 0.3 M sodium acetate, 0.5% SDS, 2 mM EDTA, 1 µg/ml tRNA). Protein was removed by phenol:chloroform:isoamyl alcohol (25:24:1 v/v/v) extraction. Nucleic acids were isolated by ethanol precipitation and dried under vacuum with low heat for 1 h. The nucleic acids were resuspended in RNase-free water with loading dye (98% formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue), denatured at 90°C for 10 min and resolved using 7% denaturing PAGE with 8 M urea at 2000 V for ∼2 h. The gel was dried and exposed to a phosphorimager screen. ImageQuant software (Molecular Dynamics; Piscataway, NJ) was used to quantify the radiolabeled transcripts.
Single round transcription assays were performed at 30°C as described for multiple round assays, but the reactions did not contain GTP. After a 10 min incubation, heparin was added to a final concentration of 80 µg/ml, the samples were heated at 30°C for 10 min and GTP was added to a final concentration of 400 µM. The transcription reaction was resumed for another 40 min and the samples were analyzed as described for multiple round transcription reactions.
In order to determine the molar quantity of RNA produced, the values obtained from the imaging software were related to the radioactivity incorporated into the nascent RNA. A range of dilutions of radiolabeled RNA was resolved by denaturing PAGE. The intensity of each band in the gel was measured using ImageQuant software. Each band of RNA was then excised from the gel and the radioactivity in each gel slice was quantified by scintillation counting (LS3800 Scintillation Counter; Beckman Instruments, Fullerton, CA). A linear correlation between radioactivity and phosphorimager band intensity was observed and this relationship, along with the [32P]phosphate specific activity, was used to determine the amount of transcript produced.
Molecular modeling of BPhDE-modified DNA in eukaryotic RNA polymerase II
The X-ray crystal structure of Saccharomyces cerevisiae RNA polymerase II complexed to a 13 base template DNA and 9 base RNA transcript (PDB i.d. 1I3Q) (29,30) was used as the starting structure for models containing (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA. Molecular modeling was carried out using the BUILDER module of InsightII (Accelrys, San Diego, CA). The template base was remodeled as adenine and the (+)-1R- and (–)-1S-trans-anti-BPhDE were docked to the N6 position of the template base. The carcinogen moiety structure was obtained from the high resolution NMR data found for each adduct when it is present in duplex DNA (8,9).
For each adduct, two conformations were modeled, one that could cause polymerase blocking by preventing nucleotide incorporation opposite the adduct, which we called the blocking conformation, and one that could allow transcriptional bypass of the adduct, which we referred to as the bypass conformation. For the blocking conformations, the uridine partner to the adduct was removed and the transcript was terminated one base prior to the damaged base. This structure was used to model the adduct in a conformation that would block the entrance of a nucleotide into the active site opposite the [BPh]-dA adduct. In the bypass conformations, the partner nucleotide was retained to model a conformation that would allow the persistence of a Watson–Crick base pair between the damaged adenine and its partner uridine. In each case, the torsion angles α′ and β′ (Fig. 1a) that govern the position of the carcinogen moiety were adjusted to fit the carcinogen moiety within the active site of the polymerase with minimal steric crowding.
RESULTS
[BPh]-dA adducts are stable in site-specifically modified DNA oligomers
In order to investigate transcription past site-specifically modified DNA templates, the lesion to be studied must be stable. The BPhDE-modified 11mers used in this work were prepared and purified as reported elsewhere (28). The BPhDE-modified and unmodified oligodeoxynucleotides exhibit different mobilities during electrophoresis, with the bulky carcinogen moiety causing the damaged molecules to migrate more slowly than their undamaged counterparts. To confirm the purity of the modified oligomers that contained the (+)-1R- or (–)-1S-trans-anti-[BPh]-N6-dA adduct, each 11mer was labeled with [32P]phosphate at the 5′-end and resolved using denaturing 20% PAGE. The results are presented in Figure 1b. Oligodeoxynucleotides with (+)-1R-trans-anti-[BPh]-N6-dA contained no detectable unmodified oligomer. The 11mer modified with (–)-1S-trans-anti-[BPh]-N6-dA was contaminated with a trace amount of material that had a mobility similar to the unmodified 11mer, but the quantity of the undamaged oligodeoxynucleotide was less than 0.02% of the total sample.
BPhDE-modified and unmodified DNA templates were synthesized
Stereochemically pure (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA adducts were site-specifically incorporated into DNA templates downstream from the human CMV immediate-early enhancer/promoter element (27). This promoter was chosen because it is a strong, constitutively active promoter for human RNA polymerase II (31). In addition, the same promoter was supplied with the HeLaScribe® Nuclear Extract In Vitro Transcription System and, thus, a comparison with the manufacturer’s control was feasible.
During promoter escape, the RNA polymerase II transcription complex is structurally unstable, resulting in the production of very short, abortive transcripts that would interfere with the study if the lesion were placed too close to the transcription start site. Hence, the adduct was incorporated 266 bases downstream from the +1 transcriptional start site, sufficiently far from the promoter to prevent abortive transcripts from interfering with analysis, as well as to ensure that the polymerase was in elongation mode when it encountered the adduct. Furthermore, the lesion was placed far enough upstream from the 5′-end of the transcribed strand so that truncated transcripts arising from the polymerase stalling at the lesion could be easily distinguished from full-length, run-off transcripts.
