Abstract
Fetuin-A is an hepatic protein whose mRNA transiently falls during the inflammatory acute phase via unknown transcriptional mechanisms. Various FETUA promoter/cat constructs transiently transfected in the Hep3B hepatoma cell line allowed us to identify four NF-1 and C/EBP binding sites (N, C) arranged in a 5′-N2-C2-N1-C1-3′ order and required for basal promoter activity. Mutant constructs demonstrated that C1 and C2 but not N1 nor N2 are required for the cytokine-driven down-regulation of the promoter. A variable spacing between C2 and N1 showed that the alignment of the (C1+N1) and (C2+N2) areas is critical for the promoter activity in quiescent but not cytokine-stimulated cells. Co-transfection of a plasmid only producing either a long or short C/EBPβ isoform prevented FETUA regulation by cytokines. Electromobility shift assays with liver nuclear extracts showed that during the acute phase the complexes formed over N1 and N2 are not modified whereas short C/EBPα and -β isoforms replace the long isoforms bound to C1 and C2 in the quiescent liver. Therefore the basal promoter activity requires an interaction between the long C/EBP isoforms bound to C1 and C2 whereas the inflammation-induced down-regulation results from the loss of interaction between the cytokine-induced, short C/EBP isoforms.
INTRODUCTION
α2-HS glycoprotein is a major human plasma protein of hepatic origin and a member of the cystatin superfamily of protease inhibitors (1,2). It has alternatively been designated as phosphoprotein of 63 kDa (pp63) in rat, countertrypin in mouse or fetuin in other mammals (2–5) and is now referred to as Fetuin-A since a paralog called Fetuin-B has been recently identified (6). Beyond its importance during development, hence the name Fetuin that was coined accordingly, Fetuin-A is a key protein in several metabolic pathways. First, Fetuin-A modulates some insulin-driven and kinase-mediated signal transduction pathways, possibly in a tissue-specific fashion (7). Specifically, Fetuin-A inhibits the insulin receptor autophosphorylation (8–10) and is a critical partner in insulin-dependent metabolism (11). Secondly, Fetuin-A is involved in osteogenesis and bone resorption and in the control of calcium salt precipitation in blood (12–14). In keeping with this, Fetuin-A accumulates in bone matrix (15) and counteracts a transforming growth factor-β activity required for bone mineralization (16,17). Thirdly, the phosphorylated form of rat Fetuin-A inhibits hepatocyte growth factor binding to its hepatic receptor (18). Fourthly, Fetuin-A is an anti-inflammatory mediator that participates in macrophage deactivation. Specifically, Fetuin-A enhances the cellular uptake of cationic inhibitors of pro-inflammatory cytokine synthesis by macrophages and hence it prevents the morbid sequelae of infection and trauma that would result from overproduction of pro-inflammatory cytokines (19–21). In agreement with this view, the plasma protein and hepatic mRNA levels for both human and rat Fetuin-A transiently fall during the acute phase of a systemic inflammation (22–25) which classifies Fetuin-A as a negative acute-phase protein (APP) (26). This event mainly results from an interleukin-1β (IL-1β)-induced down-regulation of its hepatic mRNA level (9,22,23), which further classifies Fetuin-A as a type-1 APP (26).
A strong basal transcription of the Fetuin-A-encoding gene (FETUA, formerly AHSG) takes place in liver under physiological conditions and is controlled by a series of binding sites for the C/EBP and NF-1 transcription factors (TF) in the gene promoter, a feature that is conserved from rat to human (27,28). Given the important role of Fetuin-A as an acute phase regulator, as stressed above, it is further desirable to understand the molecular events that drive the negative regulation of this protein and its mRNA during the acute phase. It has been shown that this down-regulation involves, at least partly, a transcriptional step (9,29) but the molecular mechanism that takes place at the FETUA gene level in liver during the acute phase has never been elucidated in any species. We now report that in quiescent cells transcription of the FETUA promoter depends on a concerted activity of long C/EBPα isoforms at two binding sites. This mechanism is lost when these sites are bound by the short C/EBPα and -β isoforms produced during the acute phase.
MATERIALS AND METHODS
Animals
Sprague–Dawley rats and C57/BL6 mice were housed in our animal facility according to standard ethical guidelines. Prior to any experiment, they were kept at rest under a 12 h/12 h light/dark cycle and were given chaw and water ad libitum for at least one week. An acute, systemic inflammation was induced by intraperitoneal injection of an Escherichia coli LPS solution (Sigma) in 0.15 M NaCl (2.5 mg LPS/kg of body weight). Control animals were given the vehicle alone. At the appropriate time, the mice were killed by cervical dislocation whereas the rats were anesthesized with an intra-peritoneal injection of 0.2 ml pentothal, a liver fragment was collected and the rats were finally sacrificed by section of the vena cava.
Reagents
Restriction enzymes and DNA modification enzymes were obtained from New England Biolabs. Taq polymerase was from Finnzyme. [γ-32P]ATP (5000 Ci/mmol) and [3H]acetyl Coenzyme A (5 Ci/mmol) were from Amersham. Sterile plasticware for tissue cultures was from Falcon. Dulbecco’s modified Eagle’s medium (DMEM), trypsin/EDTA, antibiotics and all other culture grade chemicals were purchased from Invitrogen. Dulbecco’s phosphate buffered saline (PBS) and BES were from Sigma. Fetal calf serum (FCS) was from Bio-Whittaker. Rabbit anti-C/EBPα, -β or -δ antisera raised against the C-terminal 4/5 of C/EBPα, the C-terminal 18 amino acid residues of C/EBPβ or the entire C/EBPδ protein, respectively, were a kind gift from Professor S. L. McKnight (Tularik Inc., San Francisco, CA). Rabbit anti-NF-1 antibodies directed against a N-terminal motif found in every NF-1 isoform (ref. sc-870X) and anti-C/EBPβ antibodies (sc-150X) were from Santa Cruz Biotechnologies.
Oligonucleotides
All oligonucleotides were purchased from Genset. Unless specified otherwise, all sequences below refer to the human FETUA gene whose transcription start site is numbered +1. For plasmid constructions, sense oligos included: oli–200/–182 (5′-CTATCAAGCTTCCCCCACAGCAGCATGGAC-3′) covering the FETUA –200/–182 sequence and added with a 5′ tail containing a Hind III site (underlined); olimC1, olimN1, olimC2 and olimN2 (5′-CCAAAGGAGAAAT CATCCTGTATCCGACTAGTGTTCTTCCGGCAGGCTCC AACAGATAAATAAAGCC-3′; 5′-CTGATGTTTGCAGG GTGTTTTTTTTTTTCTTGACTAGTGAAGGAGAAATCA TCCTGTATCCTTATGC-3′; 5′-GACTTTGGCAGATTTC TTGGGGACCAGCGAGACTAGTGCCTGTTTGCTTTTC CAGGGCTGATGTTTGC-3′; 5′-GGAGCATCTCCCCCAC AGCAGCATGGACTTGACTAGTGTTCTTGGGGACCAGCGATGTCCTAACC-3′) covering the FETUA –91/–25, –127/–61, –184/–117 and –209/–145 sequence, respectively, and containing an internal stretch of eight mutated nucleo tides (italicized) with a SpeI site (underlined); oliΔ5 and oliΔ10 (5′-GGGCTGATGTTTGCAGGGTGTTTTTTCTTT TGAACCAAAGG-3′; 5′-GGGCTGATGTTTGCAGGGTTT CTTTTGAACCAAAGG-3′) covering FETUA from –130 to –85 and deleted of the –107/–103 or –111/–102 sequence, respectively. Two antisense oligos were olipBL (5′-CAAG CTCCTCGAGATCCAGATCTGG-3′) covering the pBLcat6 polylinker and oli–61/–43 (5′-ATCGCGGATCCGAGCC TGCCGGAAGAATTG-3′) covering the –61/–43 area in FETUA and added with a BamHI site (underlined). For electromobility shift essays (EMSA), double-stranded probe C1 (5′-CTGTATCCTTATGCAATTCTTCCGGCA-3′) or C2 (5′-AGCGATGTCCTAACCTGTTTGCTTTTC-3′) with a C/EBP binding site covered the –76/–50 or –159/–133 FETUA sequence, respectively. Probe N1 (5′-TTTTTCTTTTGA ACCAAAGCAGAAATC-3′) or N2 (5′-CATGGACTTTG GCAGATTTCTTGGGGA-3′) with a NF-1 binding site covered –104/–78 or –188/–162, respectively. Mutant competitors designated mC1, mC2, mN1 and mN2 harboured an 8 bp mutant motif made of a SpeI site bracketed with two Gs at –66/–59 (mC1), –154/–147 (mC2), –96/–89 (mN1) or –179/ –172 (mN2). Other double-stranded oligos included proven binding sites for NF-1 (28) or C/EBP (5′-CTAGGGC TTGCGCAATCTATATTCG-3′; Geneka catalog number 1200009). Real-time, quantitative reverse transcription–PCR (Q-RT–PCR) was done with pairs of mouse fetuin-A-specific (5′-TTGCTCAGCTCTGGGGCT-3′ and 5′-GGCAAGTGG TCTCCAGTGTG-3′) or Gapdh-specific (5′-GAGCCAAAA GGGTCATCATC-3′ and 5′-CCATCCACAGTCTTCTGG GT-3′) oligos.
