Abstract
‘10-23’ DNAzymes can be used to cleave any target RNA in a sequence-specific manner. For applications in vivo, they have to be stabilised against nucleolytic attack by the introduction of modified nucleotides without obstructing cleavage activity. In this study, we optimise the design of a DNAzyme targeting the 5′-non-translated region of the human rhinovirus 14, a common cold virus, with regard to its kinetic properties and its stability against nucleases. We compare a large number of DNAzymes against the same target site that are stabilised by the use of a 3′-3′-inverted thymidine, phosphorothioate linkages, 2′-O-methyl RNA and locked nucleic acids, respectively. Both cleavage activity and nuclease stability were significantly enhanced by optimisation of arm length and content of modified nucleotides. Furthermore, we introduced modified nucleotides into the catalytic core to enhance stability against endonucleolytic degradation without abolishing catalytic activity. Our findings enabled us to establish a design for DNAzymes containing nucleotide modifications both in the binding arms and in the catalytic core, yielding a species with up to 10-fold enhanced activity and significantly elevated stability against nucleolytic cleavage. When transferring the design to a DNAzyme against a different target, only a slight modification was necessary to retain activity.
INTRODUCTION
DNAzymes of the 10-23 type are catalytically active nucleic acids that cleave complementary RNA molecules with high activity in a reaction dependent on divalent cations (reviewed in 1). Consisting of a catalytic core of 15 nt and two substrate-binding arms of variable length and sequence, they bind the target RNA in a sequence-specific manner and cleave it between a paired pyrimidine base and a free purine base (2,3). Cleavage efficiency is highest at AU and GU sites, but Cairns et al. (4) recently demonstrated that DNAzymes targeted against AC and GC sites can be improved by the introduction of inosine bases. Since their generation by Santoro and Joyce in 1997 using in vitro selection technology (2), 10-23 DNAzymes have been employed successfully to inhibit the expression of a number of viral and endogenous target genes in cell culture studies and in vivo (reviewed in 5). DNAzymes can be designed to cleave any desired target sequence with high specificity, discriminating against one base mismatches (6). They are often more active than ribozymes consisting of RNA (7), the requirements for the cleavage site are minimal, and as they are composed of DNA, they are relatively easy to synthesise and handle. For these reasons, DNAzymes hold great promise for diagnostic and therapeutic application.
Similar to other antisense molecules, DNAzymes are prone to nucleolytic degradation in body fluids. To prolong the half-life of oligonucleotides for in vivo usage, modified nucleotides are usually incorporated. In recent years, great progress has been achieved by the development of novel chemically modified nucleotides with improved properties like enhanced biostability, low toxicity and high target affinity (8). The introduction of modified nucleotides into catalytically active oligonucleotides is especially challenging, since it might influence their three dimensional structure and thereby lead to a reduction of cleavage activity. Beigelman et al. performed a systematic study for the hammerhead ribozyme that led to an optimised design with five unmodified ribonucleotides, two 2′-C-allyl uridines and 2′-O-methyl RNAs at all remaining positions (9).
A frequently used modification to stabilise DNAzymes is an inverted thymidine (Fig. 1B) at the 3′-end (10–14). In analogy to antisense molecules, phosphorothioate bonds have also widely been used for the stabilisation of DNAzymes (12,13,15–20) although they are known to decrease substrate affinity and tend to have toxic side-effects (21). 2′-O-methyl modifications were also successfully used to protect DNAzymes from nucleolytic degradation (18,19). Recently, the introduction of locked nucleic acids (LNA) into the binding arms has been reported (22,23).
Figure 1.
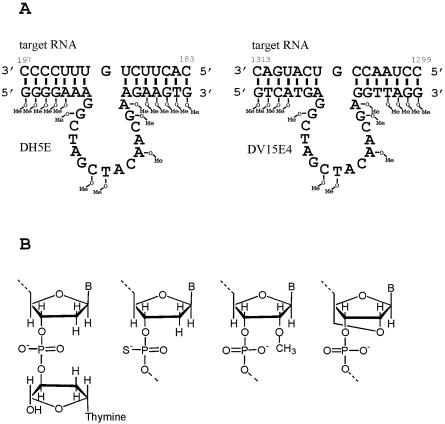
Sequences of the optimised DNAzymes and structures of modified nucleotides used in this study. (A) Sequences of DNAzymes against the 5′-NTR of HRV14 (DH5, left) and against the VR1 mRNA (DV 15, right) and their substrate RNAs. Positions of the target RNAs bound by the DNAzymes are indicated and nucleotides are marked at which 2′-O-methyl RNA monomers were introduced into the DNAzymes optimised in the present study. (B) Nucleotide modifications applied: 3′-inverted thymidine, phosphorothioate, 2′-O-methyl RNA and LNA.
In the present study, we examine the influence of the different modifications listed above on the stability and activity of a 10-23 DNAzyme (Fig. 1A). A large number of individually modified DNAzymes against the same target site in the 5′-non-translated region (NTR) of the human rhinovirus type 14 (HRV14) were compared and analysed with respect to their kinetics and stability against nucleases. HRV14 is one of about 100 human rhinoviruses causing the common cold. Human rhinoviruses, together with enteroviruses such as polio-, coxsackie- and echoviruses, belong to the picornavirus family (reviewed in 24). These viruses are small non-enveloped viruses with a positive sense single stranded RNA genome. The genomes of all human viruses and enteroviruses with a length ranging from 7200 to 7400 nt contain a 5′-NTR of approximately 630 nt for rhinoviruses and 740 nt for enteroviruses. The secondary structure of this 5′-NTR is conserved among the human rhinoviruses and enteroviruses and plays a major role in the regulation of viral transcription and translation. Therefore this sequence was selected as target site for the construction of specific DNAzymes. A second DNAzyme targeting the vanilloid receptor subtype 1 (VR1) mRNA was used as an independent control. This receptor, also designated as Transient Receptor Potential Channel V1, is a cation channel, which can be activated by capsaicin, heat and protons (25). It plays an important role in pain perception and is thought to be a new target for pain therapy.
While nucleotide modifications of DNAzymes published so far have usually been restricted to the substrate binding arms, we also introduced 2′-O-methyl modifications into the catalytic core of the DNAzyme. This region is expected to have high requirements for a correctly folded tertiary structure in order to be functional. Our recent findings, however, that a number of nucleotides in the catalytic core can be replaced by other naturally occurring or modified nucleotides (26), change the perspective toward a less strictly conserved catalytic centre, allowing for modifications on some of the nucleotides. We optimised the set-up of a DNAzyme by adjusting arm length and the number of modified nucleotides to achieve a design that led to a DNAzyme with substantially enhanced catalytic activity and increased stability against nucleolytic degradation.
MATERIALS AND METHODS
Deoxyribozyme synthesis
Unmodified oligodeoxynucleotides and phosphorothioates were purchased from MWG-Biotech AG (Ebersberg, Germany). Oligonucleotides containing 2′-O-methyl and 3′-3′ inverted thymidine modifications as well as RNA-oligonucleotides were obtained from IBA GmbH (Göttingen, Germany). LNA-containing oligonucleotides were purchased from Proligo (Boulder, CO). For the sequences of the optimised DNAzymes and their target molecules, see Figure 1A.