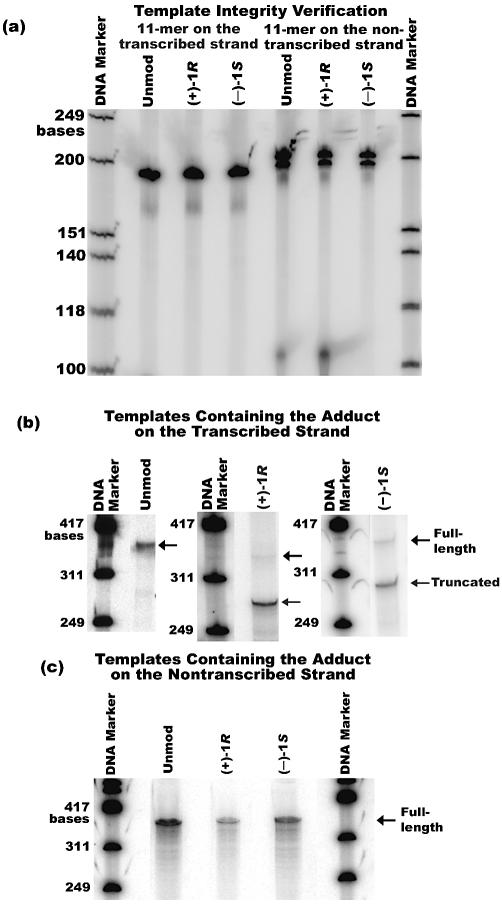
A schematic diagram of the assembled templates containing the 11mer on the transcribed or non-transcribed strand is shown in Figure 2. Following template synthesis, I-PpoI digestion was used to gauge template integrity. When a DNA adduct was located on the transcribed strand and ligation proceeded to completion, digestion with I-PpoI produced discernable fragments 189 and 193 bases in size; in contrast, when ligation was incomplete, I-PpoI digestion produced DNA fragments that were 110 and 118 bases in length. Similarly, when the adduct was placed on the non-transcribed strand, I-PpoI digestion yielded fragments that were 200 and 204 bases in length when complete ligation occurred, but gave fragments that were 110 and 118 bases long when ligation was incomplete. Synthesis of unmodified DNA templates was checked for proper ligation in the same way. Figure 3a shows the results for I-PpoI digestion of unmodified templates, as well as those containing an adduct on the transcribed or non-transcribed strand. No trace of unligated promoter was present in templates containing the damaged base on the transcribed strand or in their unmodified control counterparts. Likewise, the template with the (–)-1S adduct located on the non-transcribed strand was fully ligated, as evidenced by the absence of a band in the vicinity of 110–118 bases. For the (+)-1R and unmodified templates with the 11mer on the non-transcribed strand, trace amounts of unligated promoter were observed; however, the unligated contaminants in these samples were less than 0.01% of the total.
Figure 3.
(a) PAGE analysis of the ligated templates used in the transcription assays following digestion with I-PpoI. The unligated contaminants in the samples containing adducts on the non-transcribed strand were <0.01% and they did not result in any detectable truncated transcripts, as indicated by transcription assays shown in (c). (b) PAGE analysis of 60 min multiple round transcription reactions using templates containing adducts on the transcribed strand, as well as the control template. (c) PAGE analysis of 60 min multiple round transcription reactions using templates containing adduct on the non-transcribed strand, as well as the control template. The contents are marked above each lane and the positions of truncated and full-length transcripts, when produced, are marked with an arrow on the right in (b) and (c).
The (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA adducts impede RNA polymerase II elongation when located on the transcribed strand of the template
In order to examine RNA polymerase II behavior at [BPh]-dA lesions in DNA templates, in vitro transcription assays were performed with HeLa cell nuclear extract. Transcription of templates containing either a (+)-1R- or (–)-1S-trans-anti-[BPh]-N6-dA lesion on the transcribed strand resulted in the formation of both truncated and full-length run-off RNA, although the smaller, truncated species was far more abundant than the full-length polymer (Fig. 3b). Transcription of a DNA sequence-equivalent unmodified control template resulted in the formation of full-length, run-off transcripts and no truncated RNA. The RNA bands were quantified and the relative lesion bypass was calculated from these values (Table 1). Errors are reported as standard error of the mean for 38 trials in the case of the (+)-1R adduct and 30 trials for the (–)-1S isomer. These data indicate that the stereochemistry of the adduct–DNA linkage has a modest but reproducible influence on the behavior of the transcription machinery at [BPh]-N6-dA lesions.
Table 1. Percent bypass of BPhDE adducts in multiple and single round studies.
| Adduct | Multiple round bypass (%) | Single round bypass (%) |
|---|---|---|
| (+)-trans-anti-[BPh]-N6-dA | 8.3 ± 0.1 | 9.1 ± 0.7 |
| (–)-trans-anti-[BPh]-N6-dA | 13.7 ± 0.3 | 22.3 ± 0.3 |
The results showing that (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA lesions pose strong blocks to transcription when located on the transcribed strand lead directly to the following question: do such lesions impede transcription elongation when present on the non-transcribed strand of template DNA? To test this, templates were assembled with site-specific (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA lesions situated on the non-transcribed strand. PAGE analysis from transcription assays with templates containing the adduct on the non-transcribed strand are shown in Figure 3c. No detectable truncated transcripts were observed when either (+)-1R- or (–)-1S-trans-anti-[BPh]-N6-dA was present in the template, demonstrating that neither lesion poses a block to RNA synthesis when it is located on the non-transcribed strand.