Plasmids and wild type or mutant constructs
pCH110 (Pharmacia) contains the β-galactosidase (β-gal) gene under control of the early promoter/enhancer from SV40. pSV2cat is a cat plasmid under control of the early promoter/enhancer from SV40, pBLcat6 is a low background, promoterless cat plasmid and pBLcat5 is the counterpart with the cat gene driven by the thymidine kinase (tk) promoter from Herpes simplex virus (28). pC/EBPα and pC/EBPδ are expression plasmids with a full-length rat C/EBPα or -δ cDNA, respectively, cloned into pHD (30). pC/EBPβ is an expression plasmid with a full-length human C/EBPβ cDNA cloned into pGEM-7 and two mutants of pC/EBPβ are designated pC/EBPβMT20 and pC/EBPβAUG3 (31,32). Two pro-inflammatory cytokine-responsive and control plasmids included pApoCIIIΔDcat and pGAPDHcat: pApoCIIIΔDcat with the cat gene driven by the –890/+24 promoter of the human APOCIII gene was used as a control for a gene that is down-regulated by pro-inflammatory cytokines (30) and pGAPDHcat with the –488/+21 promoter of the human GAPDH gene (33) was used as a negative control for cytokine-modulated genes. The pBLcat6-derived plasmids p-57/48cat, p-171/48cat, p-273/48cat and p-3320/48cat that all retain 48 bp of human FETUA sequence on the 3′ side of the transcription start site and a variably deleted sequence on its 5′ side (hence the negative numbering) have been previously detailed (28). A further p-200/48cat plasmid was constructed by inserting a PCR product obtained with oli–200/–182 and olipBL in the XhoI and HindIII sites of pBLcat6 polylinker. The –200/–43 segment amplified by PCR with oli–200/–182 and oli–61/–43 was inserted in sense orientation relative to cat at the HindIII and BamHI sites of pBLcat5 to provide pFETUAtkcat. Several mutant plasmids were prepared from p-273/48cat with the ‘GeneEditor in vitro site-directed mutagenesis’ system (Promega) and the oligos olimC1, olimN1, olimC2 or olimN2 noted above. This resulted in pmC1cat, pmN1cat, pmC2cat and pmN2cat in which the –66/–59, –96/–89, –154/–147 and –179/–172 sequence in human FETUA were respectively replaced by a SpeI site bracketed with 2 Gs. Finally, two mutants plasmids were prepared from p-200/48cat by the megaprimer procedure (34) and the oligos oliΔ5, oliΔ10, olipBL and oli–200/–182 noted above. This resulted in pΔ5-200/48cat and pΔ10-200/48cat in which 5 (–107/–103) or 10 nt (–111/–102) were respectively deleted from the human FETUA promoter. All final constructs were verified by sequencing.
Cell cultures, transfections and reporter gene assays
Blood fractions enriched in peripheral blood mononuclear cells (PBMC) were obtained from Etablissement Français du Sang (Boisguillaume, France) and the PBMCs were isolated as described by Wang et al. (20). The cells were depleted from contaminating lymphocytes by a 1-h incubation (2 × 108 cells/75 ml plastic flask) in 10 ml DMEM supplemented with penicillin (50 µ/ml), streptomycin (50 µ/ml), 2 mM Gln and 10% (v/v) FCS at 37°C under a 5% CO2-enriched atmosphere, followed by gentle washing of the non-adherent cells with sterile 0.15 M NaCl at room temperature. The adherent PBMCs were then cultured for 16 h in FCS-free DMEM supplemented as above. Under such conditions, the final, post-culture medium was used as a negative, control medium (NCM). Conversely, when the medium used for culture of adherent PBMCs was further added with bacterial LPS (10 µg/ml) prior to the 16-h incubation period, the cytokine-rich medium obtained post-incubation was referred to as the conditioned medium (CM). Various pro-inflammatory cytokines were measured in these NCM and CM media with Quantikine immunoassay kits (R&D Systems) and only CM contained significant amounts of IL-1β (10 versus 1700 pg/ml), IL-6 (61 versus 13800 pg/ml) and IL-8 (undetectable versus 68600 pg/ml). Also, it was verified that CM, but not NCM, could modulate the endogenous synthesis of various positive or negative APPs in Hep3B cells (not detailed).
The human HepG2 and Hep3B hepatoma cell lines (ATCC refs HB-8065 and HB-8064) were propagated as a monolayer in 75 cm2 flasks containing DMEM/10% FCS supplemented as above, at 37°C under a 5% CO2-enriched and water-saturated atmosphere. Transfection and stimulation of these cells was done in 6-cm dishes with a FCS-free DMEM added with 5% (v/v) CM or NCM. This medium was added to the plasmid transfection medium at 30 min after the start of transfection and the final incubation lasted for 36 h as detailed below. The HepG2 cells at 40% confluency were transfected with a mixture of cat (8 µg) and pCH110 (1 µg) plasmids. The latter were purified onto Nucleobond columns PC2000 (Macherey-Nagel), diluted in 250 mM CaCl2 (250 µl final volume), added dropwise to 250 µl BES-buffered saline (BES 50 mM, pH 7.05, NaCl 280 mM, Na2HPO4 1.5 mM) and left at room temperature for 20 min; 450 µl of the resulting CaPO4/DNA precipitate were added dropwise onto the cells, left for 16 h and finally replaced by a fresh medium for a further 20 h. The cells were washed with PBS, incubated for 5 min in PBS with 5 mM EDTA, harvested by gentle scraping and immediately processed for reporter gene assays. The Hep3B cells at 80% confluency were transfected with a mixture of pcat (4.5 µg) and pCH110 (1.5 µg) plasmids purified as above, diluted in 2 ml antibiotic-free DMEM/2 mM Gln containing 36 µl GenePorter (Gene Therapy System) and pre-incubated for 40 min at room temperature. When a co-transfection experiment further included a pC/EBP expression plasmid (Results), this plasmid was brought up to a maximum amount of 0.5 µg and the empty counterpart of this plasmid was added as required to have a constant amount of plasmid in every sample to be compared. The pre-incubated plasmid mixture was layered onto the cells for 30 min, 100 µl of undiluted CM or NCM was next brought into the medium and the resulting mixture was further incubated for 3.5 h. A further 2 ml DMEM solution with 2 mM Gln, penicillin (100 µ/ml), streptomycin (100 µ/ml) and 5% CM or NCM was added and the cells were finally incubated for a further 32-h period, washed and harvested as above. We verified that pCH110 activity was not modulated by a pC/EBP plasmid co-transfection or by any CM input (data not shown) and therefore pCH110 co-transfected with every cat plasmid allowed for a β-gal-based normalization of Cat activities between culture dishes. A simultaneous measurement of β-gal and Cat activities in cell extracts was performed and the normalized Cat activities were expressed as c.p.m. [3H]acetyl-chloramphenicol/β-gal unit (28). All final values are the mean ±SD of at least three independent experiments with triplicates per experiment.
Nuclear extracts from rat liver
Rat liver nuclear extracts were prepared by rinsing a liver fragment in cold PBS and homogenizing it in a hypotonic lysis buffer enriched with a protease inhibitor cocktail (HEPES 20 mM pH 7.9, NaCl 10 mM, EDTA 1 mM, DDT 1 mM, pefabloc 200 µg/ml, aprotinin 0.5 µg/ml, leupeptin 5 µg/ml, pepstatin 5 µg/ml) and used at 3 ml/0.1 g liver in a chilled dounce-A instrument. Following a 15 min incubation on ice, the liver mixture was centrifuged at 850 g for 10 min at 4°C and the pellet was resuspended in the same lysis buffer as above (750 µl/0.1 g liver), incubated on ice for 15 min, added with 0.1% Nonidet P-40, vortexed for 10 s and centrifuged at 11 000 g for 30 s at 4°C. The pellet was resuspended in an extraction buffer (25 µl/0.1 g liver) consisting of HEPES 20 mM pH 7.9, NaCl 420 mM, glycerol 25% (v/v), DTT 1 mM and protease inhibitors, vortexed for 10 s, incubated on ice with continuous shaking for 30 min, vortexed again for 30 s and finally centrifuged at 11 000 g for 10 min at 4°C. The resulting supernatant was finally stored at –80°C and used as a nuclear extract for protein electrophoresis and EMSAs.
Protein electrophoresis and immunodetection
SDS–polyacrylamide gel electrophoresis (PAGE) of nuclear extracts was done in pre-cast 8 × 8 cm 10% polyacrylamide gels (NuPAGE Bis-Tris gel, Invitrogen). The SeeBlue Plus2 prestained standard (Invitrogen) was used as a size marker. Protein denaturation and loading (20 µg/lane), Coomassie blue staining of gels with the SimplyBlue Safestain (Invitrogen) and destaining in distilled water were done exactly as recommended by Invitrogen. An XCell SureLock Mini-Cell (Invitrogen) was used for both SDS–PAGE and electro-blotting. The proteins were transferred onto a nitrocellulose sheet (Optitran BA-S85 from Schleicher-Schüell) in the appropriate Invitrogen buffer added with 20% methanol. The nitrocellulose was next saturated with a Tris buffer saline (TBS) mixture (Tris 20 mM pH 7.5, NaCl 500 mM) containing 5% skimmed milk and 0.075% Tween 20 for 4 h at room temperature. Rabbit anti-TF antibodies (listed above) were diluted 1/1000 in the TBS/milk/Tween solution above and incubated with the electro-transferred proteins at 4°C overnight. Subsequent washes were done in TBS. Peroxidase-labelled goat IgGs directed against rabbit IgGs used as a secondary antibody were diluted in TBS and incubated for 1 h at room temperature prior to washes with TBS/Tween and a final wash with TBS alone. The antibody–protein complexes were revealed with an ECL detection kit (AP Biotech) and the membrane was exposed onto an X-ray film for 30 s to 2 min.
