Abstract
Two groups of anaerobic genes (genes induced in anaerobic cells and repressed in aerobic cells) are negatively regulated by heme, a metabolite present only in aerobic cells. Members of both groups, the hypoxic genes and the DAN/TIR/ERG genes, are jointly repressed under aerobic conditions by two factors. One is Rox1, an HMG protein, and the second, originally designated Rox7, is shown here to be Mot3, a global C2H2 zinc finger regulator. Repression of anaerobic genes results from co-induction of Mot3 and Rox1 in aerobic cells. Repressor synthesis is triggered by heme, which de-represses a mechanism controlling expression of both MOT3 and ROX1 in anaerobic cells; it includes Hap1, Tup1, Ssn6 and a fourth unidentified factor. The constitutive expression of various anaerobic genes in aerobic rox1Δ or mot3Δ cells directly implies that neither factor can repress by itself at endogenous levels and that stringent aerobic repression results from the concerted action of both. Mot3 and Rox1 are not essential components of a single complex, since each can repress independently in the absence of the other, when artificially induced at high levels. Moreover, the two repression mechanisms appear to be distinct: as shown here repression of ANB1 by Rox1 alone requires Tup1–Ssn6, whereas repression by Mot3 does not. Though artificially high levels of either factor can repress well, the absolute efficiency observed in normal cells when both are present—at much lower levels—demonstrates a novel inhibitory synergy. Evidently, expression levels for the two mutually dependent repressors are calibrated to permit a range of variation in basal aerobic expression at different promoters with differing operator site combinations.
INTRODUCTION
Yeast adapt to the absence of oxygen by expressing several groups of genes. Two of the anaerobic regulons are negatively regulated by heme, which is produced only in the presence of oxygen. The members of one heme-repressed gene group are referred to as ‘hypoxic genes’ (1,2) and encode intracellular proteins dedicated to more efficient utilization of oxygen. They are all regulated by the Rox1 repressor (3,4), which is induced during aerobic growth by heme(3), resulting in ‘aerobic repression’ of the regulon. In the absence of oxygen, expression of Rox1 ceases, partly through inhibition by Hap1 (5), and Rox1 is degraded (6), de-repressing transcription. The widely varying efficiency of Rox1 repression of various hypoxic genes depends on the number and fidelity of operator sites (1).
The second group of heme-repressed genes are the DAN/TIR/ERG genes, which are transcriptionally activated through a common response element (7) by Mox4/Upc2, a binucleate zinc cluster protein (8,9). Eight of these genes encode the Dan/Tir cell wall proteins (10–12), three of which are essential for anaerobic growth (12); others are involved in sterol synthesis (13) or transport (8). Regulators controlling expression of this group include the oxygen-inhibited Mox4/Upc2 activator, and two repressors bound at neighboring sites responsible for aerobic repression (13). One of these is Rox1 (9). The second, originally designated Rox7, was identified as a repressor of DAN1 and some of the other DAN/TIR genes, as well as of the hypoxic gene ANB1 (9). As we show here, Rox7 is Mot3. It was previously identified as a zinc finger regulator which functions either as a repressor or activator of a diverse group of genes (14–16), acting through a consensus site (14,15) shown to be important for aerobic repression of ANB1 (17). Through its versatility as an activator and repressor, Mot3 participates in a cell wall remodeling process (12) in which anaerobic and aerobic cell wall proteins [the CWP proteins (18)] are alternately expressed in response to oxygen.
Anaerobic genes encode proteins serving several different purposes. Presumably, it is advantageous for the regulatory system to permit gene-to-gene variation in basal and induced expression as well as cross-talk by other regulatory pathways. Thus, repression in aerobic cells ranges from moderate, for some genes, e.g. those whose basal expression is indispensable, like HEM13 and OLE1, to highly stringent, for genes like DAN1 and ANB1. Aerobic repression depends on heme signaling through multiple mechanisms (7,12). We show here that expression of Mot3, like Rox1 (3,5), is induced to inhibitory levels by heme. It acts to de-repress a mechanism which includes Hap1, Tup1–Ssn6 and a fourth unidentified factor. Induction levels for Mot3 and Rox1 appear to be calibrated to establish a range of responses to oxygen levels for each target gene, as determined by the number of operator sites in each promoter.