Cloning and in vitro transcription of the human rhinovirus RNA
The 5′-NTR of HRV14 was derived from the plasmid pWR3 (27; courtesy of Gail Ferstanding Arnold, Rutgers University, Piscataway, NJ), encoding for an infectious full-length HRV14 cDNA clone. For PCR amplification of the 5′-NTR the following primers were used: HRV14(1–22+T7): TAATACGACTCACTATAGGTTAAA ACAGCGGATGGGTATCC; HRV14(835–814): ATTCAA TGCTGGTGCACCC. The PCR fragment was cloned into the pCR2.1 vector (Stratagene, La Jolla, CA). Prior to transcription, the plasmid was linearised with BamH1. In vitro transcription was performed with the RiboMAX Large Scale Production System from Promega (Madison, WI) according to the manufacturer’s instructions. The transcript obtained consisted of 876 bases, comprising the 5′-NTR and part of the polyprotein sequence of HRV14 plus 41 vector-derived nucleotides at the 3′-end.
Determination of melting temperatures
The melting temperatures of the DNAzyme-target helices were determined using an HP-Spectrophotometer 8452 A. DNAzyme (1.5 µM) and the same amount of complementary 19 nt RNA were incubated at 95°C for 5 min in a buffer containing 10 mM sodium phosphate pH 7.0, 100 mM sodium chloride and 0.1 mM EDTA. Subsequently, the solution was cooled slowly to 12°C. The extinction at 260 nm was measured over a temperature range from 12 to 90°C. The melting temperature corresponds to the temperature at which the first derivative of the melting curve reaches its maximum. Values given are the means of at least one heating curve and one cooling curve, with a variation <1.5°C.
Radioactive labelling of short target RNA
Short target RNA (1 pmol) consisting of 19 nt was labelled with 10 µCi of [γ-32P]ATP using 5 U of T4 polynucleotide kinase (Promega, Madison, WI) for 90 min at 37°C. The radioactive probe was purified by electrophoresis on a 12% denaturing polyacrylamide gel. The band was eluted from the gel with 0.3 M sodium acetate, pH 5.5, for 45 min at 80°C. RNA was precipitated from the solution by adding 2.5 vol of 100% ethanol and keeping the tube at –20°C overnight.
DNAzyme cleavage reaction
To assess the target cleavage of DNAzymes with modifications in the catalytic core, 1 µM of short target RNA was mixed with 10 000 c.p.m. radioactively labelled RNA and incubated with 10 nM DNAzyme in 10 µl of ribozyme buffer (10 mM MgCl2, 50 mM Tris–HCl pH 7.5) at 37°C. Prior to mixing enzyme and target RNA, both solutions were denatured separately for 5 min at 85°C. After 20 min, the cleavage reactions were stopped by addition of EDTA to a final concentration of 83 mM and cooling on ice. Samples were denatured for 5 min at 65°C, and substrate RNA and cleavage products were separated on a 20% denaturing polyacrylamide gel. For analysis of the amount of cleavage, a Molecular Dynamics Storm 840 Phosphoimager was used. The results were confirmed with long target RNA molecules. Experiments were performed similarly except that 100 nM target RNA were used and cleavage activity was determined with agarose gels and ethidium bromide staining. The Quantity One software (Bio-Rad, Munich, Germany) was used to evaluate the gels. All values given are means ±SD of at least three independent experiments.
Kinetic analysis with the long target RNA
Kinetic experiments with the long target RNA were performed in 50 mM Tris–HCl, 10 mM MgCl2 and 1 U/µl RNasin to prevent unspecific RNA degradation. DNAzymes were denatured for 2 min at 65°C and then cooled to 37°C. Reactions were started by adding DNAzyme to 100 nM long target solution. Enzyme concentrations for single and multiple turnover experiments were 1 µM and 10 nM, respectively. Aliquots were taken after defined intervals during the first 10% of the reaction in the case of multiple turnover conditions and during a prolonged period for single turnover investigations. The reaction was stopped by adding 83 mM EDTA and cooling on ice. The cleavage reactions were analysed by agarose gel electrophoresis and ethidium bromide staining. Band intensities were quantified with the Quantity One software (Bio-Rad, Munich, Germany). Data were further analysed by fitting either linearly to obtain the initial velocity vinit for substrate excess experiments or with a single exponential decay function to obtain the observed cleavage rate for enzyme excess experiments using Origin (Microcal Software, Northampton, MA). Values given are means ±SD of at least three independent experiments.
Stability assays
The resistance against nucleolytic degradation of different DNAzymes in cell culture medium was evaluated. DNAzyme (1 µM) was incubated in DMEM (Cytogen, Sinn, Germany) containing 10% FCS (PAA laboratories, Linz, Austria) at 37°C. Samples were removed at defined time points between 0 and 72 h and all ongoing reactions were quenched by addition of an equal amount of 9 M urea in TBE and subsequent freezing in liquid nitrogen. Oligonucleotides were phenol extracted and precipitated at –20°C overnight by addition of sodium acetate, pH 5.2, to a final concentration of 0.3 M and addition of 2.5 vol of ethanol. The precipitate was washed with 70% ethanol and resuspended in a suitable amount of water. After denaturation for 5 min at 85°C, full-length DNAzyme and degradation products were separated on a 20% denaturing polyacrylamid gel. Further analysis was done using the Quantity One program (Bio-Rad, Munich, Germany). The half-lives of DNAzymes were obtained by fitting the amount of full-length oligonucleotide at different time points to a first-order exponential function using Origin (Microcal Software, Northampton, MA).
To assess DNAzyme stability against endonucleolytic degradation, 2 µM oligonucleotide were incubated with 0.4 U S1 endonuclease (Promega, Madison, WI) per 100 pmol DNAzyme in the manufacturer’s buffer (50 mM sodium acetate, pH 4.5, 280 mM NaCl, 4.5 mM ZnSO4). Aliquots were drawn after defined intervals between 0 and 180 min. The reactions were quenched by heating to 98°C for 3 min and subsequent freezing in liquid nitrogen. Oligonucleotides were ethanol precipitated and treated further as described above for stability assays in cell culture medium. Values given are means ±SD of at least three independent experiments.
RESULTS
A number of 10-23 DNAzymes against the 5′-NTR of the human rhinovirus 14 were tested and the DNAzyme targeted against bases 181–199 was found to be most efficient (data not shown). This DNAzyme was designated DH5. Figure 1A shows a shortened version of DH5 (see below) in complex with nucleotides 183–197 of the target RNA. The aim of the present study was to optimise the catalytic activity and biostability of DH5 by the introduction of modified nucleotides. Therefore, different types of modifications were used to stabilise the substrate recognition arms. Subsequently, the catalytic activity was optimised by adjusting arm length and content of modified nucleotides. In addition, the catalytic core of the DNAzyme was protected by the introduction of modified nucleotides. The optimised design was transferred to another DNAzyme, previously found to efficiently cleave the mRNA of the VR1 (7). Stability of the DNAzymes was analysed in cell culture medium and with S1 endonuclease.
For all kinetic investigations, we decided to perform experiments with long substrate RNAs, which are more likely to represent the situation inside cells than experiments with short target RNAs. These target molecules consisted of the first 835 bases of the HRV14 genome plus 41 nt derived from the vector sequence, and the 2614 nt VR1-mRNA, respectively. Since cleavage rates with long RNA molecules are several orders of magnitude lower than those for short substrate molecules, assays with long target RNAs under multiple turnover conditions are known to be difficult or even impossible (28). We therefore employed highly efficient DNAzymes and used the initial velocity as a measure for their activity. Salt concentrations in the buffers matched those commonly used for investigations on ribozymes. A further kinetic characterisation is impracticable for a large number of enzymes or even technically impossible. In single-point measurements comparing the activities of DNAzymes bearing modifications in the catalytic core, a 19mer short target RNA complementary to the substrate recognition arms was employed in addition.