EMSA
Probes and competitors made of HPLC-purified double-stranded oligos (listed above) were used as follows. The probes were end-labelled with [γ-32P]ATP by phosphorylation and purified with a Nick-column (Amersham-Pharmacia Biotech). A nuclear extract and competitor at a 200-fold molar excess over the probe were incubated at room temperature for 7 min in a binding buffer (HEPES 12 mM pH 7.9, Tris 4 mM pH 7.9, MgCl2 5 mM, EDTA 1 mM, KCl 25 mM, DTT 1 mM, poly-dIdC 0.04 µg/µl, glycerol 12% v/v) prior to probe addition (1 ng/reaction) and a further incubation for 5 min at room temperature. Supershift experiments were done by pre-incubating antisera or antibodies (listed above) with the nuclear extract on ice for 30 min prior to probe addition. The DNA/protein complexes were resolved in non-denaturing polyacrylamide gels (28) that were revealed with a Phosphorimager (Molecular Dynamics, Sunnyvale, CA).
Real-time Q-RT–PCR
Total RNAs were extracted from livers and 2 µg RNAs were reverse-transcribed in 60 µl MMLV/RT buffer (Promega) containing 1 mM dNTP, 120 U RNAsin (Promega), 400 U MMLV RT (Promega) and 500 pmol random hexamer primers (Pharmacia) at 37°C for 60 min and heat denatured at 95°C for 5 min. Two microlitres of the resulting mixture were used for Q-RT–PCR which was done with pairs of primers (see above) and the ‘FastStart DNA Master SYBR Green I’ kit in a Light Cycler instrument (Roche Diagnostics, Mannheim, Germany) exactly as recommended by the manufacturer. An arbitrary internal standard was made from a mouse liver RNA solution. In every sample the final Fetuin-A mRNA concentration was normalized with the Gapdh mRNA concentration.
RESULTS
FETUAcat construct activity and TF abundance in the HepG2 vs Hep3B cells
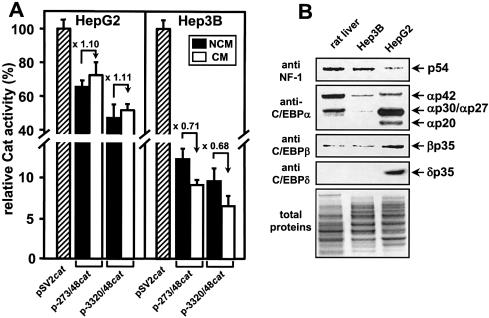
Initial experiments with a large (–3320/+48) or minimal segment (–273/+48) of the human FETUA promoter in the p-3320/48cat and p-273/48cat constructs revealed that this promoter was 7-fold more active in the hepatoma HepG2 cells than Hep3B cells when the viral SV40 promoter was taken as a reference for normalization between cell lines (Fig. 1A). However, culturing the HepG2 cells in a cytokine-enriched CM did not promote any down-regulation of the FETUA constructs whereas the Hep3B cells were permissive for a limited down-regulation that reached 70% of the reference activity seen with Hep3B cells cultured in a cytokine-free NCM (Fig. 1A). In keeping with this, the level of endogenous FETUA mRNA was down-regulated 2-fold in CM-stimulated Hep3B cells whereas it was not modified in CM-stimulated HepG2 cells (data not shown). We then wondered whether a different TF level in the HepG2 versus Hep3B cells could account for the above difference in FETUA promoter response to cytokines. As this promoter is driven by NF-1 and C/EBP (Introduction), we compared the relative amounts of these TFs in nuclei from rat liver, HepG2 and Hep3B cells (Fig. 1B). Similar amounts of the hepatic NF-1 p54 isoform (35) in the rat liver and Hep3B cells contrasted with low amounts in the HepG2 cells. Conversely, the HepG2 cells were strongly enriched in various isoforms of C/EBPα, -β and -δ including C/EBPα of 42 kDa (αp42), αp30, αp27, αp20, βp35 and δp35 (36,37). The Hep3B cells only contained low amounts of αp42 and trace amounts of αp30. The Hep3B cells as well as the rat liver contained low amounts of βp35 and no δp35. Given that αp30, αp27, αp20 and δp35 are hallmarks of the proinflammatory cytokine-challenged hepatocyte (26,36,37), we envisioned that the enrichment of these isoforms in the resting HepG2 cells could account for a block of CM responsiveness of our FETUA constructs in these cells. This will be further discussed later.
Figure 1.
Activity of FETUAcat constructs and TF abundance in human hepatoma cells. (A) The basal activity of p-3320/48cat and p-273/48cat in NCM as well as their responsiveness to CM was tested by transient transfection in HepG2 or Hep3B cells. Every Cat activity is relative to the activity (100%) of the pSV2cat plasmid (hatched bar) transfected in NCM-treated cells. For every cat construct, the CM-induced activation or repression relative to the reference activity in NCM-cultured cells is indicated by a broken arrow and a value (folds). (B) Amounts of NF-1 and C/EBP family members in nuclear extracts from Hep3B or HepG2 cells or rat liver. Total nuclear proteins (15 µg/lane) were separated by SDS–PAGE, electro-transferred and immunodetected with the antisera noted on the left. Each protein band is identified on the right with a p followed with the size (kDa) of an expected NF-1 or C/EBP isoform (35–37). A Coomassie blue staining of total proteins before electro-transfer is shown in the lower panel. At the bottom is the anode.
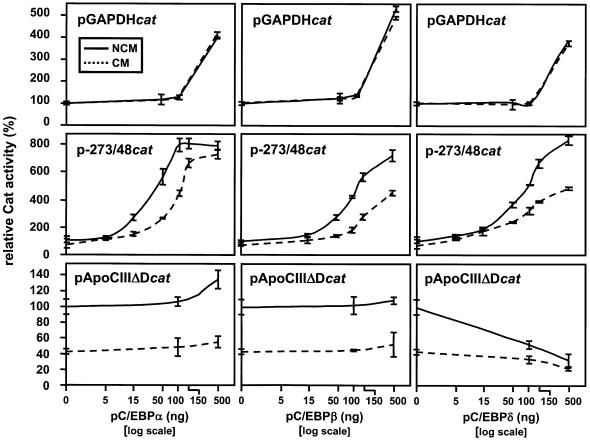
We then selected the cytokine-permissive Hep3B cells for a study of our FETUAcat constructs and we wondered whether an input of some exogenous C/EBP proteins could possibly reinforce the low activity of our constructs in these cells. This was tested by co-transfecting one of various pC/EBP expression plasmids along with a cat construct, as previously done for studies of some other promoters (38–40). As seen in Figure 2, every pC/EBPα, -β or -δ plasmid increased the basal activity of p-273/48cat up to 8-fold. Most importantly, co-transfecting pC/EBPα, -β or -δ in the 50–150 ng range enhanced the extent of CM-induced down-regulation of the FETUA promoter whose activity was half of that seen in NCM-cultured cells. On such grounds, all further studies of our pFETUAcat constructs were carried out in Hep3B cells co-transfected with 100 ng pC/EBPα. Control experiments with other reporter plasmids were also performed (Fig. 2), as follows. High amounts of every pC/EBP plasmid up-regulated 4-fold or more the basal activity of pGAPDHcat but the latter was not CM-sensitive, as expected from the GAPDH promoter that is not acute phase-regulated (24,41). Likewise, pApoCIIIΔDcat was repressed by pC/EBPδ, as expected from the APOCIII promoter that is down-regulated by the IL-1β/C/EBPδ pathway (30). Also, pApoCIIIΔDcat was down-regulated 2.5-fold by CM but this down-regulation was almost abolished by a co-transfection of pC/EBPδ. In conclusion, the pattern of FETUA promoter activity seen in Figure 2 appeared to be promoter specific.
Figure 2.
Effect of exogenous C/EBP proteins upon the basal and CM-induced response of a FETUAcat plasmid. The FETUAcat plasmid p-273/48cat and either of various pC/EBP expression plasmids were co-transfected in the Hep3B cell line cultured in the presence of CM (dotted line) or NCM (solid line). The amounts of pC/EBPα, -β or -δ plasmids used for co-transfection are indicated on the abcissa. Every Cat activity is relative to the activity (100%) obtained in NCM-treated Hep3B cells that were transfected with an empty expression plasmid. The activities of pApoCIIIΔDcat and pGAPDHcat used as controls for a gene that is down-regulated by CM (APOCIII) or is not CM-responsive (GAPDH) are also shown.
Localization of the cytokine-responsive elements in the FETUA promoter
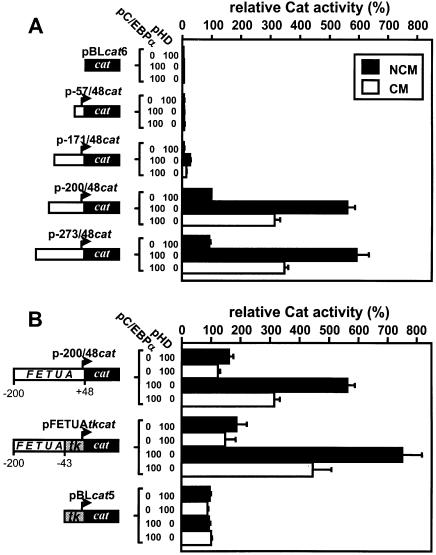
The CM-induced down-regulation of p-3320/48cat and p-273/48cat was identical whether in the absence or presence of a pC/EBPα co-transfection (Fig. 1A and data not shown), indicating that the inflammation-responsive element(s) of the FETUA gene is located in the proximal promoter. Therefore, we focused our study on the proximal FETUA promoter and deletion mutants from –273 to the transcription start site were used to map the CM-responsive area. The data in Figure 3A indicate that any construct retaining at least the proximal 200 bp in this promoter was fully active, could be strongly up-regulated by an exogenous input of C/EBPα and retained the CM responsiveness. We then tested whether these 200 bp of the FETUA promoter were able to confer a CM responsiveness to a heterologous promoter, namely that of the Herpes simplex tk gene (Fig. 3B). While pBLcat5 used as a control for tk activity could be neither up-regulated by a pC/EBPα co-transfection nor modulated by CM, the activity of pFETUAtkcat that harbours the –200/–43 segment of FETUA next to the tk promoter was enhanced by C/EBPα and down-regulated by CM in a manner very much like p-200/48cat. Therefore the FETUA element(s) that mediates the negative response to CM was ascribed to the –200/–43 area.