The requirement for both Mot3 and Rox1 for inhibition of aerobic expression might suggest each is a necessary component of a single complex. Arguing against this, we show that either repressor can almost fully compensate for the lack of the other when artificially expressed at high levels. However, at the low levels present in aerobic cells, Mot3 and Rox1 act synergistically to achieve stringent repression. Though functioning in concert, the two mechanisms are distinct, as shown by the observation that Rox1, but not Mot3, requires the Tup1–Ssn6 co-repressor for its function.
MATERIALS AND METHODS
Strains and plasmids
Strains used were FY23 (19) and RZ53 (3). A mot3::HIS3 disruption was introduced into FY23 by transformation with a PCR fragment amplified from FY1628 [provided by Fred Winston (15)], using primers homologous at –964 and +1880. This cassette was introduced into FY23his3:DAN1/URA3, which was obtained by disruption of HIS3 with a fusion of the DAN1 promoter to the URA3 gene. For the construction of the his3:DAN1/URA3 allele, a region of HIS3 from –560 to +1087 was amplified using primers homologous at those sites (5′: gagagaattcactggaacttggatttatggc; 3′: cctccatggagctcagctgtcgacgtagttgctactgttatttctggc) digested at EcoRI and SalI sites in the primer segments, and inserted into the same sites in pBS-SK (Stratagene), generating pBS-HIS3; next the DAN1/URA3 fusion was excised from YCpDAN1/URA3 (9) by digestion with EcoRI, end-filling with Klenow, and digestion with SpeI; it was then inserted into the end-filled BstBI site and the NheI site of pBS-HIS3, replacing the segment of HIS3 from –42 to +509, and generating pBS-his3:DAN1/URA3. A SpeI–XhoI fragment excised from this plasmid was used to obtain FY23his3:DAN1/URA3 by transformation of FY23 and selection on anaerobic –ura plates (the DAN1/URA3 fusion is only active in the absence of oxygen). FY23mot3Δ(HIS3) was obtained by transformation of FY23his3:DAN1/URA3 with the mot3:HIS3 disruption cassette and selection for the HIS3 marker. MOT3 disruptants were confirmed by Southern blot, and since these were prototrophic for uracil under aerobic conditions (due to loss of Mot3 repression of DAN1/URA3), the ura3 genotype was regenerated by selection for a spontaneous auxotrophic mutant on FOA. FY23mot3:Kan-Mx was generated by transformation of FY23 with a PCR fragment derived from the proprietary mot3::KanMx strain derived from derivative of strain BY4741 (Resgen), using the recommended primers (tgaattcatcaagagatttgaaaca and ctccgtctggatttactaaactttg).
FY23rox1Δ was generated as described (3), as were FY23hap1Δ and FY23hap2Δ (9). FY23tup1Δssn6Δ was constructed by serial disruption of TUP1 and SSN6. TUP1 was disrupted as described (9). FY23tup1Δssn6Δ was constructed by transformation of FY23tup1Δ with an XbaI–SphI fragment excised from pssn6:LEU2 [obtained from R. Zitomer (6)]. FY23tup1Δssn6Δ isolates were confirmed by Southern blot and the ura3 genotype was regenerated as described above.