The (–)-1S-trans-anti-[BPh]-N6-dA stereoisomer is bypassed to a greater extent by human RNA polymerase II in single round relative to multiple round transcription experiments
Multiple rounds of transcription can result from re-initiation events on a given DNA template and RNA polymerase complexes can carry out a second round of transcription during a given experiment, or a combination of both can occur (32,33). In fact, DNA templates can be transcribed by more than one transcription complex at a time in in vitro assays utilizing RNA polymerase II (34). Hence, the effect of (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA on RNA synthesis was determined under conditions when RNA polymerase II could initiate only a single round of transcription. To achieve this, transcription was initiated with a ribonucleotide mix that lacked GTP; as a consequence, RNA polymerase II initiated transcription and elongated through the G-less cassette until the first cytosine was reached in the transcribed strand. Heparin, which is a negatively charged polymer that binds tightly to free RNA polymerase II but does not interact with polymerases that are part of elongation complexes, was added to prevent transcription initiation by unbound RNA polymerase (35–37). After the addition of heparin, GTP was added to the reaction and complexes that were stalled at the first cytosine continued to elongate, but once they were released, the free polymerases were bound by heparin, thus preventing re-initiation events.
The results for single round transcription experiments are shown in Table 1. A single round of transcription past (+)-1R-trans-anti-[BPh]-N6-dA on the transcribed strand gave results similar to those observed under multiple round conditions. In contrast, (–)-1S-trans-anti-[BPh]-N6-dA posed less of a barrier to elongating RNA polymerase II complexes during a single round of transcription when compared to multiple rounds. These data also indicate that RNA polymerase II is able to bypass (–)-1S-trans-anti-[BPh]-N6-dA lesions more easily than (+)-1R-trans-anti-[BPh]-N6-dA lesions on the transcribed strand of a DNA template. In addition, the (–)-1S-trans-anti-[BPh]-N6-dA adduct is bypassed a greater percentage of time in the single round experiment than in multiple rounds.
Time–course experiments show that (+)-1R-trans-anti-[BPh]-N6-dA blocks RNA polymerase II elongation much more readily in the early rounds of transcription than does the (–)-1S isomer
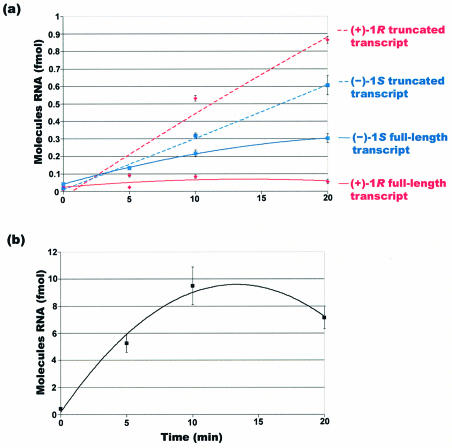
Time–course experiments for multiple rounds of transcription were carried out to measure nascent RNA accumulation over time and lend insight into time-dependent changes in bypass of each adduct. The transcription time–course experiment was performed as described earlier for multiple rounds of RNA synthesis. For these studies, [32P]phosphate-radiolabeled ΦX174-HinfI DNA was added to the reaction mix as a standard to control for potential gel loading errors during PAGE analysis. During incubation, an aliquot of the reaction mix was removed at the desired time point and the resulting transcripts were analyzed as described earlier.
The average quantities of RNA obtained in eight separate trials are given in Table 2. Figure 4 shows the time–course results plotted as a best fit interpolation of the data normalized for potential loading errors using the internal DNA marker. During the first 20 min, multiple rounds of transcription with a template containing (+)-1R-trans-anti-[BPh]-N6-dA resulted in a steady increase in RNA, both as truncated molecules and the full-length, run-off species. However, the accumulation of full-length RNA was far less than that observed for the truncated species, further supporting the notion that this stereoisomer poses a strong block to transcription elongation. In contrast, reactions performed with (–)-1S-trans-anti-[BPh]-N6-dA showed that both truncated RNA and full-length transcripts were formed almost equally during the first 20 min of transcription, indicating that RNA polymerase II was able to bypass the lesion more easily during the first few rounds of transcription.
Table 2. Amount of transcripts produced during time–course experiments.
| Time(min) | Control full-length transcript (fmol) | (+)-1R truncated transcript (fmol) | (+)-1R full-length transcript (fmol) | (–)-1S truncated transcript (fmol) | (–)-1S full-length transcript (fmol) |
|---|---|---|---|---|---|
| 0 | 0.42 ± 0.11 | 0.01 ± 0.00 | 0.03 ± 0.01 | 0.02 ± 0.00 | 0.04 ± 0.01 |
| 5 | 5.3 ± 0.7 | 0.09 ± 0.01 | 0.02 ± 0.01 | 0.13 ± 0.00 | 0.14 ± 0.01 |
| 10 | 9.5 ± 1.4 | 0.53 ± 0.02 | 0.09 ± 0.01 | 0.32 ± 0.02 | 0.22 ± 0.02 |
| 20 | 7.1 ± 0.8 | 0.87 ± 0.02 | 0.06 ± 0.01 | 0.61 ± 0.06 | 0.30 ± 0.02 |
Figure 4.
Best fit linear analysis of the time–course experiments using (a) templates containing the adduct on the transcribed strand and (b) the control template. The amount of RNA reported was normalized for possible loading errors.
The (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA adducts modeled into the active site of eukaryotic RNA polymerase II in both the bypass and blocking conformations
The (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA adducts were modeled into the active site of S.cerevisiae RNA polymerase II (29) to assist in elucidating why these adducts block transcription elongation in some cases and permit bypass in others. The modeling of the adducts was primarily guided by the steric constraints of the enzyme active site, together with the preferred domains for the important torsion angle β′ (Fig. 1a), which primarily governs the orientation of the carcinogen moiety in each stereoisomer (38,39). The torsion angles for the [BPh]-N6-dA orientations are given in Table 3. The most favored regions for the β′ torsion when the damaged adenine base is situated in the anti conformation are +90 ± 25° and –90 ± 25° for the (+)-1R and (–)-1S adducts, respectively (38,39).