Figure 3.
Identification of a CM-responsive area in the FETUA promoter. (A) A series of cat plasmids with various deletions of the FETUA promoter were co-transfected with the expression plasmid pC/EBPα (100 ng) or an empty control plasmid pHD (100 ng) in Hep3B cells cultured with CM versus NCM. All values are relative to p-200/48cat activity (100%) in the presence of pHD and NCM. (B) p-200/48cat, pFETUAtkcat and pBLcat5 that harbour various segments of the FETUA and/or tk promoters as depicted, were co-transfected with pC/EBPα or pHD. The numbering –200, –43 and +48 refers to the FETUA gene. All values are relative to pBLcat5 activity (100%) in the presence of pHD and NCM.
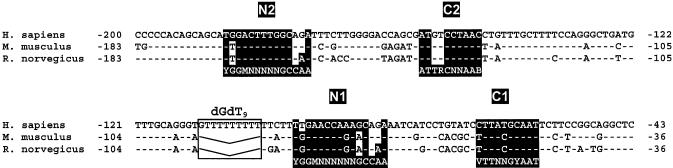
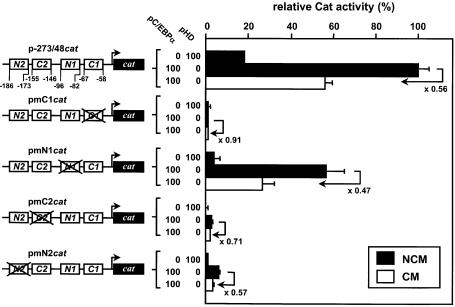
As shown in Figure 4, the human, rat and mouse FETUA promoters exhibit high similarities, including conserved NF-1 binding sites (N1 and N2) and C/EBP binding sites (C1 and C2). The involvement of all four sites in the basal transcription of the rat fetua gene has been previously shown (27). Moreover, with the program MatInspector combined with the TRANSFAC database no further potential TF binding sites could be found in this area (28 and data not shown). Accordingly, each of these four sites was mutated to identify whether it could possibly participate in the CM responsiveness. As shown with the pmN2cat and pmN1cat constructs in Figure 5, mutating the N2 site dramatically decreased the FETUA promoter activity in agreement with our deletion experiments (see p-171/48cat in Fig. 3A), whereas mutating the N1 site decreased this activity to a lower extent. However, mutating either N1 or N2 did not prevent the CM-promoted down-regulation (2-fold) of the corresponding constructs. Mutating the C2 site in pmC2cat largely abolished the basal activity of the promoter. A pC/EBPα-enhanced activity of this construct could still be obtained (by virtue of the remaining C1 site) but the CM-induced down-regulation of the promoter was barely retained (×0.71). Finally, mutating the C1 site in pmC1cat completely abolished the FETUA promoter activity and its pC/EBPα responsiveness and hence a further, CM-promoted down-regulation could not be observed. Overall, N1 and N2 did not appear to be involved in the CM responsiveness of the FETUA promoter whereas C1 and/or C2 and the cognate C/EBP proteins appeared to be the likely mediators of this event.
Figure 4.
Alignment of human, mouse and rat FETUA sequences. The human sequence (GenBank accession number AB038689) is taken as a reference and the nucleotides conserved in both mouse (AJ002146) and rat (M36547) are noted with hyphens. The nucleotide numbering is species-specific and refers to the transcription start site +1. Four conserved NF-1 and C/EBP binding sites are referred to as N1, N2, C1 and C2, respectively. Their sequences are written over a black background when they fit the consensus for a NF-1 or C/EBP binding site whereas they are written in a reversed background when they depart from this consensus. The consensus (51) sequences are written beneath the actual FETUA sequences. A dGdT9 stretch found in the human gene only is boxed and the corresponding gap in rodents is depicted with a V-shaped line.
Figure 5.
Identification of CM-responsive boxes in the CM-responsive area. Wild type p-273/48cat or various mutants with an extensive sequence change within the C1, N1, C2 or N2 box (mutated boxes are crossed out) were co-transfected with pC/EBPα or pHD. All values are relative to p-273/48cat activity (100%) in the presence of pC/EBPα and NCM. For every cat construct the CM-induced repression relative to the reference activity in NCM-cultured cells is indicated by a broken arrow and a value (folds). Other details are as in Figure 3.
Importance of a helix turn between (N2+C2) and (N1+C1)
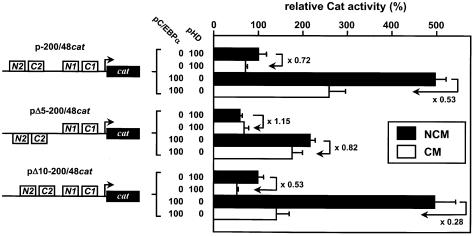
A GTTTTTTTTT stretch (dGdT9) that exactly spans one turn of DNA helix between N1 and C2 is found in the human gene (boxed in Fig. 4) but is absent in rodents. We then wondered whether this dGdT9 could have any influence upon the basal activity and/or acute phase-dependent down-regulation of the FETUA gene. To address this, two mutant constructs pΔ5-200/48cat or pΔ10-200/48cat, in which the human FETUA promoter was deleted over 5 or 10 nt within dGdT9, were designed to narrow down the distance between the (C1+N1) and (C2+N2) areas by half a turn or a full turn of DNA helix, respectively. The resulting promoter activities are shown in Figure 6. In the presence of NCM, i.e. in quiescent cells, the activity of pΔ5-200/48cat was reduced 2-fold as compared to p-200/48cat whereas pΔ10-200/48cat remained fully active. This was seen whether a pC/EBPα expression plasmid was co-transfected or not. These data point to a concerted, phasing-dependent effect of the (C1+N1) and (C2+N2) areas upon the basal FETUA transcription. As the modified structure of the human promoter in pΔ10-200/48cat is similar to that naturally found in rodents, this phasing-dependent activity is most likely retained in rodents. Moreover, the CM-induced down-regulation seen with p-200/48cat was abolished in pΔ5-200/48cat but was fully retained in pΔ10-200/48cat. In fact, in CM-stimulated cells the relative activities of all three constructs without pC/EBPα co-transfection were similar and these activities still varied moderately when pC/EBPα was present. This indicates that the importance of the (C1+N1)/(C2+N2) phasing upon the FETUA promoter activity as seen in quiescent cells is lost in the presence of pro-inflammatory cytokines.
Figure 6.
Phasing of NF-1 and C/EBP boxes in the FETUA promoter and CM-induced responsiveness. Two mutants of p-200/48cat lacking either 5 Ts (pΔ5-200/48cat) or 10 nt, namely dGdT9 (pΔ10-200/48cat) disrupt or retain the phasing between (N1+C1) and (N2+C2), as depicted. These mutants were co-transfected with pC/EBPα or pHD. Promoter activities are relative to p-200/48cat activity (100%) in the presence of pHD and NCM. For every cat construct the CM-induced repression relative to the reference activity in NCM-cultured cells is indicated by a broken arrow and a value (folds). Other details are as in Figure 3.
Importance of the size of C/EBPβ isoforms for FETUA/cat activity
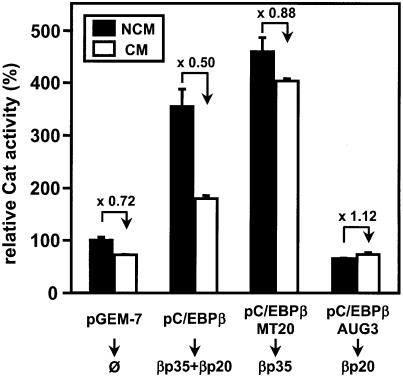
Owing to several translation start sites in the C/EBPα and -β mRNAs, long C/EBP isoforms with a complete transactivation domain (e.g. αp42 and βp35) are present in quiescent hepatocytes whereas short C/EBPβ isoforms (mostly βp20) lacking most of the transactivation domain predominate in proinflammatory cytokine-stimulated hepatocytes (31,32,37,42). Therefore, we investigated a possible importance of long versus short C/EBPβ isoforms for FETUA promoter activity. This was done by co-transfecting p-200/48cat with (i) pC/EBPβ that, similar to the C/EBPβ mRNA in vivo, is able to give rise to both βp35 and βp20, or (ii) its mutants pC/EBPβMT20 or pC/EBPβAUG3, which respectively produce βp35 or βp20 only (31,32). The data obtained in NCM- or CM-stimulated Hep3B cells are shown in Figure 7. Co-transfecting p-200/48cat with pC/EBPβ resulted in an enhanced Cat activity when compared to co-transfection with the control, empty plasmid and this activity was down-regulated 2-fold in CM-stimulated cells. In contrast, co-transfection of pC/EBPβMT20 enhanced even further p-200/48cat activity but prevented the CM-induced down-regulation. Finally, co-transfection of pC/EBPβAUG3 resulted in a lack of regulation of p-200/48cat activity in either NCM- or CM-stimulated cells. All these data are consistent with an activation of the FETUA promoter by the long βp35 isoform as opposed to a lack thereof when the short βp20 isoform predominated in the transfected cells. However, βp20 did not behave as a genuine repressor as the promoter activities measured in cells transfected with either pCMV or pC/EBPβAUG3 were similar.
