Abstract
The malachite green-molybdate reagent was used for a colorimetric assay of pure Mg2+-dependent phosphatidate phosphatase activity. This enzyme plays a major role in fat metabolism. Enzyme activity was linear with time and protein concentration, and with the concentration of water-soluble dioctanoyl phosphatidate. The colorimetric assay was used to examine enzyme inhibition by phenylglyoxal, propranolol, and dimethyl sulfoxide. Pure enzyme and a water-soluble phosphatidate substrate were required for the assay, which should be applicable to a well-defined large-scale screen of Mg2+-dependent phosphatidate phosphatase inhibitors (or activators).
Keywords: phosphatidate, Mg2+-dependent phosphatidate phosphatase, colorimetric assay, malachite green-molybdate reagent
Mg2+-dependent PA1 phosphatase (PAP1, 3-sn-phosphatidate phosphohydrolase, EC 3.1.3.4) catalyzes the dephosphorylation of PA yielding diacylglycerol and Pi [1–5]. PAP1 generates the diacylglycerol used for the synthesis of triacylglycerol, and the diacylglycerol used for the synthesis of phosphatidylethanolamine and phosphatidylcholine via the Kennedy pathway [4,5]. Recent studies have identified human lipin 1 as a PAP1 enzyme [6]. In a mouse model, lipin 1 deficiency prevents normal adipose tissue development that results in lipodystrophy (i.e., loss of body fat) and insulin resistance, whereas excess lipin 1 promotes obesity and insulin sensitivity [7,8]. That human lipin 1 is a PAP1, the penultimate enzyme in the pathway to synthesize triacylglycerol from PA, provides a mechanistic basis for how lipin 1 regulates fat metabolism in mammalian cells. Accordingly, PAP1 activity may represent an important pharmaceutical target for the control of body fat in humans.
A large-scale search of inhibitors (or activators) of PAP1 activity requires a sensitive and convenient enzymatic assay. The radioactive assays [9,10] currently used to measure PAP1 activity are not conducive to a high-throughput screen of potential drugs to control enzyme activity. Aside from the radioactive nature of these assays, they require a chloroform-methanol-water phase partition to separate water-soluble 32Pi from chloroform-soluble [32P]PA or the separation (e.g., thin-layer chromatography) of [3H]diacylglycerol from [3H]PA [9,10]. We have developed of non-radioactive colorimetric PAP1 assay based on orthophosphate analysis using the malachite green-molybdate reagent [11,12]. The reagent forms a colored complex with orthophosphate that can be measured spectrophotometrically. This colorimetric assay is not suitable for use with cell extracts or with crude PAP1 preparations because of a high phosphate background; hence the reason for using the radioactive assays [9,10]. However, the colorimetric assay was suitable for measuring the activity of pure PAP1, and the assay was amenable to screening enzyme inhibitors.
Saccharomyces cerevisiae PAH1-encoded PAP1 [6] was expressed and purified to homogeneity as described by O’Hara et al. [13]. The yeast enzyme was used a model PAP1 to develop the colorimetric assay. Pure PAP1 (12 ng) was used in a standard reaction mixture that contained 50 mM Tris-HCl buffer (pH 7.5), 1 mM MgCl2, and 0.2 mM DiC8 PA (Avanti Polar Lipids) in a total volume of 0.1 ml. Unless otherwise indicated, all enzyme assays were conducted in triplicate for 20 min at 30 °C. The malachite green-molybdate reagent [11,12] was prepared as described by Mahuren et al. [14]2. The reaction was terminated by the addition of 200 μl of the malachite green-molybdate reagent. 30 μl of 1% polyvinyl alcohol was then added to the reaction to stabilize the color complex [14]. The reaction mixture was vortexed briefly and the absorbance of the solution was measured with a spectrophotometer at 660 nm. The color was stable for at least 1 h. The amount of orthophosphate formed was quantified from a standard curve using 0.5–4 nmol of potassium phosphate. The enzyme reactions and standard curve were performed in new plastic test tubes. This obviated the concern of interfering phosphates from tubes that have been washed with detergent [14]. Statistical analyses were performed with SigmaPlot software.