Table 3. Torsion angles governing the orientation of the BPhDE moiety in the modeled structuresa.
| Adduct | Bypass conformation | Blocking conformation | ||
|---|---|---|---|---|
| α′ (°) | β′ (°) | α′ (°) | β′ (°) | |
| (+)-1R-trans-anti-[BPh]-N6-dA | 217 | 92 | 103 | 114 |
| (–)-1S-trans-anti-[BPh]-N6-dA | 179 | 245 | 295 | 257 |
aχ values remain unchanged from those in the RNA polymerase crystal structure.
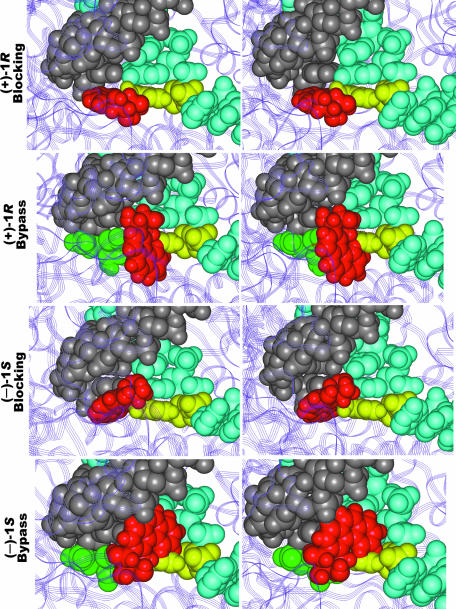
Figures 5 and 6 show the resulting structures for [BPh]-N6-dA in the yeast RNA polymerase II active site. In the modeled blocking conformation, the BPhDE moiety for the (+)-1R-trans-anti-[BPh]-N6-dA adduct is intercalated from the 5′-side of the damaged adenine, similar to that observed in the high resolution NMR structure (8), occupying the space in the active site that would otherwise be filled by the partner of the template base, thus preventing further RNA elongation. In the bypass conformation model for (+)-1R-trans-anti-[BPh]-N6-dA, the phenanthrenyl moiety resides on the major groove side of the damaged adenine and its partner uridine, the aromatic rings point towards the 5′-side of the damaged adenine and the Watson–Crick hydrogen bonding between the damaged adenine and its partner uridine remains undisturbed.
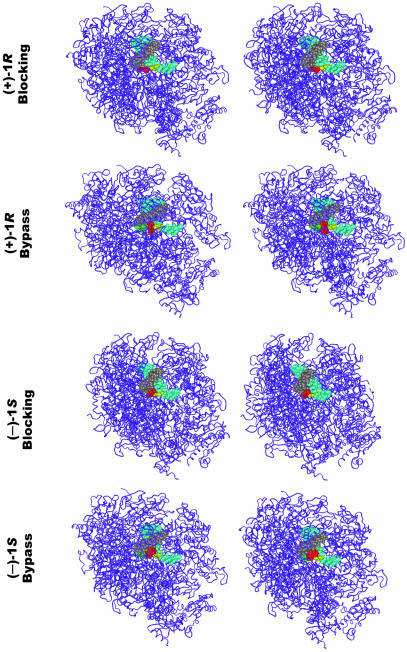
Figure 5.
Stereo views of molecular models of the (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA adducts within the active site of S.cerevisae RNA polymerase II (29) in both the proposed blocking and the bypass-permissive conformations. The identity of the model is given on the left-hand side. The figure was made for viewing with a stereo viewer. Color code: purple (ribbon model), protein; gray, RNA; cyan, DNA template; yellow, damaged adenine; red, BPhDE moiety; green, thymine partner to modified adenine in bypass conformations.
Figure 6.
Close-up stereo views of the bypass and blocking conformations of (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA adducts within the active site of S.cerevisae RNA polymerase II. The identity of the model is given on the left-hand side. The figure was made for viewing with a stereo viewer. Color code: purple (ribbon model), protein; gray, RNA; cyan, DNA template; yellow, damaged adenine; red, BPhDE moiety; green, thymine partner to modified adenine in bypass conformations.
In contrast to the (+)-1R-trans-anti-[BPh]-N6-dA blocking conformation, the phenanthrenyl moiety in the model (–)-1S-trans-anti-[BPh]-N6-dA blocking conformation intercalates from the 3′-side of the damaged adenine, as in the solution structure (9). However, similar to the (+)-1R isomer, the phenanthrenyl moiety of 1S-trans-anti-[BPh]-N6-dA also occupies the space that would otherwise accommodate a partner nucleotide opposite the template base, thus thwarting further RNA synthesis. Similarly, the phenanthrenyl moiety of the modeled (–)-1S-trans-anti-[BPh]-N6-dA bypass conformation is oriented opposite to the (+)-1R-trans-anti-[BPh]-N6-dA in its bypass conformation, with the BPhDE moiety also in the major groove of the active site heteroduplex, but with its phenanthrenyl group pointing towards the 3′-side of the damaged adenine. This conformation of (–)-1S-trans-anti-[BPh]-N6-dA also permits intact Watson–Crick hydrogen bonding between the damaged adenine and its partner uridine, like that observed for the (+)-1R adduct.
DISCUSSION
(+)-1R- or (–)-1S-trans-anti-[BPh]-N6-dA on the transcribed strand of a DNA template partially blocks human RNA polymerase II elongation
Current models propose that RNA polymerase II arrested at a DNA adduct initiates TCR of the lesion. The work presented here shows that (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA lesions located on the transcribed strand of a DNA template pose significant blocks to human RNA polymerase II elongation, although a small amount of lesion bypass is observed. These results are consistent with the finding that [BPh]-induced lesions are preferentially cleared from the transcribed strand of active genes in CHO cells, an event that has been linked to strand-biased mutations that are associated with exposure to this agent (26).