Figure 7.
Co-transfection of a FETUAcat plasmid with various expression plasmids for long and/or short C/EBPβ isoforms. p-200/48cat was co-transfected with pC/EBPβ (100 ng) that gives rise to βp35 and βp20 or two mutants pC/EBPβMT20 (100 ng) or pC/EBPβAUG3 (100 ng) that produce βp35 or βp20, respectively. Promoter activities are relative to p-200/48cat activity (100%) co-transfected with an empty pGEM-7 plasmid (100 ng) in the presence of NCM. The CM-induced repression relative to the reference activity in NCM-cultured cells is indicated by a broken arrow and a value (folds).
EMSAs for NF-1 binding sites
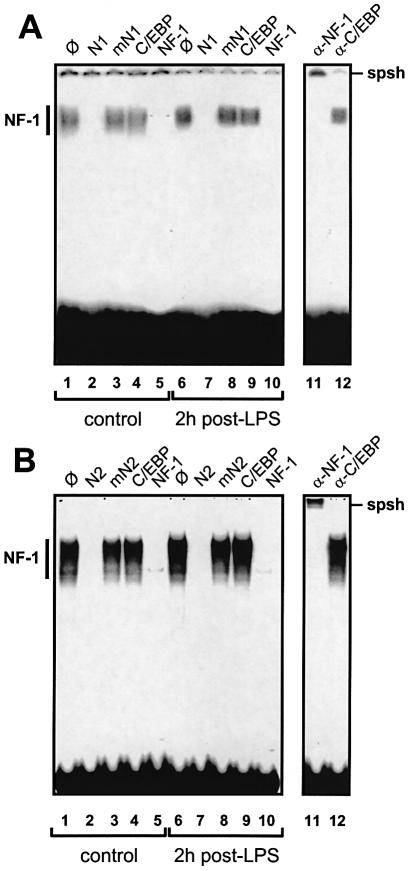
All four (N1, N2, C1 and C2) binding sites were studied by EMSA to further assess their relative importance in the basal versus acute phase-mediated transcription of the FETUA gene. As shown in Figure 8, the N1 and N2 binding sites were studied with two separate probes called N1 or N2 and liver nuclear extracts from normal or LPS-challenged rats. When using a normal liver extract, the patterns obtained with either N1 or N2 probe (Fig. 8A or B, lanes 1–5) were identical and made of multiple, specific bands in close proximity as expected for NF-1 complexes (28). With either probe, binding specificity was assessed from the band disappearance that was induced by an autocompetitor (lane 2) or a competitor for NF-1 (lane 5) but not by an autocompetitor mutated in the NF-1 site (lane 3) or a control competitor for an unrelated TF binding site (lane 4). Super-shift experiments with anti-NF-1 antibodies (lane 11) or control antibodies (lane 12) confirmed these data. Interestingly, the intensity of the [N1 probe/NF-1] complexes was much weaker than that of the [N2 probe/NF-1] complexes, despite similar specific activities of these probes (see legend to Fig. 8 for relative exposure times). This indicates a relatively weak affinity of NF-1 for the N1 site. When using nuclear extracts from LPS-challenged rats the same EMSA patterns and competitions as above were obtained with the N1 or N2 probe (Fig. 8A or B, lanes 6–10). We concluded that the binding of NF-1 to the N1 and N2 sites remained unchanged during the acute phase.
Figure 8.
EMSAs for the NF-1 binding sites N1 and N2. (A) A labelled probe N1 and liver nuclear extracts from control rats (lanes 1–5) or animals at 2 h post-LPS (lanes 6–10) were used. A set of NF-1-induced complexes is collectively referred to as NF-1. The unlabelled probe N1, a mutant probe mN1 and oligos with a C/EBP or NF-1 consensus sequence were used as competitors (∅, no competitor) as noted above the lanes. Super-shifts were made with anti-NF-1 or anti-C/EBPβ antibodies (used as a negative control) noted α- above lanes 11 and 12 and supershifted bands are referred to as spsh. (B) Probe N2, all other details are as in (A). The autoradiographic exposure was done for 10 (A) or 2 days (B).
EMSAs for C/EBP binding sites
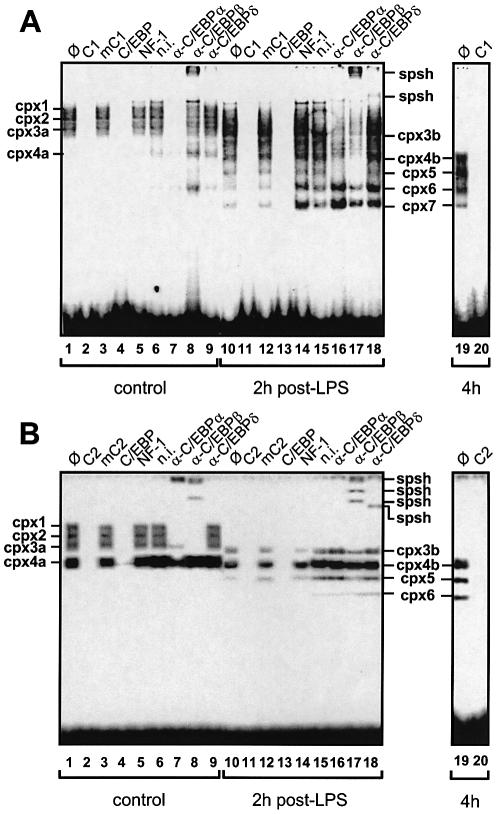
EMSAs allowed us to verify which C/EBP isoforms were bound to the C1 and C2 sites in quiescent versus cytokine-stimulated hepatocytes. When using a nuclear extract from normal rat liver, the C1 probe formed a set of four specific complexes, cpx1 to cpx4a (Fig. 9A, lane 1), as expected from a study with another promoter (36). Again, band specificity was proven with an autocompetitor (lane 2), a mutant autocompetitor in which the C1 box was abolished (lane 3) and competitors for C/EBP (lane 4) or an unrelated TF binding site (lane 5). Anti-C/EBPα (lane 7) and -β antisera (lane 8) but neither an anti-C/EBPδ antiserum (lane 9) nor a non-immunized rabbit serum (lane 6) induced either a band disappearance (lane 7) or a super-shift (lane 8), thus indicating that C/EBPα and -β but not C/EBPδ is responsible for the pattern seen in lane 1. When using a rat liver nuclear extract obtained at 2-h post-LPS (lanes 10–18), a weakening of cpx1 and cpx2 and a slightly increased mobility of the other two complexes, now called cpx3b and cpx4b, were observed (lane 10 versus 1) along with the appearance of three complexes of fast mobility referred to as cpx5, cpx6 and cpx7 (lane 10). At 4 h post-LPS, only cpx4b to cpx7 were observed (lane 19). All three (cpx5, cpx6 and cpx7) were competed out by a C/EBP oligo (lanes 13 and 20). Also, cpx5 was abrogated by the anti-C/EBPα and -β antisera (lanes 16–17) and cpx6 and cpx7 were weakened by the anti-C/EBPβ antiserum when compared to the anti-C/EBPα or -δ antisera (lane 17 versus 16 and 18). A faint supershift induced by the anti-C/EBPδ antiserum was also observed (lane 18) but it could not be specifically ascribed to a single complex. The immunoreactivities of cpx1 to cpx7 are summarized in Table 1.
Figure 9.
EMSAs for the C/EBP binding sites C1 and C2. (A) A labelled probe C1 and liver nuclear extracts from control rats (lanes 1–9) or animals at 2 (lanes 10–18) or 4 h post-LPS challenge (lanes 19 and 20) were used. The unlabelled probe C1, a mutant probe mC1 and oligos with a C/EBP or NF-1 consensus sequence were used as competitors (∅, no competitor) as noted above lanes 1–5, 10–14 and 19 and 20. Super-shifts were made with various antisera noted α- above lanes 6–9 and 15–18 (n.i., non immunized rabbit serum) and supershifted bands are referred to as spsh. Lanes 1–18 and 19 and 20 are from two separate gels. (B) Probe C2, all other details are as in (A). In (A) and (B), various C/EBP complexes are noted cpx1 to cpx7. Differently migrating cpx3 bands are noted cpx3a or cpx3b; likewise, cpx4 is noted cpx4a or cpx4b. The autoradiographic exposure was done for 2 (A) or 10 days (B).
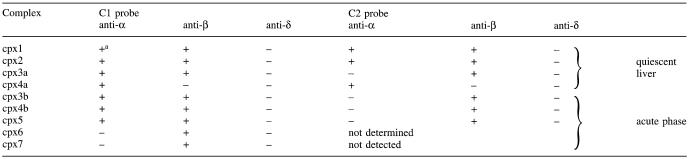
Table 1. Immunoreactivity of the complexes formed with the C1 or C2 probe.
aThe intensity of a given complex in EMSA was weakened/suppressed (+) or remained unchanged (–) with the indicated anti-C/EBP antiserum. These data summarize the supershifts shown in Figure 9.