Nucleotide modifications in the binding arms
In order to compare the influence of different nucleotide modifications in the binding arms on the efficiency of the DH5 DNAzyme, time course experiments on the long target RNA were performed under enzyme excess and substrate excess conditions. Values were obtained for the observed cleavage rate (kobs) in single turnover experiments and the vinit in a multiple turnover set-up. Initial measurements were performed with the unmodified enzyme, the species with an inverted thymidine at the 3′-end, DH5 with phosphorothioate linkages throughout the binding arms, and two species with four 2′-O-methyl nucleotides and four LNA-nucleotides on either side of the enzyme, respectively (Table 1).
Table 1. List of DNAzymes used in this study.
| DNAzyme | Arm length | Modifications | Tm (°C) | kobs (min–1) | vinit (nM min–1) |
|---|---|---|---|---|---|
| DH5-9/0 | 9 | None | 32 | 0.057 ± 0.005 | 0.21 ± 0.03 |
| DH5-7/0 | 7 | None | n.d. (<25) | 0.033 ± 0.002 | n.d. (<0.05) |
| DH5-iT | 9 | Inverted T at the 3′-end | 33 | 0.052 ± 0.005 | 0.19 ± 0.01 |
| DH5-Thio | 9 | All-phosphorothioate binding arms | 27 | 0.009 ± 0.001 | n.d. (<0.05) |
| LH5-9/4 | 9 | 4 LNA end-blocks | 63 | 0.24 ± 0.02 | 0.08 ± 0.02 |
| LH5-7/3 | 7 | 3 LNA end-blocks | 48 | 0.45 ± 0.01 | 1.2 ± 0.1 |
| LH5-7/4 | 7 | 4 LNA end-blocks | 61 | 0.44 ± 0.05 | 0.45 ± 0.09 |
| DH5-OMe9/4 | 9 | 4 OMe end-blocks | 47 | 0.11 ± 0.01 | 1.2 ± 0.1 |
| DH5-OMe8/4 | 8 | 4 OMe end-blocks | 44 | 0.29 ± 0.01 | 2.8 ± 0.5 |
| DH5-OMe7/3 | 7 | 3 OMe end-blocks | 32 | 0.12 ± 0.03 | 3.8 ± 0.3 |
| DH5-OMe7/4 | 7 | 4 OMe end-blocks | 37 | 0.31 ± 0.01 | 4.7 ± 0.4 |
| DH5-OMe7/5 | 7 | 5 OMe end-blocks | 39 | 0.5 ± 0.1 | 4.7 ± 0.9 |
| DH5-OMe7/6 | 7 | 6 OMe end-blocks | 44 | 0.23 ± 0.06 | 1.9 ± 0.5 |
| DH5-OMe7/7 | 7 | 7 OMe end-blocks | 47 | 0.027 ± 0.006 | 0.21 ± 0.02 |
| DH5-OMe6/5 | 6 | 5 OMe end-blocks | 26 | 0.17 ± 0.01 | 1.1 ± 0.2 |
| DH5-CM6 | 9 | 6 OMe in the catalytic core | 31 | 0.06 ± 0.01 | 0.09 ± 0.01 |
| DH5 E | 7 | 5 OMe end-blocks; 6 OMe in the catalytic core | 39 | 0.57 ± 0.07 | 2.0 ± 0.2 |
| DV15 9/0 | 9 | None | 37 | 0.9 ± 0.1 | 0.5 ± 0.1 |
| DV15-OMe 7/5 | 7 | 5 OMe end-blocks | 40 | 0.83 ± 0.07 | 1.7 ± 0.2 |
| DV15-CM6 | 9 | 6 OMe in the catalytic core | 36 | 0.43 ± 0.03 | 0.8 ± 0.2 |
| DV15 E | 7 | 5 OMe end-blocks; 6 OMe in the catalytic core | 37 | 0.05 ± 0.01 | n.d. (<0.05) |
| DV15 E4 | 7 | 4 OMe end-blocks, 6 OMe in the catalytic core | 36 | 0.31 ± 0.08 | 1.3 ± 0.2 |
Figure before the dash, number of nucleotides in each binding arm; figure after dash, number of modified nucleotides in each binding arm. OMe, 2′-O-methyl modification; iT, 3′-inverted thymidine; Thio, all-phosphorothioate binding arms; L, LNA modification. Melting temperatures (Tm) of the target/enzyme duplexes, observed cleavage rates (kobs) in single turnover experiments and initial velocities (vinit) in multiple turnover set-ups (both with long target RNA) are given for each DNAzyme.
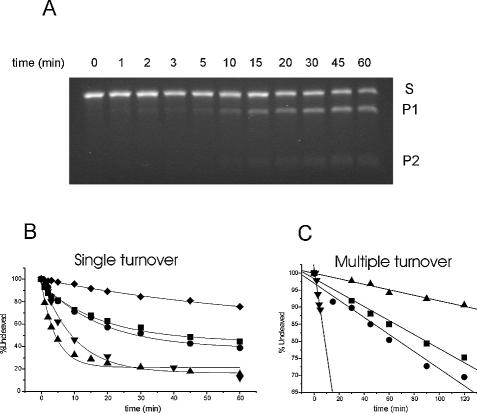
Substrate cleavage by the DNAzyme with an inverted thymidine at the 3′-end is shown as an example (Fig. 2A), kinetic traces of the experiments under single and multiple turnover conditions are shown in Figure 2B and C, respectively. Addition of an inverted thymidine at the 3′-end did not alter the kinetic behaviour of the enzyme significantly (Table 1). Introduction of phosphorothioates had an adverse effect on cleavage efficiency, decelerating the observed cleavage rate to one-fifth of that of the unmodified enzyme under single turnover conditions and to virtually zero in a substrate excess experiment. 2′-O-methyl modifications and LNA nucleotides are known to increase binding affinity of the oligonucleotide towards its target. In agreement with literature, we found the melting temperature of the DNAzyme/target hybrid helix to be increased by ∼4°C per LNA nucleotide and by ∼1.9°C with every 2′-O-methyl nucleotide introduced (Table 1). This enhanced affinity is reflected in significantly faster reaction kinetics under single turnover conditions with an increase of kobs for DH5-OMe9/4 and LH5-9/4 by a factor of 2 and 4, respectively. In a substrate excess situation, however, the introduction of four LNA nucleotides on either end decelerated the reaction dramatically, probably due to a slower product release step. The DNAzyme modified with 2′-O-methyl nucleotides had a 6-fold higher initial velocity than the unmodified species.
Figure 2.
Cleavage kinetics of DNAzymes with modified substrate recognition arms. (A) Substrate cleavage by the DNAzyme with a 3′-3′ inverted thymidine is shown as an example. S, substrate band; P1 and P2, product bands. Kinetic behaviour of DNAzymes was investigated under single turnover conditions with a 10-fold enzyme excess (B) and under multiple turnover conditions with a 10-fold substrate excess (C). Substrate recognition arms were always 9 nt long. Unmodified DNAzyme (DH5-9/0), squares; DNAzyme with an inverted thymidine (DH5-iT), circles; DNAzyme with thioate linkages (DH5-Thio), diamonds; DNAzyme with four LNA monomer end-blocks (LH5-9/4), triangles; DNAzyme with four 2′-O-methyl RNA end-blocks (DH5-OMe9/4), inverted triangles. DH5-Thio showed hardly any detectable activity under multiple turnover conditions.