A number of DNA adducts significantly block RNA polymerase II elongation, including cyclobutane pyrimidine dimers (23) and adducts derived from benzo[a]pyrene diol epoxide (BPDE) (40). We report here that (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA adducts can be added to the list of lesions that impede transcription elongation. However, unlike (+)-1S- and (–)-1R-trans-anti-N2-BPDE-dG, which pose absolute blocks to human RNA polymerase II (40), (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA allow some bypass, which could be attributed to the different carcinogen–base linkage site, carcinogen topology and/or conformational preferences of the adducts. If an adduct were to block RNA polymerase II less readily, it may be repaired more slowly by TCR and persist longer in the genome, which increases the likelihood that it will disrupt transcription and/or replication, causing aberrant transcripts or mutations. Decreased processing of trans-anti-[BPh]-N6-dA adducts by TCR could also contribute to the high tumorigenicity of BPh as compared to other PAHs. In addition, greater bypass of (–)-1S-trans-anti-[BPh]-N6-dA by RNA polymerase II, compared to the (+)-1R isomer, may contribute to the even greater tumorigenicity of (–)-anti-[BPh] when compared to (+)-anti-[BPh](6).
In prior reports we have shown that the stereochemistry of PAH-derived DNA adducts has profound consequences on their ability to block transcription (24,40,41), an observation that the data reported here further support. (–)-1S-trans-anti-[BPh]-N6-dA permits more bypass than does its (+)-1R counterpart. If the polymerase complex associates tightly with [BPh]-dA lesions, which would be analogous to results observed for RNA polymerases that stall at cyclobutane pyrimidine dimers (42), the penalty to the cell would be severe. In fact, it has been proposed that TCR has evolved to assist the cell in coping with the potential consequences of stalled transcription elongation complexes that could interfere with gene expression (16,43,44). We propose that the RNA polymerase complexes that succeed at bypassing a DNA lesion would dissociate from the template DNA, making RNA polymerase and other transcription factors in the complex available for additional rounds of transcription. In contrast, adducts that block transcription would titrate out such complexes and the associated factors unless the stalled polymerases were released and the damage was repaired.
The [BPh]-dA lesions studied here do not interfere with transcription elongation when the adduct is located on the non-transcribed strand. In contrast, lesions derived from BPDE adducts, such as (+)-10S- and (–)-10R-trans-anti-BPDE-N2-dG, enhance the ability of innate pause sites to inhibit RNA polymerase II elongation. (40). While BPDE lesions may increase pausing because the carcinogen moiety interacts with the polymerase enzyme far removed from the active site (40), it is possible that the tendency of the [BPh]-N6-dA carcinogen ring system to shield its aromatic rings between the damaged adenine and its adjacent base may preclude its interaction with the polymerase protein when it is located on the non-transcribed strand.
The effects of trans-anti-[BPh]-N6-dA adducts on RNA polymerase II elongation is of particular interest because these lesions are poor substrates for NER, leaving TCR as an important pathway for their removal (26,45). (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA lesions induce relatively less perturbations to double-stranded DNA compared with the analogous adducts derived from benzo[a]pyrene (46); this fact, coupled with the absence of damaged duplex destabilization and optimized stacking of the stereoisomeric adducts with the DNA, explains why NER recognizes these lesions poorly when they are present in the genome (46,47).
It is unlikely that the effects of the [BPh]-dA adducts on transcription are a result of adduct instability. If the phenanthrenyl moiety had been cleaved from the oligomer, leaving an intact adenine at the site, transcriptional bypass of the adducts on the transcribed strand would be observed. However, our data suggest that this is improbable because [BPh]-dA lesions are still present on oligomers that have been heated and denatured, as shown in Results. Furthermore, the kinetic data for transcription past the (–)-1S isomer on the transcribed strand support the notion that the adduct is stable. Single rounds of transcription permit more read-through past the lesion than do multiple rounds. If such bypass were due to lesion instability during the reaction, the relative amounts of full-length transcript should increase over time, not decrease as our data indicate.
The conformational preferences of stereoisomeric (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA adducts explain their effects on RNA polymerase II elongation
Molecular modeling of (+)-1R- and (–)-1S-trans-anti-[BPh]-N6-dA into the active site of S.cerevisiae RNA polymerase II (29) assisted in our elucidation of why these two adducts pose partial blocks to transcription, most often blocking elongation but still permitting a measurable amount of read-through. The phenanthrenyl moiety in both the (+)-1R and (–)-1S adducts can be accommodated within the RNA polymerase active site. In the blocking conformations, the phenanthrenyl moieties are shielded from solvent through interactions with the RNA polymerase active site amino acid residues and the DNA:RNA heteroduplex, and they protrude toward the nucleotide entry channel, thus acting as a physical barrier to NTP entrance into the active site and impeding RNA chain growth. These conformations exist with minimal steric crowding and are reminiscent of those adopted by each adduct in duplex DNA, with the phenanthrenyl ring system intercalated toward the 5′-side of the modified adenine in the (+)-1R adduct and toward the 3′-side of the modified adenine in the (–)-1S adduct (8,9).
In contrast to the blocking conformations, the modeled bypass conformations permit ribonucleotide incorporation into the transcript opposite the damaged base in the template DNA. In the bypass conformation, the phenanthrenyl rings reside on the major groove side of the heteroduplex, but the modified adenine is exposed in such a way as to be available for hydrogen bonding with an incoming UTP. This model leads to the prediction that full-length transcripts should contain uracil in the position that the [BPh]-N6-dA adduct encodes. Preliminary studies from our laboratory using RT–PCR to sequence the full-length transcripts suggest that this is indeed the case.