YEpGAL1/ROX1 was constructed by cloning an XbaI–HindIII fragment containing ROX1 (–433 to +1592) from the plasmid clone (3) into the same sites in pBS-M13+ (Stratagene). The EcoRI–BglII fragment from plasmid YCpGZ-15-Bg (1) containing the region from –830 to –268 of the GAL1 promoter was inserted 5′ to the ROX1 gene between the EcoRI and BamHI sites of the pBS-M13+ polylinker. The resulting fusion joined the ROX1 gene at –433 to the GAL1 promoter at –268; it was excised with EcoRI and HindIII and inserted into the same sites of the 2µ vector, YEpLac195 (20), generating YEpGAL1/ROX1. YCpGAL1/MOT3 was constructed in two steps: first a plasmid containing the GAL1 promoter region (–820 to –9) was constructed by inserting a PCR fragment amplified from the GAL1 promoter with primers homologous at those sites (5′: gagaggtaccgaattcgacaggttatcagcaac; 3′: gagaggatccttctccttgacgttaaagtatagagg); the fragment was digested at EcoRI and BamHI sites in the primer segments and inserted into YCpLac33 (20) at the same sites, generating YCppGAL1. Next a fragment (–1 to +2389) carrying the MOT3 ORF (which extends from +1 to +1971) was amplified by PCR using primers homologous at those sites (5′: gagaggatccacaataatgaatgcggaccatcac; 3′: gaaaatctgtccccttagcg ); it was digested at a BamHI site contained in the 5′ primer segment and at an EcoRV site at +2227 and ligated to YCppGAL1 which had been linearized with HindIII, end-filled, and digested with BamHI. A MOT3/lacZ fusion plasmid was constructed in two steps: first the MOT3 promoter region (–978 to +5) was amplified using primers (5′: tttacttcattcatgcttacagag; 3′: gagaggatcctcgttcattgctcaaatatgatatgtc) homologous at those sites, and cloned into the TA cloning vector pCRII (Invitrogen). It was then excised by digestion at an EcoRV site in the polylinker and a BamHI site in the 3′ primer segment, and ligated to the BamHI site and the end-filled EcoRI site of YCpDAN1/lacZ (9); this replaced the DAN1 promoter fragment with the MOT3 promoter region, generating YCpMOT3/lacZ.
Cloning ROX7
ROX7 was cloned by complementation of the rox7-1 mutation in FY23MSrox7-1, which carries integrated DAN1–URA3 and DAN1–lacZ fusions (9) and which is prototrophic for uracil due to the constitutive phenotype of the recessive rox7 allele. FY23MSrox7-1 was transformed with a centromeric library (9), and cells transformed with the wild-type ROX7 gene were selected on FOA plates for loss of uracil prototrophy resulting from complementation.
Cell growth and analysis of gene expression
Cells were grown under aerobic or anaerobic conditions in YPD or SC media. Anaerobic cultures were bubbled with high purity nitrogen. Anaerobic cultures for RNA analysis were harvested in late log phase after 90 min of anaerobic growth. RNA was extracted and analyzed as described (21). Anaerobic cultures for lacZ assay were harvested after 7 h and the activity quantified as described (22). For expression of MOT3 and ROX1 FY23 cells carrying YCpGAL1/MOT3 or YCpGAL1/ROX1 were grown in SD-ura overnight, diluted 40-fold into SD-ura, grown for 3 h into log phase, washed twice in SD-ura-raffinose (1%)–galactose (1%) and grown for 4 h in the same medium before harvesting for RNA analysis.
RESULTS
ROX7 identified as MOT3
The ROX7 locus was defined by recessive mutations in one of three complementation groups (9) selected for constitutive expression of DAN1, which is normally repressed during aerobic growth. rox7 mutations also de-repressed the hypoxic gene ANB1. We cloned ROX7 from a centromeric library by complementation in cells carrying an integrated DAN1–URA3 fusion (see Materials and Methods). Complementing plasmids carried the YMR070W open-reading frame, previously identified as MOT3, encoding a zinc finger protein that regulates expression of a number of different genes (14,15), but which had not been associated with oxygen regulation.
Synergistic aerobic repression of ANB1 and DAN1 by Mot3 and Rox1
As expected from the constitutive phenotype of the rox7 alleles, disruption of MOT3 caused partial loss of repression of ANB1 and DAN1 (Fig. 1), indicating that Mot3 functions along with Rox1 as an aerobic repressor of both genes. We focused on the roles of the two repressors on ANB1 expression, since the regulation of DAN1 is complicated by the contributions of two other oxygen-sensitive factors (7,9) and shows a lesser dependence on Rox1 and Mot3. Disruption of ROX1 caused an apparent full loss of repression of the ANB1 gene: expression in the rox1Δ strain was equal under aerobic and anaerobic conditions (Fig. 1); moreover, expression in aerobic rox1Δ cells was equal to that in anaerobic mot3Δrox1Δ cells (Fig. 1B), representing the fully de-repressed output of the promoter. Hence, Mot3 has no repression function on its own under these conditions, and even though Rox1 represses weakly by itself, the efficient repression observed in normal aerobic cells occurs only when the two proteins function in concert, i.e. ‘synergistically’. Negative synergy is a less well defined concept than positive, but a reasonable index for synergisic inhibition is a comparison of the residual activities when one or both repressors are present. These were determined to be 100 or 20–40%, respectively, with Mot3 or Rox1 alone, versus <1% when both are functioning. Hence, the residual expression with both repressors present is much less than that with either one alone, i.e. the combined inhibitory effects are clearly more than multiplicative (see Discussion).