Optimisation of arm design
The affinity of any antisense molecule towards its complementary sequence can be optimised by adjustment of the number of modified nucleotides and by fine-tuning the length of the substrate binding arms. A large portion of modified nucleotides on both ends of the DNAzyme is desirable, as it should convey higher stability against exonucleolytic degradation in body fluids. At the same time, the velocity of the reaction can be increased if modifications are employed that raise the affinity towards the target. Too high an affinity, however, interferes with product release and slows down the reaction rate.
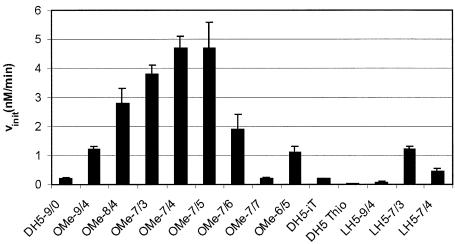
Arm length adjustments were made for 2′-O-methyl modified DNAzymes and for enzymes containing LNAs. For practical reasons, we changed the length of both binding arms uniformly, rather than adjusting the length of each arm individually. The initial velocity of cleavage of the long target RNA was measured under multiple turnover conditions, as those are considered closest to the situation in the cell. Shortening of the substrate recognition arms led to an increase of the reaction rate for both types of modifications (Fig. 3). For LNA modifications, the initial velocity could be increased by a factor of six employing 7-nt arms with three LNA nucleotides on either end. Introduction of a fourth LNA led to a drop in activity. For 2′-O-methyl modified enzymes, an immense increase in maximum activity was achieved with 7-nt arms and four or five modified nucleotides on either side. A decrease of velocity occurred on introduction of further modified nucleotides. One might speculate that this design interferes with the correct folding of the catalytic centre.
Figure 3.
Optimisation of DNAzyme activity. Initial velocity of the unmodified DNAzyme and DNAzymes with modified substrate recognition arms was measured under multiple turnover conditions with a long RNA target. See Table 1 for nomenclature.
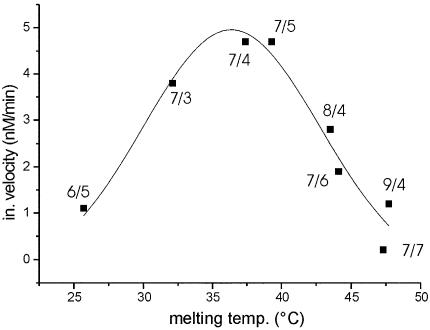
Thus, by employing the optimal design for the substrate-binding arms with 7-nt arms and four or five 2′-O-methyl modifications at the 5′- and 3′-end, we were able to enhance the activity of the DNAzyme by a factor >20. To assess the affinity of the respective DNAzyme for its target sequence, we determined the melting temperatures of the enzyme/target heteroduplex. For DNAzymes containing 2′-O-methyl end-blocks, we found a correlation resembling a Gaussian distribution between the initial velocity in multiple turnover experiments and the melting temperature with a maximum of activity for DNAzymes with a Tm around the working temperature of 37°C (Fig. 4).
Figure 4.
Correlation between melting temperature and initial velocity of DNAzymes. Initial velocity of DNAzymes with 2′-O-methyl modifications in the binding arms is plotted as a function of their melting temperature. Arm length and number of modified nucleotides on each side of the DNAzymes are given for each data point.
Nucleotide modifications in the catalytic core
Recently, we reported the finding that a number of nucleotides within the catalytic core of DNAzymes are not highly conserved and can be substituted by other naturally occurring or modified nucleotides (26). This result prompted us to investigate the possibility of introducing modified nucleotides into the catalytic core and by this means to increase protection of the DNAzyme against endonucleolytic cleavage further without impeding catalytic activity. Each of the 15 nt in the catalytic centre was individually replaced by its 2′-O-methyl modified counterpart. The DNAzyme species bearing these modifications were allowed to cleave the 19mer target sequence for 20 min under multiple turnover conditions (100-fold substrate excess).
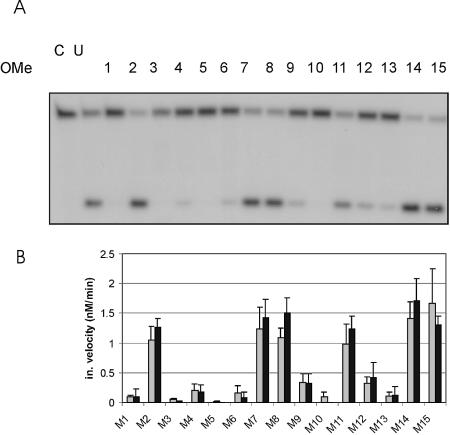
As can already be seen on the gel (Fig. 5A), modification of nine of the nucleotides virtually abolished cleavage of the short target, but a number of modifications did not appear to compromise catalytic activity. Quantitative evaluation confirmed that 2′-O-methyl groups at positions 2, 7, 8, 11, 14 and 15 do not interfere with the correct folding of the catalytic centre (Fig. 5B, grey columns). We therefore designed a DNAzyme that bore 2′-O-methyl modifications at all six feasible positions of the core. The resulting deoxyribozyme was found to be active, cleaving the long target sequence with a catalytic rate decreased to half its original value (Table 1). Similar results for the individual nucleotides were obtained with the long target RNA (Fig. 5B, black columns).
Figure 5.
Target cleavage of DNAzymes with 2′-O-methyl modifications in the catalytic core. (A) A 100-fold excess of the short, 19-nt target RNA was incubated with DNAzymes at 37°C for 20 min. The uncleaved substrate (upper band) and the 5′ cleavage product (lower band) can be seen. Lane 1, control without enzyme; lane 2, unmodified DH5; lanes 3–17, DNAzymes with 2′-O-methyl RNA at position 1–15, respectively. (B) Quantitative evaluation of the cleavage activity of DNAzymes with modifications in the catalytic core. Percentage of target RNA cleavage of DNAzymes with single 2′-O-methyl modifications at position 1–15 are shown for the short (grey columns) and long (black columns) target molecules. Values given are means ±SD of at least three independent experiments.
DNAzyme with modified nucleotides in the catalytic core and in the substrate recognition arms
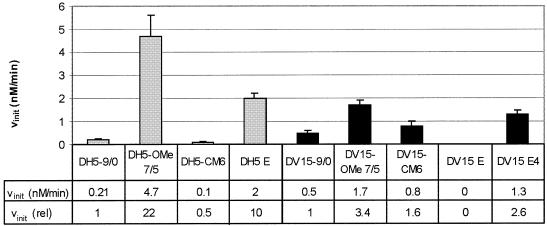
To obtain the optimal design for a DNAzyme with respect to activity and stability, we subsequently combined our findings of the optimisation of the arm design and the modification study of the catalytic core to yield an enhanced DNAzyme designated DH5 E (Fig. 1A, left). This DNAzyme was found to cleave the long target sequence under multiple turnover conditions with a cleavage rate 10 times that of the unmodified enzyme, arithmetically corresponding to a 20-fold increase of activity found for the optimised arm design and a decrease in velocity by 2 due to the modifications in the catalytic core (Fig. 6, grey columns).
Figure 6.