It is likely that the more solvent-exposed surface area of the hydrophobic phenanthrenyl moiety that is found in the bypass conformations contributes to the factors that make this conformation less favorable than the blocking intercalated conformation. It is also plausible that the (+)-1R adduct less frequently adopts the bypass conformation compared to the (–)-1S adduct because the 5′-oriented phenanthrenyl moiety of the (+)-1R isomer encounters significant steric crowding with protein residues. In contrast, there are no protein residues with which the (–)-1S adduct must contend and the phenanthrenyl moiety encounters a much less sterically crowded region towards the 3′-side of the modified adenine. Therefore, the more permissive accommodation of (–)-1S-trans-anti-[BPh]-N6-dA in the major groove conformation could account for the more frequent bypass of this adduct by RNA polymerase II. Hence it is possible that an intercalated adduct conformation can permit base accommodation opposite the damaged adenine through a small conformational motion, and such a scheme is illustrated in Figure 7.
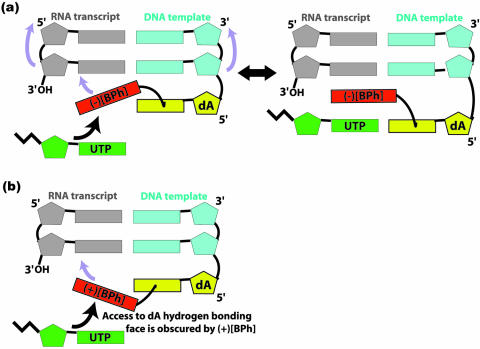
Figure 7.
(a) Schematic representation of a possible small conformational motion of the intercalated blocking conformation of (–)-1S-trans-anti-[BPh]-N6-dA, intercalated from the 3′-side of the modified adenine, that could allow bypass by RNA polymerase II. The directions and positions of the motions are indicated by the purple arrows. The entrance path of the incoming UTP is indicated by the single-headed black arrow. (b) Schematic demonstration of how the intercalated conformation of (+)-1R-trans-anti-[BPh]-N6-dA, intercalated from the 5′-side of the modified adenine, blocks the hydrogen bonding edge of the damaged adenine and would prevent an analogous motion to that hypothesized for the (–)-1S isomer.
Conclusions
Many factors influence the ability of a DNA adduct to block RNA polymerase progression, including lesion size, the sequence context of the adduct, the linkage site of the carcinogen moiety, the stereochemistry of the adduct and the specific RNA polymerase being studied. In the case of [BPh]-N6-dA lesions, the stereochemistry of the lesion plays a role in how well the adduct is bypassed during transcription elongation. Computer modeling sheds light on how the conformation of each trans-anti-[BPh]-N6-dA adduct within the active site of RNA polymerase II could influence its effect on bypass and suggests possible future experiments, including the examination of the effect of N2-[BPh]-dG on transcription and more detailed structural studies. Molecular dynamics with full-scale solvation would be very useful in the future to explore these models further. However, such a huge system, requiring simulation of ∼300 000 or more atoms for 1 ns or longer, is currently beyond our reach. Nevertheless, with the ever increasing availability of computational resources, such studies will become feasible. Future studies that couple biochemical, cellular and computer modeling approaches should allow us to understand the degree to which stereochemistry, carcinogen–DNA linkage site and conformational preference of DNA adducts influence their effects on cellular processes, including transcription, and provide insight as to how they are removed from the genome.
Acknowledgments
ACKNOWLEDGEMENTS
R.A.P. expresses appreciation for support from the American Cancer Society, Postdoctoral Fellowship no. PF-01-108-01-CNE. This work was supported by grants from the NIH, ES10581 (D.A.S.), CA28038 (S.B.) and CA76660 (N.E.G.).
REFERENCES
- 1.Friedberg E., Walker,G. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- 2.Conney A. (1982) Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons. Cancer Res., 42, 4875–4917. [PubMed] [Google Scholar]
- 3.Thakker D.R., Levin,W., Yagi,H., Yeh,H.J., Ryan,D.E., Thomas,P.E., Conney,A.H. and Jerina,D.M. (1986) Stereoselective metabolism of the (+)-(S,S)- and (–)-(R,R)-enantiomers of trans-3,4-dihydroxy-3,4-dihydrobenzo[c]-phenanthrene by rat and mouse liver microsomes and by a purified and reconstituted cytochrome P-450 system. J. Biol. Chem., 261, 5404–5413. [PubMed] [Google Scholar]
- 4.Mushtaq M. and Yang,S.K. (1987) Stereoselective metabolism of benzo[c]phenanthrene to the procarcinogenic trans-3,4-dihydrodiol. Carcinogenesis, 8, 705–709. [DOI] [PubMed] [Google Scholar]
- 5.Wood A.W., Chang,R.L., Levin,W., Thakker,D.R., Yagi,H., Sayer,J.M., Jerina,D.M. and Conney,A.H. (1984) Mutagenicity of the enantiomers of the diastereomeric bay-region benzo(c)phenanthrene 3,4-diol-1,2-epoxides in bacterial and mammalian cells. Cancer Res., 44, 2320–2324. [PubMed] [Google Scholar]
- 6.Levin W., Chang,R.L., Wood,A.W., Thakker,D.R., Yagi,H., Jerina,D.M. and Conney,A.H. (1986) Tumorigenicity of optical isomers of the diastereomeric bay-region 3,4-diol-1,2-epoxides of benzo(c)phenanthrene in murine tumor models. Cancer Res., 46, 2257–2261. [PubMed] [Google Scholar]