Figure 1.
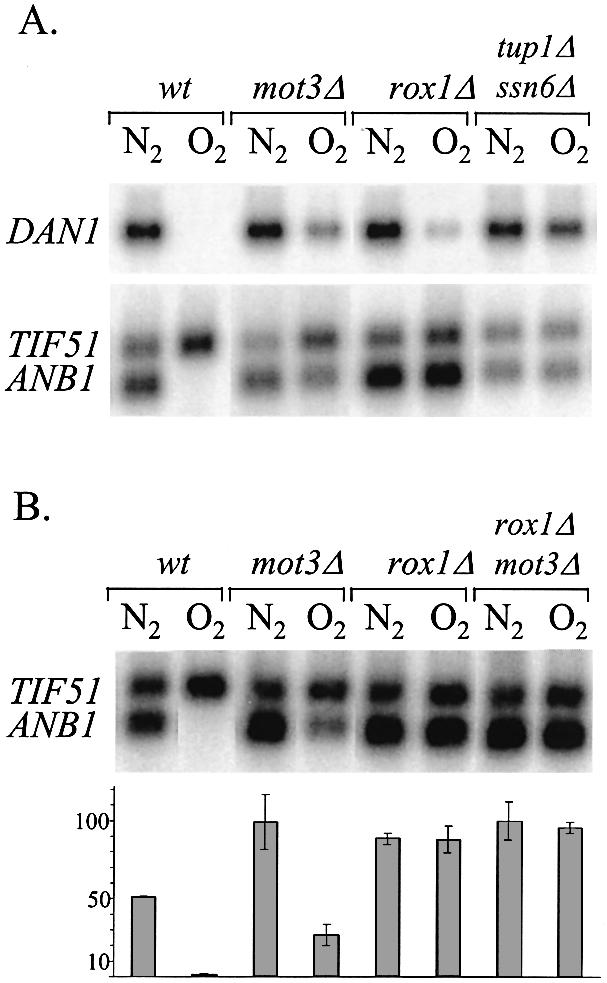
Effect of regulatory mutations on expression of anaerobic genes. (A) Strains of FY23 carrying disruptions of MOT3 (mot3::HIS3), ROX1, or a double disruption of TUP1 and SSN6 were grown under aerobic and anaerobic conditions as described in Materials and Methods; RNA was extracted and subjected to northern blot analysis using probes prepared from the DAN1, ANB1 and ROX1 genes. The ANB1 probe hybridizes to ANB1 mRNA and also to the homologous TIF51 transcript. (B) Cells of strains FY23, FY23mot3Δ(mot3::HIS3), FY23rox1Δ and FY23rox1Δmot3Δ (mot3::Kan-Mx) grown under the same conditions and analyzed as in (A). Duplicate northern blots were quantified by phosphorimager analysis with values (and standard deviations) for ANB1 mRNA levels shown in units of percentage of the maximum value in the sample set (anaerobic FY23rox1Δmot3Δ cells). The value for aerobic expression of ANB1 in wild-type cells is statistically indistinguishable from zero, at the limit both of phosphorimager quantification and direct visualization.
Differences in the expression of DAN1 and ANB1 in these mutants reveals a variable degree of participation in repression by the two factors at different promoters. The full loss of repression of ANB1 in the rox1Δ strain compared with partial loss in the mot3Δ strain implies that Rox1 plays a more important role than Mot3 at this promoter, presumably reflecting the preponderance of Rox1 sites (1). In contrast, aerobic repression of DAN1 was more affected by loss of Mot3 than of Rox1, also consistent with the relative numbers of Mot3 and Rox1 sites (1,7). The greater overall sensitivity of ANB1 to both rox1 and mot3 mutations compared with DAN1 may reflect the fact that DAN1 is also repressed by at least two other oxygen-sensitive mechanisms in aerobic cells (7,9). The results show constitutive expression of both genes in the absence of the Tup1–Ssn6 complex. The fact that expression in anaerobic tup1Δssn6Δ cells is lower than in wild type (Fig. 1A) may result from de-repression of ROX1or MOT3, which are both regulated by the heterodimer (6, and see below); the same could hold for the relatively lower expression in aerobic tup1Δssn6Δcells compared with rox1Δ cells (see Discussion).