When using a nuclear extract from normal rat liver, the C2 probe formed three very faint but specific complexes and a major one (Fig. 9B, lane 1). As above, band specificity was checked with an autocompetitor (lane 2), a mutant thereof (lane 3) and a competitor oligo for C/EBP (lane 4) or an unrelated TF (lane 5), which identified these complexes as made of C/EBP proteins. Their general pattern and relative migrations argue for the presence of counterparts of the cpx1, cpx2, cpx3a and cpx4a complexes described above. However, the intensity of cpx1, cpx2 and cpx3a with the C2 probe was much weaker than what was seen with the C1 probe (see legend to Fig. 9 for relative exposure times). This difference may be accounted for by mismatches between the C2 sequence and the C/EBP consensus (Fig. 4). An anti-C/EBPα antiserum promoted a band retention in the loading well and a concomitant disappearance or decrease of all complexes but cpx3a (lane 7). An anti-C/EBPβ antiserum also induced supershifted bands and the disappearance of cpx1, cpx2 and cpx3a (lane 8). An anti-C/EBPδ antiserum had no effect (lane 9). When using a rat liver nuclear extract obtained at 2 h post-LPS (lanes 10–18), the disappearance of cpx1 and cpx2 and a slightly increased mobility of the other two complexes, now called cpx3b and cpx4b, were observed (lane 10 versus 1), as with the C1 probe. At 2 h post-LPS, and even more so at 4 h, two further, fast complexes appeared (lane 19). Given their relative mobilities with respect to cpx4b they are referred to as cpx5 and cpx6, as above with the C1 probe. From the band weakening and supershifts induced by the anti-C/EBPβ (lane 17) but not by the anti-C/EBPα antiserum (lane 16) we further concluded that all three cpx3b, cpx4b and cpx5 contain C/EBPβ isoforms and no C/EBPα, a feature that was not seen with a normal liver (lanes 7–9). A faint supershift with the anti-C/EBPδ antiserum (lane 18) suggested that some of these complexes also contained C/EBPδ. The immunoreactivities of cpx1 to cpx6 are summarized in Table 1.
Taken together, the EMSA results pointed to shared properties of the C1 and C2 binding sites, including: (i) an LPS-induced, transient disappearance of cpx1, cpx2 and cpx3 as well as a change in the mobility of cpx4a towards a smaller size cpx4b; (ii) an LPS-induced change in the nature of cpx4a mostly made of C/EBPα towards cpx4b mostly made of C/EBPβ; and (iii) an LPS-induced appearance of cpx5 and cpx6, with cpx5 being comprised of short C/EBPα and -β isoforms (C1) or short C/EBPβ isoforms only (C2).
LPS-induced down-regulation of the hepatic fetua mRNA in mouse
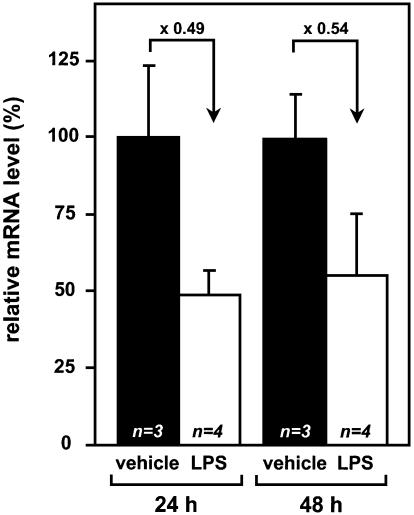
All the above features pointed to shared properties of the human, rat and mouse promoters but the mouse fetua gene has so far been considered to lack any responsiveness to acute phase (Discussion). Therefore, we questioned whether the mouse fetua mRNA indeed lacks an acute phase-associated down-regulation. Real-time Q-RT–PCR of fetua mRNA in mouse livers (Fig. 10) clearly demonstrated that LPS-challenged mice down-regulate 2-fold this mRNA level, in keeping with what is known in human and rat.
Figure 10.
Down-regulation of the hepatic Fetuin-A mRNA level in LPS-challenged mice. The Fetuin-A mRNA level was measured by real-time Q-RT–PCR at 24 or 48 h post-challenge in mice given either LPS in an aqueous NaCl solution (n = 4) or the vehicle alone as a control (n = 3). At each time point, the LPS-induced mRNA levels are relative to the average level in mice given the vehicle alone.
DISCUSSION
We have found that two C/EBP (C1 and C2) and two NF-1 (N1 and N2) binding sites account for the basal activity of the human FETUA promoter, which is in keeping with a former study in the rat (27). On the basis of the residual Cat activity of mutant constructs, the relative basal activities of these four sites can be approximately ranked N1 << N2 = C2 < C1 in human. In keeping with this, N2 harbours mismatches with only one side of the bi-partite NF-1 consensus and provides a strong signal in EMSA whereas N1 harbours mismatches on both sides and provides a limited signal in EMSA. Interestingly, reverse features apply for the rodent N1 and N2 sequences, which fits the relative importance N1 > N2 reported in rat (27). It has been proposed that the hepatic NF-1 p54 isoform is a negative sensor of cellular stress (43) and some NF-1 isoforms have been regarded as transcriptional co-regulators because of their location next to other TF binding sites (44). Although the N1, N2, C1 and C2 sites are close to each other, our data argue against any significant involvement of N1 and N2 in the acute phase-induced down-regulation of the FETUA promoter. Indeed, our mutant cat plasmids with an abolished N1 or N2 site fully retained a capacity to be down-regulated in CM-stimulated Hep3B cells and the EMSA patterns of the N1 and N2 probes did not change when using liver nuclear extracts from LPS-challenged rats.
In the hepatocyte, the C/EBP isoforms αp42, αp30, αp20, βp38, βp35, βp20 and δp35 all bind C/EBP binding sites as homo- or hetero-dimers of various sizes (36,37,45). The overall pattern of the cpx1 to cpx5 complexes seen herein by EMSA is quite similar to what has been previously observed in the context of another APP in mouse, namely orosomucoid (36). In the orosomucoid promoter, cpx1 and cpx2 have been tentatively identified as (αp42+αp42), cpx3 as (αp42+βp35) and cpx4 as (βp20+βp20) (36). We anticipate that quite similar complexes form with the C1 and C2 sites in the FETUA promoter although our supershifts suggest some minor discrepancies between the subunit composition of the C/EBP dimers bound to the FETUA and orosomucoid promoters. Such differences are most likely to be ascribed to different promoter contexts. During the acute phase the short αp30 and βp20 isoforms as well as C/EBPδ replace the large αp42, βp38 and βp35 isoforms in the hepatocyte nucleus (26,36,37). In the present study, the disappearance of the slow cpx1, cpx2 and cpx3a complexes, the changes in the mobility of cpx3a and cpx4a towards different, smaller size heterodimers within cpx3b and cpx4b, and the appearance of the fast cpx5 to cpx7 are all in keeping with such a kinetic view. We conclude that in the quiescent hepatocyte, both C1 and C2 binding sites are able to form slowly migrating complexes that involve large C/EBPα and -β isoforms whereas after an LPS challenge, the cpx1 to cpx4a complexes are transiently replaced by others of faster migration that involve short C/EBPα and -β isoforms. Taken together, our EMSA patterns and the relative Cat activities of various wild type and mutant cat constructs all argue for the C/EBP binding sites C1 and C2 being central to the phasing-dependent activity of the FETUA promoter in the quiescent hepatocyte as well as to its acute phase-promoted down-regulation. Accordingly, the FETUA transcription appears to take place as follows. In the quiescent cell, long C/EBP isoforms are bound to C1 and C2 that together activate the transcription in a phase-dependent fashion. Whether this phasing acts at the C/EBP binding stage or involves a direct or cofactor-mediated contact between the bound C/EBP proteins remains to be seen. In the acute phase-triggered hepatocyte, short C/EBP isoforms mostly made of C/EBPα and -β replace the long isoforms and a weakened activity results from a loss of phase-dependent interaction between these short isoforms. A similar loss of interaction was created in pΔ5-200/48cat which, accordingly, did not respond to cytokines. Also, a weak or even absent transactivating capacity of most short C/EBP isoforms is well established (37) and this was also observed in our study when the βp20-producing expression plasmid was used in a co-transfection experiment. Therefore the lack of transactivation of short C/EBPβ isoforms likely participates in the reduced activity of the FETUA promoter during the acute phase but it is in no way the single event at this stage as it adds to the loss of cooperation between C1 and C2. Finally, our comparison of Hep3B versus HepG2 cells brings further evidence for the above mechanisms. In the HepG2 cells, the FETUA down-regulation under cytokine challenge was prevented by the high, possibly saturating level of the short αp30, αp27 and αp20 isoforms already found in the resting cells. Likewise, in the permissive Hep3B cells a block of FETUA down-regulation could be obtained when the cells were enriched in either long or short β isoforms produced by plasmid co-transfection.
The overall sequence of the FETUA promoter, the number and relative arrangement of the four NF-1 and C/EBP binding sites and the phasing along the DNA helix are conserved from human to rat and mouse. Yet, both human and rat FETUA mRNAs are down-regulated by the acute phase (9,22–25) whereas the mouse fetua gene has been considered to lack any responsiveness to inflammation (46). In fact, we have demonstrated that LPS-treated mice down-regulate 2-fold their hepatic fetua mRNA level at 1 and 2 days post-challenge. Therefore, we now conclude that the above mechanism of hepatic FETUA transcription in the quiescent or acute phase-triggered hepatocyte is shared between man, rat and mouse.