Activity of DNAzymes against the 5′-NTR of HRV14 and the VR1 mRNA. Initial velocity with the long target RNA under multiple turnover conditions for unmodified DNAzymes, DNAzymes with 2′-O-methyl stabilised substrate recognition arms and cores, respectively, and optimised DNAzymes with modifications of the arms and the catalytic core. See Table 1 for nomenclature. Initial velocities are given as well as catalytic activities normalised to the respective unmodified enzyme.
Transferability of the design
To investigate whether the optimised design of a DNAzyme with a protected core and enhanced catalytic activity by modified binding arms is also applicable to DNAzymes of different specificity, we employed the DNAzyme DV15 against the mRNA of the VR1, which we reported on earlier (7). DV15 is known to cut its 2614 nt target RNA with a high reaction rate. A modular approach was followed, determining first the cleavage rates of DV15-OMe7/5 modelled after the optimal substrate recognition arm design found for DH5, and then for DV15-CM6, bearing six 2′-O-methyl modified nucleotides in the catalytic centre, according to the findings for DH5. All measurements were conducted on the long target RNA. As expected, the 2′-O-methyl modifications in the binding arms were found to increase the initial velocity of the cleavage reaction substantially (∼3.4-fold, Fig. 6, black columns), although to a lesser extent than found for DH5. Application of the modified core design surprisingly did not only retain activity, but actually slightly enhanced the velocity of the reaction to 1.6 times that of the unmodified enzyme (Fig. 6, black columns).
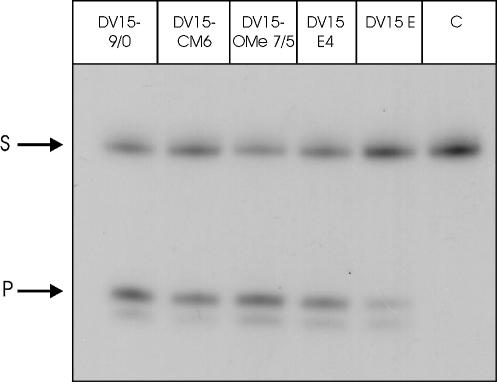
For reasons as yet unknown, a combination of the arm set-up and the core modification analogous to the DH5 E set-up did not lead to an active enzyme. Some activity was found in single turnover experiments (data not shown), but the cleavage rate in a substrate excess situation was virtually zero. To decide whether this lack of activity is due to the secondary structure of the target or due to the specific DNAzyme sequence, the different DV15 species were challenged with the short target sequence, consisting of 19 nt. As can already be seen on the gel (Fig. 7), the activity of DV15 E is diminished dramatically also towards the short target sequence. Quantification of the experiment yields only 25% activity of DV15 E as compared to the unmodified enzyme. This result shows that independent of the secondary structure of the target gene, DV15 E is severely compromised in its catalytic efficiency.
Figure 7.
Cleavage activity of differently modified DNAzymes against the 19-nt vanilloid receptor target RNA. DNAzymes were incubated with a 100-fold excess of target RNA for 20 min at 37°C. Substrate and product bands can be seen. Lane 1, unmodified DV15; lane 2, DV15 with six modified core nucleotides; lane 3, DV15 with optimised arm-design; lane 4, DV15 E4; lane 5, DV15 E; lane 6, control RNA without enzyme. See text for details.
An active species was generated, however, by combining the CM6 core design with four 2′-O-methyl modifications on either side of the DNAzyme (Fig. 1A, right), rather than using 5 nt end-blocks. The resulting enzyme, which we named DV15 E4, showed considerable enhancement of the catalytic rate with the initial velocity exceeding that of unmodified DV15 2.6-fold (Fig. 6, black columns). We therefore conclude that only slight modifications are necessary to transfer the optimised design to DNAzymes of different specificity.
Stability against nucleolytic degradation
The introduction of modified nucleotides serves the purpose of enhancing the stability of the DNAzyme against nucleolytic degradation in body fluids. To test the efficiency of the respective modification in this regard, stability assays were conducted in cell culture medium containing 10% fetal calf serum. We refrained from using the widely applied method of labelling the 5′ termini of the oligonucleotides with 32P, because phosphatases in the medium might lead to an underestimation of stability. Since ethidium bromide staining requires larger amounts of oligonucleotides, the DNAzymes were incubated in a relatively large volume of cell culture medium and subsequently concentrated by precipitation to avoid saturation of nucleases.
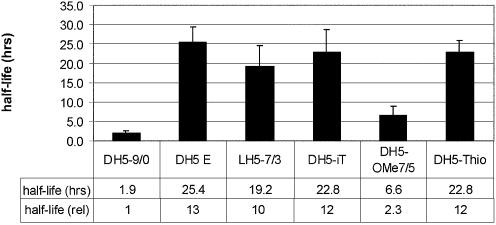
Modification of the binding arms with LNA nucleotides and phosphorothioates increased the half-life of the DNAzyme from ∼2 h to ≥20 h (Fig. 8), respectively. In accordance with literature, an inverted thymidine at the 3′-end was also found to enhance stability by a factor of 10 (11). Surprisingly, 2′-O-methyl modifications, situated exclusively in the binding arms, were clearly inferior to the other modifications, yielding a half-life of ∼6.5 h. The newly designed DNAzyme DH5 E containing 2′-O-methyl nucleotides in both the binding arms and the catalytic centre, however, showed excellent resistance against degradation. The half-life of the enzyme was increased to 25 h.
Figure 8.
Stability of modified DNAzymes in cell culture medium. DNAzymes were incubated at a final concentration of 1 µM in DMEM with 10% fetal calf serum. Aliquots were taken at appropriate time points, and the remaining amount of full-length oligonucleotides was determined by polyacrylamide gel electrophoresis. Average half-lives and half-lives normalised to the unmodified DNAzyme of three experiments are given. See Table 1 for nomenclature.
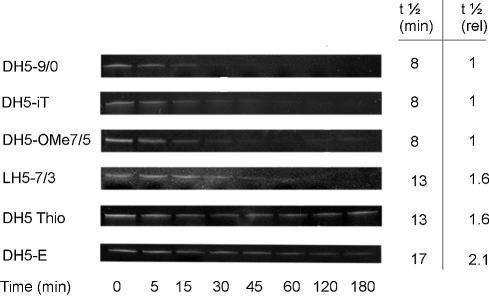
To investigate the stability against endonucleases further, the modified DNAzymes were incubated in a solution of 0.4 U S1 endonuclease per 100 pmol of enzyme. As can be seen in Figure 9, the unmodified DNAzyme as well as DNAzymes with an inverted thymidine and 2′-O-methyl RNA end-blocks are almost completely degraded after 30 min with a half-life of ∼8 min. Oligonucleotides containing LNA monomers were only weakly stained by ethidium bromide. It can still be seen that the DNAzyme that was stabilised by the introduction of LNAs into the substrate recognition arms was more stable against S1 endonuclease than the unmodified DNAzyme. DH5-Thio revealed a biphasic degradation curve, which is due to the chiral nature of the modified nucleotides. Approximately 50% of the starting amount is degraded by the endonuclease with a t1/2 of ∼13 min. The other half obeys a much slower degradation constant. The optimised DNAzyme with 2′-O-methyl monomers in the catalytic core and in the substrate recognition arms has the longest half-life of all modified enzymes investigated. Its stability is enhanced >2-fold compared to the unmodified DNAzyme.
Figure 9.
Comparison of DNAzyme stability against S1 endonuclease. DNAzymes at a concentration of 2 µM were incubated with 0.4 U S1 endonuclease per 100 pmol DNAzyme. Aliquots were removed at appropriate time points. Full-length and degraded oligonucleotides were separated on a 20% denaturing polyacrylamide gel. Half-lives and relative stability normalised to the unmodified DNAzyme are given.