Mot3 function is independent of oxygen
The role of Mot3 in aerobic repression suggested that it might be converted to a repressing form in the presence of oxygen (or heme). To test this we monitored expression of DAN1 and ANB1 in anaerobic cells expressing MOT3 under galactose control. Both genes were repressed in cells expressing high levels of Mot3 (Fig. 2), implying that inhibition of transcription by the repressor is concentration dependent and not the result of a conformational change occurring as a result of the presence of oxygen and/or heme. The same was earlier demonstrated for Rox1 (4).
Figure 2.
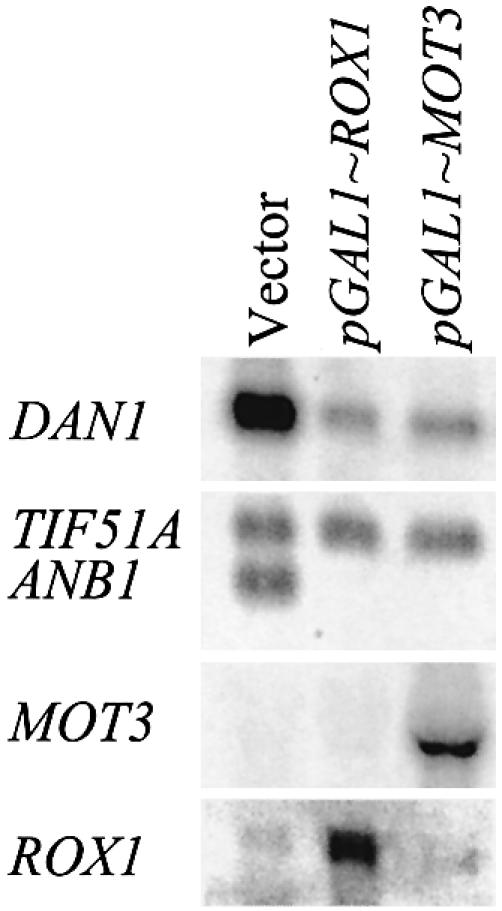
Over-expression of MOT3 represses DAN1 and ANB1 in anaerobic cells. Cells of strains FY23, FY23mot3Δ and FY23rox1Δ transformed as indicated with YEpGAL1/ROX1, YCpGAL1/MOT3 or YCpLac33(vector) were pre-grown in SD(–ura) medium to early log phase, pelleted, washed twice with SD-raf/gal(–ura) medium, grown in that medium to mid-log phase (2 h), then shifted to anaerobic growth for 6 h before harvesting for RNA extraction. Northern blots were probed with DAN1, ANB1, ROX1 and MOT3.
Induction of MOT3 by heme and repression by Hap1 and Tup1–Ssn6
Concentration-dependent repression by Mot3 suggested that heme inhibits expression by inducing MOT3 expression. This was confirmed by the observation first, that MOT3 mRNA is down-regulated in anaerobic cells (Fig. 3) and second, that expression is de-repressed by heme, in parallel with ROX1 (3).
Figure 3.
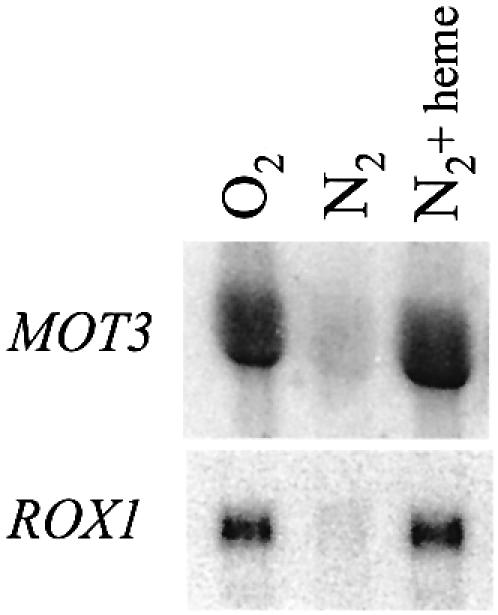
Regulation of MOT3 expression by oxygen and heme. RZ53 cells were grown under aerobic conditions and under anaerobic conditions with and without supplementation with heme at 25 µg/ml; heme was added to the culture 40 min before shifting to anaerobic growth. Northern blots were probed with MOT3 and ROX1.