- 7.Agarwal S., Sayer,J., Yeh,H.J., Pannell,L., Hilton,B., Pigott,M.A., Dipple,A., Yagi,H. and Jerina,D. (1987) Chemical characterization of DNA adducts derived from the configurationally isomeric benzo[c]phenanthrene-3,4-diol 1,2-epoxides. J. Am. Chem. Soc., 109, 2497–2504 [Google Scholar]
- 8.Cosman M., Fiala,R., Hingerty,B.E., Laryea,A., Lee,H., Harvey,R.G., Amin,S., Geacintov,N.E., Broyde,S. and Patel,D. (1993) Solution conformation of the (+)-trans-anti-[BPh]dA adduct opposite dT in a DNA duplex: intercalation of the covalently attached benzo[c]phenanthrene to the 5′-side of the adduct site without disruption of the modified base pair. Biochemistry, 32, 12488–12497. [DOI] [PubMed] [Google Scholar]
- 9.Cosman M., Laryea,A., Fiala,R., Hingerty,B.E., Amin,S., Geacintov,N.E., Broyde,S. and Patel,D.J. (1995) Solution conformation of the (–)-trans-anti-benzo[c]phenanthrene-dA ([BPh]dA) adduct opposite dT in a DNA duplex: intercalation of the covalently attached benzo[c]phenanthrenyl ring to the 3′-side of the adduct site and comparison with the (+)-trans-anti-[BPh]dA opposite dT stereoisomer. Biochemistry, 34, 1295–1307. [DOI] [PubMed] [Google Scholar]
- 10.Geacintov N., Cosman,M., Hingerty,B., Amin,S., Broyde,S. and Patel,D. (1997) NMR solution structures of stereoisomeric covalent polycyclic aromatic carcinogen-DNA adducts: principles, patterns and diversity. Chem. Res. Toxicol., 10, 111–146. [DOI] [PubMed] [Google Scholar]
- 11.Seidel A., Sun,Z., Kroth,H., Steinbrecher,T., Oesch,F. and Friedberg,E.C. (1996) DNA polymerase action on oligonucleotides from human Ha-ras proto-oncogene containing N6-deoxyadenosine adduct derived from trans addition of (+)- and (–)-anti-benzo[c]phenanthrene-3,4-dihydrodiol-1,2-epoxides at codon 61. Polycyclic Aromatic Compounds, 10, 161–170. [Google Scholar]
- 12.Alberts B. (2003) DNA replication and recombination. Nature, 421, 431–435. [DOI] [PubMed] [Google Scholar]
- 13.Friedberg E.C., Wagner,R. and Radman,M. (2002) Specialized DNA polymerases, cellular survival and the genesis of mutations. Science, 296, 1627–1630. [DOI] [PubMed] [Google Scholar]
- 14.Bigger C.A., Strandberg,J., Yagi,H., Jerina,D.M. and Dipple,A. (1989) Mutagenic specificity of a potent carcinogen, benzo[c]phenanthrene (4R,3S)-dihydrodiol (2S,1R)-epoxide, which reacts with adenine and guanine in DNA. Proc. Natl Acad. Sci. USA, 86, 2291–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carothers A.M., Mucha,J. and Grunberger,D. (1991) DNA strand-specific mutations induced by (+/–)-3 alpha,4 beta-dihydroxy-1 alpha,2 alpha-epoxy-1,2,3,4-tetrahydrobenzo[c]phenanthrene in the dihydrofolate reductase gene. Proc. Natl Acad. Sci. USA, 88, 5749–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellon I., Spivak,G. and Hanawalt,P.C. (1987) Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell, 51, 241–249. [DOI] [PubMed] [Google Scholar]
- 17.Plosky B., Samson,L., Engelward,B.P., Gold,B., Schlaen,B., Millas,T., Magnotti,M., Schor,J. and Scicchitano,D.A. (2002) Base excision repair and nucleotide excision repair contribute to the removal of N-methylpurines from active genes. DNA Repair (Amst.), 1, 683–696. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl T., Karran,P. and Wood,R.D. (1997) DNA excision repair pathways. Curr. Opin. Genet. Dev., 7, 158–169. [DOI] [PubMed] [Google Scholar]
- 19.Sugasawa K., Ng,J.M., Masutani,C., Iwai,S., van der Spek,P.J., Eker,A.P., Hanaoka,F., Bootsma,D. and Hoeijmakers,J.H. (1998) Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2, 223–232. [DOI] [PubMed] [Google Scholar]
- 20.Hoeijmakers J.H. (2001) DNA repair mechanisms. Maturitas, 38, 17–23. [DOI] [PubMed] [Google Scholar]
- 21.Choi D.J., Roth,R.B., Liu,T., Geacintov,N.E. and Scicchitano,D.A. (1996) Incorrect base insertion and prematurely terminated transcripts during T7 RNA polymerase transcription elongation past benzo[a]pyrenediol epoxide-modified DNA. J. Mol. Biol., 264, 213–219. [DOI] [PubMed] [Google Scholar]
- 22.Cullinane C., Mazur,S.J., Essigmann,J.M., Phillips,D.R. and Bohr,V.A. (1999) Inhibition of RNA polymerase II transcription in human cell extracts by cisplatin DNA damage. Biochemistry, 38, 6204–6212. [DOI] [PubMed] [Google Scholar]
- 23.Tornaletti S., Donahue,B.A., Reines,D. and Hanawalt,P.C. (1997) Nucleotide sequence context effect of a cyclobutane pyrimidine dimer upon RNA polymerase II transcription. J. Biol. Chem., 272, 31719–31724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roth R.B., Amin,S., Geacintov,N.E. and Scicchitano,D.A. (2001) Bacteriophage T7 RNA polymerase transcription elongation is inhibited by site-specific, stereospecific benzo[c]phenanthrene diol epoxide DNA lesions. Biochemistry, 40, 5200–5207. [DOI] [PubMed] [Google Scholar]
- 25.Woychik N.A. and Hampsey,M. (2002) The RNA polymerase II machinery: structure illuminates function. Cell, 108, 453–463. [DOI] [PubMed] [Google Scholar]