In our study, the pro-inflammatory cytokines that promote the down-regulation of the FETUA gene have not been considered separately as the participation of TNFα, IL-1β and IL-6 has been previously established (9,22,23). Our conclusion that C/EBP binding sites mediate the FETUA down-regulation perfectly fits the mandatory requirement for IL-1β in the acute phase-associated regulation of this gene (9,22,23). Indeed, the IL-1β transduction pathway primarily involves C/EBP family members whereas the IL-6 pathway mostly involves STAT members (26). Yet, the IL-1β/C/EBP and IL-6/STAT pathways should not be regarded as strictly independent as IL-6 can secondarily up-regulate the C/EBP genes in liver (26). Therefore an IL-6-sustained C/EBPβ synthesis may account for (i) the established need for an IL-1β + IL-6 co-stimulation to observe a maximum down-regulation of the Fetuin-A mRNA level in human hepatoma cells (22) and (ii) the relatively late down-regulation of rat fetua gene transcription and mRNA level that respectively plateau at 16 h and 1–2 days after the start of an experimental inflammation (6,29).
The mechanisms behind the down-regulated transcription of negative APP-encoding genes in liver have been elucidated in a limited number of instances (30,47–50). This is accounted for by a limited decrease in mRNA level and transcription (24,30,47,48,50) which makes a functional promoter analysis often difficult. Various mechanisms have been put forward to account for this acute phase-dependent down-regulation of negative APP-encoding genes in liver. In the TRANSTHYRETIN promoter two AP-1 and HNF-3 binding sites overlap with a predominant occupancy of the latter; however during the acute phase, occupancy of the AP-1 site by a neosynthesized Jun–Jun homodimer with a low transactivating activity is favoured over HNF-3 binding (47). In the APOCIII promoter, an acute phase-associated binding of C/EBPδ to its cognate sequence interferes with the arrangement of a set of otherwise activating TFs bound to a distant site (30). An acute phase-mediated modification of chromatin arrangement has been put forward to account for the transient lack of access of an activating GAGA box-binding protein to the rat spi2.1 promoter (48). An IL-6-driven TF has been considered to negatively interfere with the HNF-1-dependent transcription of the rat glutathione-S-transferase A2 gene (50). Finally, a TNFα-induced export of C/EBPβ out of the nucleus results in a down-regulation of the ALBUMIN gene (49). Our data now indicate yet an entirely different mechanism for the acute phase-regulated transcription of another negative APP-encoding gene, namely the transient loss of a concerted activity between long C/EBP isoforms tethered to two neighbouring binding sites. Given the important functions of Fetuin-A in inflammation, including a limitation of cytokine production by macrophages (20,21) and a protection against TNF (19), the present data pave the way for a controlled expression of this protein whose up-regulation may prove beneficial in life-threatening situations such as a septic syndrome complicated with multiple organ failure.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Dr J. L. Danan (CNRS, Meudon, France) for many helpful comments on the manuscript and to Dr J. M. Lacorte (Institut des Cordeliers, Paris) for the generous gift of various plasmids. C.G. is the recipient of a doctoral fellowship from the French Ministère de la Recherche. This work was supported in part by the University of Rouen, France.
REFERENCES
- 1.Kellermann J., Haupt,H., Auerswald,E.A. and Müller-Esterl,W. (1989) The arrangement of disulfide loops in human alpha2-HS glycoprotein. Similarity to the disulfide bridge structures of cystatins and kininogens. J. Biol. Chem., 264, 14121–14128. [PubMed] [Google Scholar]
- 2.Dziegielewska K.M., Brown,W.M., Casey,S.J., Chrisite,D.L., Foreman,R.C., Hill,R.M. and Saunders,N.R. (1990) The complete cDNA and amino acid sequence of bovine fetuin. Its homology with alpha2HS glycoprotein and relation to other members of the cystatin superfamily. J. Biol. Chem., 265, 4354–4357. [PubMed] [Google Scholar]
- 3.Le Cam A., Auberger,P., Falquerho,L., Contreres,O.J., Pages,G., Le Cam,G. and Rossi,B. (1992) pp63 is very likely the rat fetuin. Cell, 68, 8. [DOI] [PubMed] [Google Scholar]
- 4.Rauth G., Pöschke,O., Fink,E., Eulitz,M., Tippmer,S., Kellerer,M., Häring,H.U., Nawratil,P., Haasemann,M., Jahnen-Dechent,W. and Müller-Esterl,W. (1992) The nucleotide and partial amino acid sequences of rat fetuin. Identity with the natural tyrosine kinase inhibitor of the rat insulin receptor. Eur. J. Biochem., 204, 523–529. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto K. and Sinohara,H. (1993) Isolation and characterization of mouse countertrypin, a new trypsin inhibitor belonging to the mammalian fetuin family. J. Biol. Chem., 268, 17750–17753. [PubMed] [Google Scholar]
- 6.Olivier E., Soury,E., Ruminy,P., Husson,A., Parmentier,F., Daveau,M. and Salier,J.P. (2000) Fetuin-B, a second member of the fetuin family in mammals. Biochem. J., 350, 589–597. [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H., Srinivas,P.R., Cong,L.N., Li,Y., Grunberger,G. and Quon,M.J. (1998) α2-Heremans Schmid glycoprotein inhibits insulin-stimulated Elk-1 phosphorylation, but not glucose transport, in rat adipose cells. Endocrinology, 139, 4147–4154. [DOI] [PubMed] [Google Scholar]
- 8.Auberger P., Falquerho,L., Contreres,J.O., Pages,G.L.E., Le Cam,G., Rossi,B.L.E. and Le Cam,A. (1989) Characterization of a natural inhibitor of the insulin receptor tyrosine kinase: cDNA cloning, purification and anti-mitogenic activity. Cell, 58, 631–640. [DOI] [PubMed] [Google Scholar]
- 9.Akhoundi C., Amiot,M., Auberger,P., Le Cam,A. and Rossi,B. (1994) Insulin and interleukin-1 differentially regulate pp63, an acute phase phosphoprotein in hepatoma cell line. J. Biol. Chem., 269, 15925–15930. [PubMed] [Google Scholar]
- 10.Mathews S.T., Chellam,N., Srinivas,P.R., Cintron,V.J., Leon,M.A., Goustin,A.S. and Grunberger,G. (2000) alpha(2)-HSG, a specific inhibitor of insulin receptor autophosphorylation, interacts with the insulin receptor. Mol. Cell. Endocrinol., 164, 87–98. [DOI] [PubMed] [Google Scholar]
- 11.Mathews S.T., Singh,G.P., Ranalletta,M., Cintron,V.J., Qiang,X., Goustin,A.S., Jen,K.L., Charron,M.J., Jahnen-Dechent,W. and Grunberger,G. (2002) Improved insulin sensitivity and resistance to weight gain in mice null for the Ahsg gene. Diabetes, 51, 2450–2458. [DOI] [PubMed] [Google Scholar]
- 12.Jahnen-Dechent W., Schinke,T., Trindl,A., Müller-Esterl,W., Sablitzky,F., Kaiser,S. and Blessing,M. (1997) Cloning and targeted deletion of the mouse fetuin gene. J. Biol. Chem., 272, 31496–31503. [DOI] [PubMed] [Google Scholar]
- 13.Price P.A., Thomas,G.R., Pardini,A.W., Figueira,W.F., Caputo,J.M. and Williamson,M.K. (2002) Discovery of a high molecular weight complex of calcium, phosphate, fetuin and matrix gamma-carboxyglutamic acid protein in the serum of etidronate-treated rats. J. Biol. Chem., 277, 3926–3934. [DOI] [PubMed] [Google Scholar]
- 14.Heiss A., DuChesne,A., Denecke,B., Grotzinger,J., Yamamoto,K., Renne,T. and Jahnen-Dechent,W. (2003) Structural basis of calcification inhibition by alpha 2-HS glycoprotein/fetuin-A. Formation of colloidal calciprotein particles. J. Biol. Chem., 278, 13333–13341. [DOI] [PubMed] [Google Scholar]
- 15.Schinke T., Amendt,C., Trindl,A., Pöschke,O., Müller-Esterl,W. and Jahnen-Dechent,W. (1996) The serum protein α2-HS glycoprotein/fetuin inhibits apatite formation in vitro and in mineralizing calvaria cells. A possible role in mineralization and calcium homeostasis. J. Biol. Chem., 271, 20789–20796. [DOI] [PubMed] [Google Scholar]
- 16.Demetriou M., Binker,C., Sukhu,B., Tenenbaum,H.C. and Dennis,J.W. (1996) Fetuin/α2-HS glycoprotein is a transforming growth factor-β type II receptor mimic and cytokine antagonist. J. Biol. Chem., 271, 12755–12761. [DOI] [PubMed] [Google Scholar]
- 17.Szweras M., Liu,D., Partridge,E.A., Pawling,J., Sukhu,B., Clokie,C., Jahnen-Dechent,W., Tenebaum,H.C., Swallow,C.J., Grynpas,M.D. and Dennis,J.W. (2002) α2-HS glycoprotein/fetuin, a transforming growth factor-β/bone morphogenetic protein antagonist, regulates postnatal bone growth and remodeling. J. Biol. Chem., 277, 19991–19997. [DOI] [PubMed] [Google Scholar]
- 18.Ohnishi T., Nakamura,O., Arakaki,N. and Daikuhara,Y. (1997) Effect of phosphorylated rat fetuin on the growth of hepatocytes in primary culture in the presence of human hepatocyte-growth factor. Evidence that phosphorylated fetuin is a natural modulator of hepatocyte-growth factor. Eur. J. Biochem., 243, 753–761. [DOI] [PubMed] [Google Scholar]