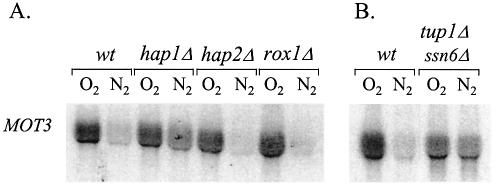
We tested factors known to mediate heme regulation for involvement in induction of MOT3. Neither Hap1 nor Hap2 were required for expression (Fig. 4A). However, rather than activating aerobic MOT3 expression, Hap1 functions by repressing in the absence of heme, as indicated by higher levels of MOT3 mRNA in hap1Δ cells during anaerobic growth. Heme regulation of MOT3 is not attributable solely to Hap1, since MOT3 mRNA levels in the hap1Δ strain were still higher in the presence of oxygen. This implicates another unknown oxygen-sensitive factor in the regulatory pathway. Anaerobic repression of MOT3 also depends on Tup1–Ssn6 more than on Hap1, as indicated by the presence of de-repressed levels of mRNA in anaerobic tup1Δssn6Δ cells (Fig. 4B). Whether the co-repressor complex is recruited by Hap1 and/or another repressor is not clear. Thus, MOT3 and ROX1 expression are co-regulated in four respects: heme induction, repression of basal expression by Hap1, regulation by a second heme-responsive system, and repression by Tup1–Ssn6.
Figure 4.
Role of Hap1, Hap2, Tup1 and Ssn6 in regulation of MOT3. (A) FY23 cells carrying disruptions of HAP1, HAP2 and ROX1 were grown under aerobic and anaerobic conditions. (B) FY23 cells carrying a double disruption of TUP1 and SSN6 were grown under aerobic and anaerobic conditions. Northern blots were probed with MOT3.
We tested for auto-repression of MOT3 analogous to that observed for ROX1 (6), but saw no increase in expression of the MOT3-lacZ reporter in the mot3Δ strain (data not shown). We also observed that neither factor regulates expression of the other (Fig. 4A and data not shown).
Rox1 requires Tup1–Ssn6, but Mot3 functions through another mechanism
Tup1 and Ssn6 are co-repressors of the hypoxic genes (23–25) and of the DAN/TIR/ERG genes (9) (see Fig. 1). It has been assumed that the heterodimer is recruited by Rox1, as with other repressors (26,27). If Tup1–Ssn6 is required for Rox1 function, loss of either the repressor or the co-repressor would be expected to cause similar levels of constitutive expression. However, it has been repeatedly observed that constitutive expression of ANB1 in aerobic tup1Δssn6 cells is significantly lower than in rox1Δ cells (Fig. 1). This difference doesn’t mean that Rox1 and Tup1–Ssn6 don’t interact, since Rox1 might be able to repress inefficiently by itself. Another explanation might be that the weaker Mot3 repressor, rather than Rox1, recruits the heterodimer. To test for these interactions, we made use of the observation that Gal4-driven expression of either factor causes efficient repression of ANB1 and DAN1 in wild-type anaerobic cells. In a tup1Δssn6 strain, Gal4-induced expression of MOT3 still caused efficient repression of ANB1, but expression of ROX1 did not (Fig. 5). Hence, Rox1 requires the Tup1–Ssn6 co-repressor in this context, while Mot3 evidently functions through a different mechanism.
Figure 5.
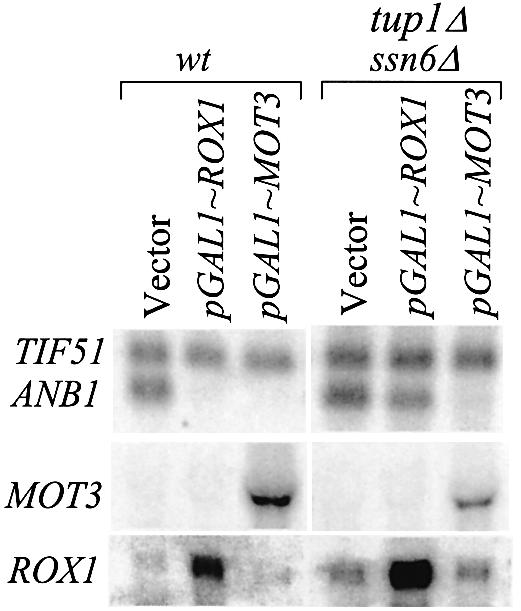
Role of Tup1–Ssn6 in repression by MOT3 and ROX1. FY23 cells and tup1Δssn6Δ derivatives carrying the indicated plasmids were grown anaerobically, as described for Figure 2. Northern blots were probed with ANB1, ROX1 and MOT3.