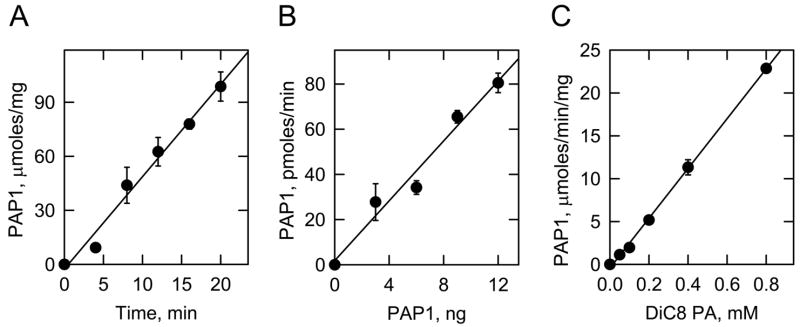
The PAP1 colorimetric assay was linear with respect to time (Fig. 1A) and enzyme concentration (Fig. 1B) indicating that the enzyme followed zero order kinetics under these reaction conditions. In addition, PAP1 activity was linear with respect the DiC8 PA substrate at concentrations between 0.05–0.8 mM (Fig. 1C). Indeed, the analysis of potential inhibitors would be best carried out at a low substrate concentration at or below (e.g., < 1 mM) the Km value for the substrate.
Fig. 1.
Time course, enzyme concentration, and substrate concentration dependencies of the colorimetric assay on pure PAP1 activity. Panel A, PAP1 activity was measured with 0.2 mM DiC8 PA and 12 ng of pure enzyme for the indicated time intervals. Panel B, PAP1 activity was measured for 20 min with 0.2 mM DiC8 PA and the indicated amounts of pure enzyme. Panel C, PAP1 activity was measured for 20 min with 12 ng of pure enzyme and the indicated amounts of DiC8 PA. The lines drawn in the panels were the result of a least-squares analysis of the data. The data shown were derived from triplicate determinations ± S.D.
For comparison, PAP1 activity was measured by following the release of 32Pi from chloroform-soluble [32P]DiC18 PA (10,000 cpm/nmol) as described by Carman and Lin [9]. In this assay, 0.2 mM [32P]DiC18PA was solubilized with 2 mM Triton X-100 to give a surface concentration of 9 mol % [9]. The specific activity (5.6 ± 0.6 μmol/min/mg) of the pure PAP1 enzyme determined with the colorimetric assay was in good agreement with the specific activity (5.2 ± 0.1 μmol/min/mg) determined with the radioactive assay. While these values were close, it is difficult to make a comparison of an enzyme activity measured with a water-soluble substrate when compared with a detergent-solubilized substrate [15].
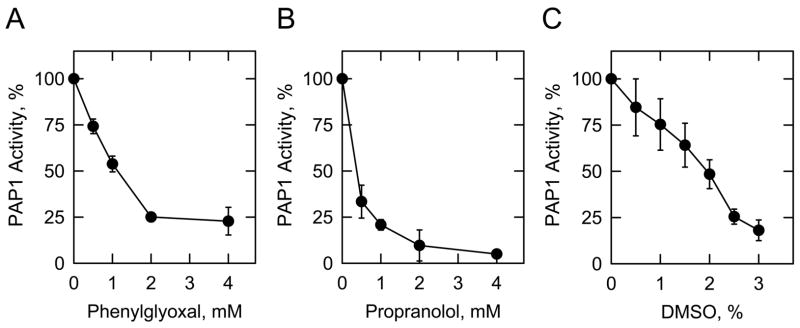
The suitability of the colorimetric assay to screen for PAP1 inhibitors was tested with two known inhibitors of the enzyme, namely phenylglyoxal and propranolol [16,17]. Phenylglyoxal is an arginine reactive compound [18], whereas propranolol is thought to interact with the Mg2+binding site of the enzyme [19]. Phenylglyoxal (Fig. 2A) and propranolol (Fig. 2B) inhibited PAP1 activity in dose-dependent manners with IC50 values of 1.3 mM and 0.2 mM, respectively. These values were in the same range determined for PAP1 activity measured by the radioactive assay with [32P]DiC18PA [17].