DISCUSSION
Our present study describes the optimisation of the 10-23 DNAzyme with regard to its catalytic activity and its stability against nucleolytic degradation. It is well known that nucleotide modifications introduced into the binding arms influence the catalytic behaviour of the enzyme as well as its stability. In addition, the introduction of modified nucleotides into the catalytic site of ribozymes is problematic, because it might diminish catalytic activity as a consequence of an impaired three-dimensional structure.
Approaching in a modular manner, we first compared the effects of various modifications in the binding arms on catalytic activity. It is expected that these effects are governed mainly by the variation in affinity towards the target molecule. All kinetic experiments were performed under multiple turnover conditions with long substrate RNA molecules to simulate intracellular conditions and to investigate the potential of differently modified DNAzymes to compete with secondary structures of the target RNA like stable helices. To ensure detectable cleavage rates, we used salt concentrations that are commonly employed in investigations on ribozymes. It must be noted, however, that under physiological conditions with lower Mg2+ concentrations, activity will be substantially lower. On the other hand, further factors exist in the cellular environment that bear influence on the catalytic activity of ribozymes. Therefore, experiments in cell culture will have to be performed in order to prove that the design optimised in vitro is suitable for biological applications.
As mentioned above, an inverted thymidine at the 3′-end of the oligonucleotide has widely been used to stabilise DNAzymes against nucleolytic degradation. We did not find a significant influence of this modification on the cleavage activity of an otherwise unmodified DNAzyme (Table 1 and Figs 1 and 2). However, a kinetic analysis of DNAzymes with different arm lengths targeting c-myc reported elsewhere revealed that the addition of an inverted thymidine might have an influence on the cleavage rate constant ranging from a slight loss of efficiency to a 4-fold increase of activity (11). When adding an inverted thymidine to a number of modified DNAzymes, we also found an unpredictable influence on cleavage activity (data not shown).
In contrast to the inverted thymidine, phosphorothioate linkages and 2′-O-methyl monomers exerted significant effects on catalytic activity. The DNAzyme with phosphorothioate linkages throughout the substrate recognition arms revealed a significantly reduced catalytic activity, which might be due to a decrease in target affinity. In contrast, four 2′-O-methyl RNA monomers as end-blocks led to a 6-fold increase of the initial velocity. Warashina et al. reported a similar tendency of the influence of phosphorothioate and 2′-O-methyl nucleotides on the catalytic activity, although the effects were much smaller, since they introduced only two modified nucleotides per substrate recognition arm (18). No significant effect of the introduction of phosphorothioate linkages on the cleavage activity was reported for a DNAzyme targeting the tumour necrosis factor α mRNA (15). These differences might be explained by the use of short substrate RNAs in the latter experiments, in which the ability of the DNAzyme to unfold secondary structures in the target RNA was not investigated.
Recently, LNA monomers were introduced into the substrate recognition arms (22,23). These LNA containing enzymes had an increased catalytic activity under single and multiple turnover conditions compared to the unmodified DNAzyme. LNAs as well as 2′-O-methyl modifications convey a RNA-like character to the oligonucleotide, pushing a helix with a complementary oligonucleotide toward an A-type (all-RNA) helix (29). This leads to an enhanced affinity to the target sequence that might be the reason for the higher catalytic rates. These results question a previous finding that DNAzymes with RNA-like substrate recognition arms have a reduced cleavage activity (30). We also found increased reaction rates for the DNAzyme containing LNA monomers under single turnover conditions. Under multiple turnover conditions, however, a high LNA content can diminish catalytic activity by impeding substrate release due to its high target affinity.
After having demonstrated that different types of modifications can be introduced into the DNAzymes without severe loss of activity, we optimised cleavage efficiency by fine-tuning arm length and content of modified nucleotides. A systematic study led to a >20-fold increase of activity for a DNAzyme with 7-nt substrate recognition arms including four or five 2′-O-methyl monomers as end-blocks. The best LNAzyme with 7-nt arms and three LNA nucleotides at both ends had an ∼6-fold increased initial velocity. For 2′-O-methyl as well as for LNA substituted DNAzymes a further increase of the content of modified nucleotides led to a reduced cleavage activity. Two explanations might be given for this finding: (i) modified nucleotides in the inner part of the substrate recognition arms might interfere with the correct folding of the catalytic centre and/or (ii) an optimal target affinity is required for high catalytic activity. Comparing the melting temperatures of the enzymes modified with 2′-O-methyl, we found the highest activity in a multiple turnover reaction to correspond to a melting temperature of the enzyme/target helix around the working temperature of 37°C. Also for DNAzymes modified with LNAs, a lowering of the melting temperature from 63 to 48°C correlated with higher reaction rates. These measurements demonstrate the pivotal role of the balance between increased substrate binding and uncompromised product release.
It should be noted that we only used DNAzymes with symmetric substrate binding arms, since it was beyond the scope of the present investigation to try out all possible combinations. In a previous study, Cairns et al. have shown that asymmetric binding arms in DNAzymes can in some cases lead to higher reaction rates (31). Therefore, an individual optimisation of each binding arm may be useful when transferring the design to DNAzymes that target other RNAs. The introduction of modified nucleotides to the substrate recognition arms protects the DNAzyme against exonucleolytic degradation. A number of lines of evidence, however, suggest that, in many cells and tissues, endonucleases play an important role in the metabolism of oligonucleotides. This finding led to the conclusion that strategies in which oligonucleotides are modified only at both ends as a means of enhancing stability have not proven to be successful (32). In the only report known to us describing the use of modified nucleotides in the catalytic core of the 10-23 DNAzyme, the introduction of phosphorothioate linkages at the pyrimidines or all nucleotides of the catalytic site led to a substantial or almost complete loss of catalytic activity (13). These results demonstrate that the effects of modified nucleotides in the catalytic core on cleavage activity have to be studied systematically in order to obtain a DNAzyme that combines high catalytic activity and stability against endonucleases. Our recent finding that a number of core nucleotides can be substituted by other nucleotides (26) encouraged us to introduce stabilising modifications into the catalytic core to achieve enhanced resistance against endonucleolytic cleavage. We found that 6 of the 15 core nucleotides can be replaced by their 2′-O-methyl modified counterparts without severely compromising catalytic activity. The fact that cytidine at position 7 and thymidine at position 8 of the core can be modified lends further proof to our observation that they are of minor importance for the catalytic moiety and can be replaced by other nucleotides (26) or can even be left out altogether without losing catalytic activity (Z.Zaborowska, S.Schubert, J.Kurreck and V.A.Erdmann, unpublished data). The same six core nucleotides could be modified in a DNAzyme of different specificity with the same result. Thus, protecting the catalytic core against endonucleolytic cleavage by the design we developed seems to be feasible irrespective of the sequence of the binding arms. The optimised design for the binding arms and the modified catalytic core were subsequently combined and yielded a DNAzyme, named DH5 E (Fig. 1A), with 10-fold enhanced activity compared to the unmodified DNAzyme.
When transferring the design to a DNAzyme of different specificity, only a slight modification had to be made. Although the modifications in the catalytic core appeared to have no adverse effect on the cleavage rate, and although 5-nt 2′-O-methyl end-blocks accelerated cleavage 3-fold, the combination of the two designs yielded a species that showed little activity in a single turnover set-up and was virtually inactive in a substrate excess situation. The activity of this species was also diminished when challenged with a short complementary RNA. Because of this finding, the lack of activity can be said to be independent of the secondary structure of the target and may point to possible interactions between the inner nucleotides of the binding arms with the catalytic core. By changing arm design and employing 4-nt end-blocks rather than five, catalytic activity could be restored and was measured as being elevated by a factor of ∼3.