Mot3 and Rox1 are independent synergizing repressors
The failure of Rox1 and Mot3 to repress by themselves is in striking contrast to the highly stringent repression observed when both factors are present (Fig. 1). The mutual dependence of Mot3 and Rox1 could arise either because both are essential parts of a single mechanism (e.g. Tup1–Ssn6) or because they act independently in a synergistic fashion. We tested whether either factor is sufficient when over-expressed, starting from the observation that Gal4-driven expression of either MOT3 or ROX1 in anaerobic cells resulted in efficient repression of both DAN1 and ANB1 (Fig. 2). This observation had implied that each factor is mechanistically sufficient for repression, since under these conditions high levels of Mot3 were paired with very low (uninduced) levels of Rox1, or vice versa. To rule out the possibility that low levels of the non-induced factor assisted the over-expressed one, we repeated the experiment in mot3Δ and rox1Δ mutants. In both cases, expression of ANB1 was still almost fully inhibited, in both aerobic and anaerobic cells (Fig. 6), each repressor compensating for the absence of the other. Thus, either one of the two independent repression mechanisms is effective at high operator occupancy. It was clear, however, that repression is significantly more efficient in aerobic wild-type cells with both factors present (Figs 1 and 6B), even at their relatively low endogenous levels. Hence, even though both Mot3 and Rox1 can repress independently at artificially high levels, stringent regulation in aerobic cells depends on a synergistic interaction.
Figure 6.
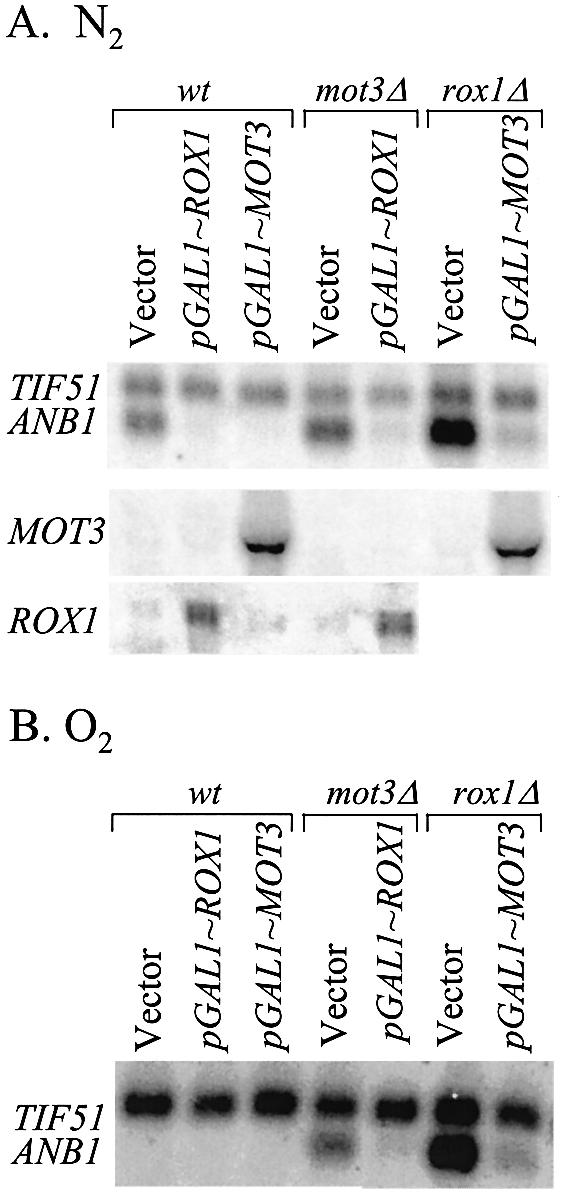
Independent repression by Mot3 and Rox1. Strains described in Figure 2 were grown as described. (A) Anaerobic growth. (B) Aerobic growth.