- 26.Carothers A.M., Zhen,W., Mucha,J., Zhang,Y.J., Santella,R.M., Grunberger,D. and Bohr,V.A. (1992) DNA strand-specific repair of (+–)-3 alpha,4 beta-dihydroxy-1 alpha,2 alpha-epoxy-1,2,3,4-tetrahydrobenzo[c]phenanthrene adducts in the hamster dihydrofolate reductase gene. Proc. Natl Acad. Sci. USA, 89, 11925–11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perlow R.A., Schinecker,T.M., Kim,S.J., Geacintov,N.E. and Scicchitano,D.A. (2003) Construction and purification of site-specifically modified DNA template for transcription assays. Nucleic Acids Res., 31, e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laryea A., Cosman,M., Lin,J.M., Liu,T., Agarwal,R., Smirnov,S., Amin,S., Harvey,R.G., Dipple,A. and Geacintov,N.E. (1995) Direct synthesis and characterization of site-specific adenosyl adducts derived from the binding of a 3,4-dihydroxy-1,2-epoxybenzo[c]phenanthrene stereoisomer to an 11-mer oligodeoxyribonucleotide. Chem. Res. Toxicol., 8, 444–454. [DOI] [PubMed] [Google Scholar]
- 29.Gnatt A.L., Cramer,P., Fu,J., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science, 292, 1876–1882. [DOI] [PubMed] [Google Scholar]
- 30.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt E.V., Christoph,G., Zeller,R. and Leder,P. (1990) The cytomegalovirus enhancer: a pan-active control element in transgenic mice. Mol. Cell. Biol., 10, 4406–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woodard R.L., Anderson,M.G. and Dynan,W.S. (1999) Nuclear extracts lacking DNA-dependent protein kinase are deficient in multiple round transcription. J. Biol. Chem., 274, 478–485. [DOI] [PubMed] [Google Scholar]
- 33.Oelgeschlager T., Tao,Y., Kang,Y.K. and Roeder,R.G. (1998) Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol. Cell, 1, 925–931. [DOI] [PubMed] [Google Scholar]
- 34.Szentirmay M.N., Musso,M., Van Dyke,M.W. and Sawadogo,M. (1998) Multiple rounds of transcription by RNA polymerase II at covalently cross-linked templates. Nucleic Acids Res., 26, 2754–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang-Keck Z.Y. and Stallcup,M.R. (1988) Optimized reaction conditions and specific inhibitors for initiation of transcription by RNA polymerase II in nuclei from cultured mammalian cells. J. Biol. Chem., 263, 3513–3520. [PubMed] [Google Scholar]
- 36.Logan K., Zhang,J., Davis,E.A. and Ackerman,S. (1989) Drug inhibitors of RNA polymerase II transcription. DNA, 8, 595–604. [DOI] [PubMed] [Google Scholar]
- 37.Tornaletti S., Maeda,L.S., Lloyd,D.R., Reines,D. and Hanawalt,P.C. (2001) Effect of thymine glycol on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. J. Biol. Chem., 276, 45367–45371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu M., Yan,S., Patel,D.J., Geacintov,N.E. and Broyde,S. (2001) Cyclohexene ring and fjord region twist inversion in stereoisomeric DNA adducts of enantiomeric benzo[c]phenanthrene diol epoxides. Chem. Res. Toxicol., 14, 1629–1642. [DOI] [PubMed] [Google Scholar]
- 39.Wu M., Yan,S., Tan,J., Patel,D.J., Geacintov,N.E. and Broyde,S. (2004) Conformational searches elucidate effects of stereochemistry on structures of deoxyadenosine covalently bound to tumorigenic metabolites of benzo[c]phenanthrene. Front Biosci., 9, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perlow R.A., Kolbanovskii,A., Hingerty,B.E., Geacintov,N.E., Broyde,S. and Scicchitano,D.A. (2002) DNA adducts from a tumorigenic metabolite of benzo[a]pyrene block human RNA polymerase II elongation in a sequence- and stereochemistry-dependent manner. J. Mol. Biol., 321, 29–47. [DOI] [PubMed] [Google Scholar]
- 41.Choi D.J., Marino-Alessandri,D.J., Geacintov,N.E. and Scicchitano,D.A. (1994) Site-specific benzo[a]pyrene diol epoxide-DNA adducts inhibit transcription elongation by bacteriophage T7 RNA polymerase. Biochemistry, 33, 780–787. [DOI] [PubMed] [Google Scholar]
- 42.Donahue B.A., Yin,S., Taylor,J.S., Reines,D. and Hanawalt,P.C. (1994) Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc. Natl Acad. Sci. USA, 91, 8502–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scicchitano D. and Mellon,I. (1997) Transcription and DNA damage: a link to a kink. Environ. Health Perspect., 105, 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tornaletti S. and Hanawalt,P.C. (1999) Effect of DNA lesions on transcription elongation. Biochimie, 81, 139–146. [DOI] [PubMed] [Google Scholar]
- 45.Buterin T., Hess,M.T., Luneva,N., Geacintov,N.E., Amin,S., Kroth,H., Seidel,A. and Naegeli,H. (2000) Unrepaired fjord region polycyclic aromatic hydrocarbon-DNA adducts in ras codon 61 mutational hot spots. Cancer Res., 60, 1849–1856. [PubMed] [Google Scholar]
- 46.Wu M., Yan,S., Patel,D.J., Geacintov,N.E. and Broyde,S. (2002) Relating repair susceptibility of carcinogen-damaged DNA with structural distortion and thermodynamic stability. Nucleic Acids Res., 30, 3422–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geacintov N.E., Broyde,S., Buterin,T., Naegeli,H., Wu,M., Yan,S. and Patel,D.J. (2002) Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers, 65, 202–210. [DOI] [PubMed] [Google Scholar]