Fig. 2.
Effects of phenylglyoxal, propranolol, and DMSO on PAP1 activity measured with the colorimetric assay. PAP1 activity was measured under standard conditions in the presence of the indicated concentrations of phenylglyoxal (panel A), propranolol (panel B), and DMSO (panel C). The data shown were derived from triplicate determinations ± S.D.
Some enzyme inhibitors are not soluble in aqueous buffers, and are commonly solubilized in DMSO. Accordingly, the effect of DMSO on PAP1 activity was tested using the colorimetric assay. The addition of DMSO to the reaction mixture resulted in a dose-dependent inhibition of PAP1 activity (Fig. 2C). A 1% concentration of DMSO is commonly used for screens of water-insoluble inhibitors, and only 25% of PAP1 activity was lost using that concentration (Fig. 2C). Thus, a significant amount of PAP1 activity would still be present in a control reaction when potential inhibitors were solubilized in 1% DMSO.
Detergents (e.g., Triton X-100 and Tween 20) that were used to solubilize water-insoluble DiC18 PA [9] caused a high background color. This problem was solved by using water-soluble DiC8 PA as substrate. That pure PAP1 was a requirement for the colorimetric assay might be considered a major limitation. However, this limitation is also a major benefit because the screen for inhibitors (or activators) should be carried out under well-defined conditions that are free from other reactions that might generate orthophosphate and interfere with the interpretation of results. Obtaining large quantities of pure PAP1 enzyme is facilitated by the overexpression and purification of yeast [13] and human proteins [6]. As advertised by commercial vendors of the malachite green-molybdate reagent, the PAP1 colorimetric assay was applicability to a 96-well format (data not shown), which should facilitate a large-scale screen of PAP1 inhibitors (or activators).
Acknowledgments
This work was supported in part by United States Public Health Service, National Institutes of Health Grant GM-28140 (to G.M.C.) and by a Wellcome Trust Career Development Fellowship in Basic Biomedical Science (to S.S.). We thank Gil-Soo Han for helpful discussions during the course of this work.
Footnotes
Abbreviations: PA, phosphatidate; PAP1, Mg2+-dependent PA phosphatase; DiC8 PA, dioctanoyl phosphatidate; DiC18 PA, dioleoyl phosphatidate; DMSO, dimethyl sulfoxide.