Following the optimisation of the catalytic properties, the nuclease resistance of the different DNAzymes in cell culture medium containing fetal calf serum was analysed (Fig. 8 and Table 2). For the DNAzyme protected by an inverted thymidine at the 3′-end we found an ∼10-fold increase of half-life from 2 to >20 h in cell culture medium. These results are in agreement with various reports in literature demonstrating an ∼10-fold increase of stability of the DNAzyme to ∼20 h in human serum (11,12). However, half-lives for DNAzymes with an inverted thymidine in cell culture medium reported in the literature vary from 6 h (13) to ∼3 days (12). These differences might either be due to a sequence dependency of the DNAzyme stability or to different experimental setups (DNAzyme concentration, detection method etc.). Therefore, the efficiency of different methods to stabilise an oligonucleotide should only be compared for identical sequences investigated under the same conditions.
Table 2. Comparison of modified DNAzymes.
| vinit | t1/2 medium | t1/2 S1-Nucl. | Performance index | |
|---|---|---|---|---|
| DH5-9/0 | 1 | 1 | 1 | 1 |
| DH5-Thio | <0.23 | 11.5 | 1.6 | <4 |
| DH5-iT | 0.9 | 11.5 | 1 | 10 |
| DH5-OMe7/5 | 22.4 | 3.5 | 1 | 78 |
| LH5-7/3 | 5.7 | 9.5 | 1.5 | 81 |
| DH5 E | 9.7 | 12.5 | 2.1 | 255 |
Initial velocity under multiple turnover conditions, stability in cell culture medium and stability against endonuclease S1 are given. All values are normalised to the unmodified DNAzyme. An index of the overall performance of modified DNAzymes is given as the product of the three values.
DNAzymes with two phosphorothioate or 2′-O-methyl substitutions at both ends were reported to enhance the biostability of DNAzymes without a detailed analysis of the half-lives (18,19). Iversen et al. reported an increase of half-life in 50% human serum from 3 h for the unmodified DNAzyme to >50 h for a DNAzyme with phosphorothioate arms (15). Interestingly, the DNAzyme with phosphorothioate modifications investigated in the present study revealed a significantly enhanced stability (>10-fold) in medium, whereas the DNAzyme with five 2′-O-methyl RNA end-blocks was only weakly (3.5-fold) stabilised. In contrast, the DNAzyme with 2′-O-methyl monomers in the arms and in the catalytic core exhibited a high biostability with a half-life in cell culture medium of 25 h. LNAs are another suitable modification to stabilise antisense oligonucleotides against nucleolytic degradation (33,34), but they have not yet been used to enhance the resistance of DNAzymes against nucleases. In the only report on the usage of LNAs in DNAzymes published to date, the modified nucleotides were introduced into the inner part of the substrate recognition arms to analyse the kinetic behaviour (22). Our experiments show that LNAs are well suited to stabilise DNAzymes against nucleolytic degradation.
Finally, we analysed the different modified DNAzymes with respect to their resistance against endonucleases (Fig. 9 and Table 2). As expected, the inverted thymidine at the 3′-end did not enhance stability in this assay. The same result was found for 2′-O-methyl end-blocks. Thioate linkages as well as LNA monomers in the substrate recognition arms enhanced stability, but the highest resistance against endonucleolytic degradation was found for the optimised DNAzyme DH5 E with modified arms and core nucleotides.
When Beigelman et al. analysed the influence of modified bases on the hammerhead ribozyme kinetics and stability, they used overall stability/activity ratios to compare various designs (9). We therefore introduced a performance index as the product of initial velocity, stability in cell culture medium and resistance against endonucleolytic degradation as a measure for each DNAzyme. As can be seen in Table 2, the DNAzyme with phosphorothioate linkages in the substrate recognition arms revealed a significantly enhanced biostability, but due to the low catalytic activity the performance index is only slightly increased compared to the unmodified DNAzyme. The inverted thymidine at the 3′-end enhanced serum stability but did not significantly influence cleavage activity and resistance against S1 endonuclease. DNAzymes with 2′-O-methyl or LNA nucleotides as end-blocks revealed enhanced activity and stability against degradation leading to a performance index approximately 80 times that of the unmodified DNAzyme. The optimised DNAzymes with 2′-O-methyl RNA nucleotides in the substrate recognition arms and in the catalytic core had a ∼10-fold enhanced catalytic activity and the highest biostability leading to a 250-fold increased performance index.
In addition to catalytic activity and nuclease resistance, efficient delivery to the target cells is a major requirement for the use of oligonucleotides in cell culture or in vivo. In a recent study, cellular uptake of labelled 10-23 DNAzymes was analysed by fluorescence microscopy and revealed that the DNAzyme was distributed primarily in punctuate structures surrounding the nucleus (12). Despite a number of successful in vivo approaches (5), a detailed pharmacokinetic investigation on the biodistribution of the DNAzyme is still lacking.
In summary, we established a new design for the 10-23 DNAzyme that yields molecules with enhanced activity and stability, particularly protected against endonucleases. Only a slight modification was necessary when transferring the design to another DNAzyme. Our work shows the feasibility of nucleotide modifications in the catalytic centre and provides further insights into the usage of locked nucleic acids in DNAzymes. More experiments in cellular systems and with long RNAs are needed in the future to show that the findings are of general application in the design of efficient nucleic acid RNases based on a DNA backbone. The optimised DNAzymes DH5 E and DV15 E4 will now be used in cell culture and in vivo to inhibit virus replication and to investigate the role of the vanilloid receptor for nociception, respectively.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by funds from the Bundesministerium für Bildung und Forschung (grant 01GG9818/0), the Deutsche Forschungsgemeinschaft (KU 1436/1-1) and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Joyce G.F. (2001) RNA cleavage by the 10-23 DNA enzyme. Methods Enzymol., 341, 503–517. [DOI] [PubMed] [Google Scholar]
- 2.Santoro S.W. and Joyce,G.F. (1997) A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA, 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santoro S.W. and Joyce,G.F. (1998) Mechanism and utility of an RNA-cleaving enzyme. Biochemistry, 37, 13330–13342. [DOI] [PubMed] [Google Scholar]