- 19.Wang H., Zhang,M., Soda,K., Sama,A. and Tracey,K.J. (1997) Fetuin protects the fetus from TNF. Lancet, 350, 861–862. [DOI] [PubMed] [Google Scholar]
- 20.Wang H., Zhang,M., Bianchi,M., Sherry,B., Sama,A. and Tracey,K.J. (1998) Fetuin (α2-HS-glycoprotein) opsonizes cationic macrophage-deactivating molecules. Proc. Natl Acad. Sci. USA, 95, 14429–14434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ombrellino M., Wang,H., Yang,H., Zhang,M., Vishnubhakat,J., Frazier,A., Scher,L.A., Friedman,S.G. and Tracey,K.J. (2001) Fetuin, a negative acute phase protein, attenuates TNF synthesis and the innate inflammatory response to carrageenan. Shock, 15, 181–185. [DOI] [PubMed] [Google Scholar]
- 22.Daveau M., Davrinche,C., Julen,N., Hiron,M., Arnaud,P. and Lebreton,J.P. (1988) The synthesis of human α2-HS glycoprotein is down-regulated by cytokines in hepatoma HepG2 cells. FEBS Lett., 241, 191–194. [DOI] [PubMed] [Google Scholar]
- 23.Daveau M., Davrinche,C., Djelassi,N., Lemetayer,J., Julen,N., Hiron,M., Arnaud,P. and Lebreton,J.P. (1990) Partial hepatectomy and mediators of inflammation decrease the expression of liver α2-HS glycoprotein gene in rats. FEBS Lett., 273, 79–81. [DOI] [PubMed] [Google Scholar]
- 24.Milland J., Tsykin,A., Thomas,T., Aldred,A.R., Cole,T. and Schreiber,G. (1990) Gene expression in regenerating and acute-phase rat liver. Am. J. Physiol., 259, G340–G347. [DOI] [PubMed] [Google Scholar]
- 25.Abiodun P.O. and Olomu,I.N. (1991) Serum alpha2-HS-glycoprotein levels in nenonatal infections. Biol. Neonate, 60, 114–117. [DOI] [PubMed] [Google Scholar]
- 26.Ruminy P., Gangneux,C., Claeyssens,S., Scotte,M., Daveau,M. and Salier,J.P. (2001) Gene transcription in hepatocyte during the acute phase of a systemic inflammation: from transcription factors to target genes. Inflamm. Res., 50, 383–390. [DOI] [PubMed] [Google Scholar]
- 27.Falquerho L., Paquereau,L., Vilarem,M.J., Galas,S., Patey,G. and Le Cam,A. (1992) Functional characterization of the promoter of pp63, a gene encoding a natural inhibitor of the insulin receptor tyrosine kinase. Nucleic Acids Res., 20, 1983–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banine F., Gangneux,C., Mercier,L., Le Cam,A. and Salier,J.P. (2000) Positive and negative elements modulate the promoter of the human, liver-specific α2-HS glycoprotein gene. Eur. J. Biochem., 267, 1214–1222. [DOI] [PubMed] [Google Scholar]
- 29.Daveau M., Jean,L., Soury,E., Olivier,E., Masson,S., Lyoumi,S., Chan,P., Hiron,M., Lebreton,J.P., Husson,A. et al. (1998) Hepatic and extra-hepatic transcription of Inter-α-Inhibitor family genes under normal or acute inflammatory conditions in rat. Arch. Biochem. Biophys., 350, 315–323. [DOI] [PubMed] [Google Scholar]
- 30.Lacorte J.M., Ktistaki,E., Beigneux,A., Zannis,V.I., Chambaz,J. and Talianidis,I. (1997) Activation of CAAT enhancer-binding protein δ (C/EBPδ) by interleukin-1 negatively influences apolipoprotein C-III expression. J. Biol. Chem., 272, 23578–23584. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh C.C., Xiong,W., Xie,Q., Rabek,J.P., Scott,S.G., An,M.R., Reisner,P.D., Kuninger,D.T. and Papaconstantinou,J. (1998) Effects of age on the posttranscriptional regulation of CCAAT/enhancer binding protein alpha and CCAAT/enhancer binding protein beta isoform synthesis in control and LPS-treated livers. Mol. Biol. Cell, 9, 1479–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong W., Hsieh,C.C., Kurtz,A.J., Rabek,J.P. and Papaconstantinou,J. (2001) Regulation of CCAAT/enhancer-binding protein-beta isoform synthesis by alternative translational initiation at multiple AUG start sites. Nucleic Acids Res., 29, 3087–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexander M.C., Lomanto,M., Nasrin,N. and Ramaika,C. (1988) Insulin stimulates glyceraldehyde-3-phosphate dehydrogenase gene expression through cis-acting DNA sequences. Proc. Natl Acad. Sci. USA, 85, 5092–5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinberg R.A. and Gorman,K.B. (1994) A high-yield method for site-directed mutagenesis using polymerase chain reaction and three primers. Anal. Biochem., 219, 155–157. [DOI] [PubMed] [Google Scholar]
- 35.Morel Y. and Barouki,R. (1998) Down-regulation of cytochrome P450 1A1 gene promoter by oxidative stress. Critical contribution of nuclear factor 1. J. Biol. Chem., 273, 26969–26976. [DOI] [PubMed] [Google Scholar]
- 36.An M.R., Hsieh,C.C., Reisner,P.D., Rabek,J.P., Scott,S.G., Kuninger,D.T. and Papaconstantinou,J. (1996) Evidence for posttranscriptional regulation of C/EBPα and C/EBPβ isoform expression during the lipopolysaccharide-mediated acute-phase response. Mol. Cell. Biol., 16, 2295–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramji D.P. and Foka,P. (2002) CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J., 365, 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubicka S., Kuhnel,F., Zender,L., Rudolph,K.L., Plumpe,J., Manns,M. and Trautwein,C. (1999) p53 represses CAAT enhancer-binding protein (C/EBP)-dependent transcription of the albumin gene. A molecular mechanism involved in viral liver infection with implications for hepatocarcinogenesis. J. Biol. Chem., 274, 32137–32144. [DOI] [PubMed] [Google Scholar]
- 39.Greenwel P., Tanaka,S., Penkov,D., Zhang,W., Olive,M., Moll,J., Vinson,C., Di Liberto,M. and Ramirez,F. (2000) Tumor necrosis factor alpha inhibits type I collagen synthesis through repressive CCAAT/enhancer-binding proteins. Mol. Cell. Biol., 20, 912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okazaki K., Li,J., Yu,H., Fukui,N. and Sandell,L.J. (2002) CCAAT/enhancer-binding proteins β and δ mediate the repression of gene transcription of cartilage-derived retinoic acid-sensitive protein induced by interleukin-1β. J. Biol. Chem., 277, 31526–31533. [DOI] [PubMed] [Google Scholar]
- 41.Essani N.A., McGuire,G.M., Manning,A.M. and Jaeschke,H. (1996) Endotoxin-induced activation of the nuclear transcription factor kappa B and expression of E-selectin messenger RNA in hepatocytes, Kupffer cells and endothelial cells in vivo. J. Immunol., 156, 2956–2963. [PubMed] [Google Scholar]
- 42.Welm A.L., Mackey,S.L., Timchenko,L.T., Darlington,G.J. and Timchenko,N.A. (2000) Translational induction of liver-enriched transcriptional inhibitory protein during acute phase response leads to repression of CCAAT/enhancer binding protein alpha mRNA. J. Biol. Chem., 275, 27406–27413. [DOI] [PubMed] [Google Scholar]
- 43.Morel Y., Coumoul,X., Nalpas,A. and Barouki,R. (2000) Nuclear factor I/CCAAT box transcription factor trans-activating domain is a negative sensor of cellular stress. Mol. Pharmacol., 58, 1239–1246. [DOI] [PubMed] [Google Scholar]
- 44.Osada S., Matsubara,T., Daimon,S., Terazu,Y., Xu,M., Nishihara,T. and Imagawa,M. (1999) Expression, DNA-binding specificity and transcriptional regulation of nuclear factor 1 family proteins from rat. Biochem. J., 342, 189–198. [PMC free article] [PubMed] [Google Scholar]
- 45.Calkhoven C.F., Müller,C. and Leutz,A. (2000) Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev., 14, 1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 46.Kalmovarin N., Friedrichs,W.E., O‘Brien,H.V., Linehan,L.A., Bowman,B.H. and Yang,F. (1991) Extrahepatic expression of plasma protein genes during inflammation. Inflammation, 15, 369–379. [DOI] [PubMed] [Google Scholar]
- 47.Qian X., Samadani,U., Porcella,A. and Costa,R.H. (1995) Decreased expression of hepatocyte nuclear factor 3 alpha during the acute-phase response influences transthyretin gene transcription. Mol. Cell. Biol., 15, 1364–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simar-Blanchet A.E., Legraverend,C., Thissen,J.P. and Le Cam,A. (1998) Transcription of the rat serine protease inhibitor 2.1 gene in vivo: correlation with GAGA box promoter occupancy and mechanism of cytokine-mediated down-regulation. Mol. Endocrinol., 12, 391–404. [DOI] [PubMed] [Google Scholar]
- 49.Buck M., Zhang,L., Halasz,N.A., Hunter,T. and Chojkier,M. (2001) Nuclear export of phosphorylated C/EBPbeta mediates the inhibition of albumin expression by TNF-alpha. EMBO J., 20, 6712–6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voss S.H., Whalen,R. and Boyer,T.D. (2002) Mechanism of negative regulation of rat glutathione S-transferase A2 by the cytokine interleukin 6. Biochem. J., 365, 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matys V., Fricke,E., Geffers,R., Gossling,E., Haubrock,M., Hehl,R., Hornischer,K., Karas,D., Kel,A.E., Kel-Margoulis,O.V. et al. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res., 31, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]