The malachite green-molybdate reagent (PiBlue™) commercially prepared by BioAssay Systems worked equally as well as the reagent prepared in the laboratory.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith SW, Weiss SB, Kennedy EP. The enzymatic dephosphorylation of phosphatidic acids. J Biol Chem. 1957;228:915–922. [PubMed] [Google Scholar]
- 2.Brindley DN. Intracellular translocation of phosphatidate phosphohydrolase and its possible role in the control of glycerolipid synthesis. Prog Lipid Res. 1984;23:115–133. doi: 10.1016/0163-7827(84)90001-8. [DOI] [PubMed] [Google Scholar]
- 3.Carman GM. Phosphatidate phosphatases and diacylglycerol pyrophosphate phosphatases in Saccharomyces cerevisiae and Escherichia coli. Biochim Biophys Acta. 1997;1348:45–55. doi: 10.1016/s0005-2760(97)00095-7. [DOI] [PubMed] [Google Scholar]
- 4.Kocsis MG, Weselake RJ. Phosphatidate phosphatases of mammals, yeast, and higher plants. Lipids. 1996;31:785–802. doi: 10.1007/BF02522974. [DOI] [PubMed] [Google Scholar]
- 5.Nanjundan M, Possmayer F. Pulmonary phosphatidic acid phosphatase and lipid phosphate phosphohydrolase. Am J Physiol Lung Cell Mol Physiol. 2003;284:L1–23. doi: 10.1152/ajplung.00029.2002. [DOI] [PubMed] [Google Scholar]
- 6.Han GS, Wu WI, Carman GM. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 8.Phan J, Reue K. Lipin, a lipodystrophy and obesity gene. Cell Metab. 2005;1:73–83. doi: 10.1016/j.cmet.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Carman GM, Lin YP. Phosphatidate phosphatase from yeast. Methods Enzymol. 1991;197:548–553. doi: 10.1016/0076-6879(91)97182-x. [DOI] [PubMed] [Google Scholar]
- 10.Martin A, Gomez-Munoz A, Jamal Z, Brindley DN. Characterization and assay of phosphatidate phosphatase. Methods Enzymol. 1991;197:553–563. doi: 10.1016/0076-6879(91)97183-y. [DOI] [PubMed] [Google Scholar]
- 11.Itaya K, Ui M. A new micromethod for the colorimetric determination of inorganic phosphate. Clin Chim Acta. 1966;14:361–366. doi: 10.1016/0009-8981(66)90114-8. [DOI] [PubMed] [Google Scholar]
- 12.van Veldhoven PP, Mannaerts GP. Inorganic and organic phosphate measurements in the nanomolar range. Anal Biochem. 1987;161:45–48. doi: 10.1016/0003-2697(87)90649-x. [DOI] [PubMed] [Google Scholar]
- 13.O’Hara L, Han GS, Peak-Chew S, Grimsey N, Carman GM, Siniossoglou S. Control of Phospholipid Synthesis by Phosphorylation of the Yeast Lipin Pah1p/Smp2p Mg2+-dependent Phosphatidate Phosphatase. J Biol Chem. 2006;281:34537–34548. doi: 10.1074/jbc.M606654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahuren JD, Coburn SP, Slominski A, Wortsman J. Microassay of phosphate provides a general method for measuring the activity of phosphatases using physiological, nonchromogenic substrates such as lysophosphatidic acid. Anal Biochem. 2001;298:241–245. doi: 10.1006/abio.2001.5402. [DOI] [PubMed] [Google Scholar]
- 15.Carman GM, Deems RA, Dennis EA. Lipid signaling enzymes and surface dilution kinetics. J Biol Chem. 1995;270:18711–18714. doi: 10.1074/jbc.270.32.18711. [DOI] [PubMed] [Google Scholar]
- 16.Jamal Z, Martin A, Gomez-Munoz A, Brindley DN. Plasma membrane fractions from rat liver contain a phosphatidate phosphohydrolase distinct from that in the endoplasmic reticulum and cytosol. J Biol Chem. 1991;266:2988–2996. [PubMed] [Google Scholar]
- 17.Morlock KR, McLaughlin JJ, Lin YP, Carman GM. Phosphatidate phosphatase from Saccharomyces cerevisiae. Isolation of 45-kDa and 104-kDa forms of the enzyme that are differentially regulated by inositol. J Biol Chem. 1991;266:3586–3593. [PubMed] [Google Scholar]
- 18.Dawson RMC, Elliott DC, Elliott WH, Jones KM. B Reagents for Protein Modification. Oxford: 1986. Biochemical Reagents; pp. 384–397. [Google Scholar]
- 19.Abdel-Latif AA, Smith JP. Studies on the effects of Mg2+ ion and propranolol on iris muscle phosphatidate phosphatase. Can J Biochem Cell Biol. 1984;62:170–177. doi: 10.1139/o84-024. [DOI] [PubMed] [Google Scholar]