- 4.Cairns M.J., King,A. and Sun,L.-Q. (2003) Optimisation of the 10-23 DNAzyme-substrate pairing interactions enhanced RNA cleavage activity at purine-cytosine target sites. Nucleic Acids Res., 31, 2883–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khachigian L.M. (2002) Cutting a path to a new class of therapeutics. Curr. Opin.Mol. Ther., 4, 119–121. [PubMed] [Google Scholar]
- 6.Cairns J.M., King,A. and Sun,L.-Q. (2000) Nucleic acid mutation analysis using catalytic DNA. Nucleic Acids Res., 28, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurreck J., Bieber,B., Jahnel,R. and Erdmann,V.A. (2002) Comparative study of DNA enzymes and ribozymes against the same full-length messenger RNA of the vanilloid receptor subtype I. J. Biol. Chem., 277, 7099–7107. [DOI] [PubMed] [Google Scholar]
- 8.Kurreck J. (2003) Antisense technologies—improvement through novel chemical modifications. Eur. J. Biochem., 270, 1628–1644. [DOI] [PubMed] [Google Scholar]
- 9.Beigelman L., McSwiggen,J.A., Draper,K.G., Gonzalez,C., Jensen,K., Karpeisky,A.M., Modak,A.S., Matulic-Adamic,J., DiRenzo,A.B., Haeberli,P., Sweedler,D., Tracz,D., Grimm,S., Wincott,F.E., Thackaray,V.G. and Usman,N. (1995) Chemical modification of hammerhead ribozymes. J. Biol. Chem., 270, 25702–25708. [DOI] [PubMed] [Google Scholar]
- 10.Santiago S.F., Lowe,H.C., Kavurma,M.M., Chesterman,C.N., Baker,A., Atkins,D.G. and Khachigian,L.M. (1999) New DNA enzyme targeting Egr-1 mRNA inhibits vascular smooth muscle proliferation and regrowth after injury. Nature Med., 11, 1264–1269. [DOI] [PubMed] [Google Scholar]
- 11.Sun L.-Q., Cairns,M.J., Gerlach,W.L., Witherington,C., Wang,L. and King,A. (1999) Suppression of smooth muscle cell proliferation by a c-myc RNA-cleaving deoxyribozyme. J. Biol. Chem., 274, 17236–17241. [DOI] [PubMed] [Google Scholar]
- 12.Dass C.R., Saravolac,E.G., Li,Y. and Sun,L.-Q. (2002) Cellular uptake, distribution and stability of 10-23 deoxyribozymes. Antisense Nucleic Acid Drug Dev., 12, 289–299. [DOI] [PubMed] [Google Scholar]
- 13.Sioud M. and Leirdal,M. (2000) Design of nuclease resistant protein kinase Cα DNA enzymes with potential therapeutic application. J. Mol. Biol., 296, 937–947. [DOI] [PubMed] [Google Scholar]
- 14.Yen L., Strittmatter,S.M. and Kalb,R.G. (1999) Sequence-specific cleavage of Huntingtin mRNA by catalytic DNA. Ann. Neurol., 46, 366–373. [DOI] [PubMed] [Google Scholar]
- 15.Iversen P.O., Nicolaysen,G. and Sioud,M. (2001) DNA Enzyme targeting TNFalpha mRNA improves hemodynamic performance in rats with postinfarction heart failure. Am. J. Physiol. Heart Circ. Physiol., 281, 2211–2217. [DOI] [PubMed] [Google Scholar]
- 16.Iversen P.O., Emanuel,P.D. and Sioud,M. (2002) Targeting Raf-1 gene expression by a DNA enzyme inhibits juvenile myelomonocytic leukemia cell growth. Blood, 99, 4147–4153. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L., Gasper,W.J., Stass,S.A., Ioffe,O.B., Davis,M.A. and Mixson,A.J. (2002) Angiogenic inhibition mediated by a DNAzyme that targets vascular endothelial growth factor receptor 2. Cancer Res., 62, 5463–5469. [PubMed] [Google Scholar]
- 18.Warashina M., Kuwabara,T., Nakamatsu,Y. and Taira,K. (1999) Extremely high and specific activity of DNA enzymes in cells with a Philadelphia chromosome. Chem. Biol., 6, 237–250. [DOI] [PubMed] [Google Scholar]
- 19.Cieslak M., Niewiarowska,J., Nawrot,M., Koziolkiewicz,M., Stec,W.J. and Cierniewski,C. (2002) DNAzymes to β1 and β3 mRNA down-regulate expression of the targeted integrins and inhibit endothelial cell capillary tube formation in fibrin and matrigel. J. Biol. Chem., 277, 6779–6787. [DOI] [PubMed] [Google Scholar]
- 20.Wu Y., Yu,L., McMahon,R., Rossi,J.J., Forman,S.J. and Snyder,D.S. (1999) Inhibition of bcr-abl oncogene expression by novel deoxyribozymes (DNAzymes). Hum. Gene Ther., 10, 2847–2857. [DOI] [PubMed] [Google Scholar]
- 21.Levin A.A. (1999) A review of issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim. Biophys. Acta, 1489, 69–84. [DOI] [PubMed] [Google Scholar]
- 22.Vester B., Lundberg,L.B., Sorensen,M.D., Babu,B.R., Douthwaite,S. and Wengel,J. (2002) LNAzymes: incorporation of LNA-type monomers into DNAzymes markedly increases RNA cleavage. J. Am. Chem. Soc., 124, 13682–13683. [DOI] [PubMed] [Google Scholar]
- 23.Petersen M. and Wengel,J. (2003) LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol., 21, 74–81. [DOI] [PubMed] [Google Scholar]
- 24.Zeichhardt H. and Grunert,H.-P. (2000) Enteroviruses. In Specter,S., Hodinka,R. and Young,S. (eds), Clinical Virology Manual. American Society for Microbiology, Washington, DC, pp. 252–269. [Google Scholar]
- 25.Caterina M.J., Schumacher,M.A., Tominaga,T.A., Rosen,T.A., Levine,J.D. and Julius,D. (1997) The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature, 389, 816–824. [DOI] [PubMed] [Google Scholar]
- 26.Zaborowska Z., Fürste,J.P., Erdmann,V.A. and Kurreck,J. (2002) Sequence requirements in the catalytic core of the ‘10-23’ DNA enzyme. J. Biol. Chem., 277, 40617–40622. [DOI] [PubMed] [Google Scholar]
- 27.Lee W.M. (1991) Human Rhinovirus 14: synthesis and characterization of a molecular cDNA clone which makes highly infectious transcripts. PhD Thesis, University of Wisconsin-Madison, WI.
- 28.Thomson J.B., Tuschl,T. and Eckstein,F. (1996) The hammerhead ribozyme. In Eckstein,F. and Lilley,D.M.J.(eds), Catalytic RNA. Springer-Verlag, Berlin, Germany, pp. 173–196. [Google Scholar]
- 29.Bondensgaard K., Petersen,M., Singh,S.K., Rajwanshi,V.K., Kumar,R. and Wengel,J. and Jacobsen,J.P. (2000) Structural studies of LNA:RNA duplexes by NMR: Conformations and implications for RNase H activity. Chem. Eur. J., 6, 2687–2695. [DOI] [PubMed] [Google Scholar]
- 30.Ota N., Warashina,M., Hirano,K., Hatanaka,K. and Taira,K. (1998) Effects of helical structures formed by the binding arms of DNAzymes and their substrates on catalytic activity. Nucleic Acids Res., 26, 3385–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cairns M.J., Hopkins,T.M., Witherington,C. and Sun.,L.-Q. (2000) The influence of arm length asymmetry and base substitution on the activity of the 10-23 DNA enzyme. Antisense Nucleic Acid Drug Dev., 10, 323–332. [DOI] [PubMed] [Google Scholar]
- 32.Crooke S.T. (2000) Progress in antisense technology: the end of the beginning. Methods Enzymol., 313, 3–45. [DOI] [PubMed] [Google Scholar]
- 33.Wahlestedt C., Salmi,P., Good,L., Kela,J., Johnsson,T., Hökfeldt,T., Broberger,C., Porreca,F., Lai,J., Ren,K., Ossipov,M., Koshkin,A., Jakobsen,N., Skouv,J., Oerum,H., Jacobson,M.H. and Wengel,J. (2000) Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl Acad. Sci. USA, 97, 5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurreck J., Wyszko,E., Gillen,C. and Erdmann,V.A. (2002) Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res., 30, 1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]