DISCUSSION
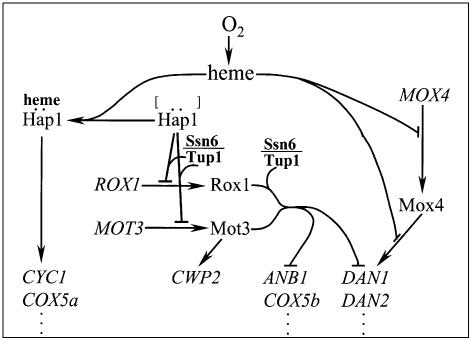
We have identified Mot3 as a repressor that functions synergistically with Rox1 to suppress aerobic expression of genes in two different anaerobic regulons. Mot3 and Rox1 are part of a complex regulatory network which mediates heme regulation through several different pathways (Fig. 7). Remarkably, heme regulation of the DAN/TIR genes, exemplified by DAN1, depends on four independent mechanisms, functioning at four different promoter sites: the Tup1–Ssn6-dependent Rox1 mechanism; the Tup1–Ssn6-independent Mot3 mechanism; an inhibitory mechanism which operates at the ‘AR1’ site through a regulatory domain in the oxygen-regulated Mox4/Upc2 activator (8,9); and a fourth anaerobic induction mechanism involving an unidentified activator operating at a second anaerobic response element (AR2) (7). Regulation through the AR1 and AR2 sites by heme is not highly stringent, and the combined action of Mot3 and Rox1 bound to their respective operators serves to enhance aerobic repression, reducing basal expression to undetectable levels. In the simpler ANB1 promoter, Mot3 and Rox1 appear to be solely responsible for heme regulation, providing a useful model for their possible interaction.
Figure 7.
The heme regulatory network controlling expression of anaerobic and aerobic genes. The scheme is consistent with evidence on the interlocked regulatory pathways controlling expression of aerobic and anaerobic genes, as described here and elsewhere (3,5,9,12). Heme regulates the anaerobic genes via several mechanisms. It de-represses Hap1-inhibited expression of the Mot3 and Rox1 repressors, which in turn block expression of several anaerobic genes during aerobic growth. A subset of anaerobic genes are activated by Mox4/Upc2, whose activation function and expression are both inhibited by heme during aerobic growth. In addition, there is a second unidentified oxygen-inhibited factor operating through the AR2 site in the DAN1 promoter (7). Heme also induces expression of a large group of genes (e.g. CYC1 and SOD1), via Hap1 in its alternate role as a heme-dependent activator. Finally, Mot3, known to function as either an activator or a repressor (14), also activates expression of the aerobically induced CWP2 mannoprotein gene (12).
The mutual dependence of Mot3 and Rox1 was originally inferred from the strongly constitutive expression of ANB1 in rox1 and mot3 mutants, which implies that neither factor can repress on its own at endogenous levels. We have demonstrated that the highly stringent repression of the ANB1 gene results from true synergy between the two repressors, i.e. through concerted action by independent mechanisms, rather than through obligatory co-participation in a single mechanism. The independent function of Mot3 and Rox1 was shown by the fact that over-expression of either can compensate for lack of the other. Strikingly, however, even when titrated by high levels of either Mot3 or Rox1, single-factor repression is still less stringent than in cells containing low levels of both repressors. One possible explanation for synergistic co-dependence might be cooperative binding, though in vitro studies have indicated that this does not occur on naked DNA (17). Perhaps more likely is the possibility that the two factors act through different targets in the PolII initiation complex, to produce concerted inhibition analogous to numerous examples of synergistic activation. A two-target model is strengthened by the indication that Mot3 and Rox1 utilize different mechanisms, with only the latter requiring Tup1–Ssn6 for its function.
This regulatory system has solved the problem of allowing some anaerobic genes to be expressed at low or moderate levels in aerobic cells while others are stringently repressed. Apparently, during evolution of the regulatory pathway, the levels of Mot3 and Rox1 have been calibrated in such a way that on some promoters they are able to fully repress, and at others, less so, depending on the number and quality of operator sites [which vary considerably (1)], and possibly on the type and strength of activation mechanism driving expression of each gene.
The response to oxygen depends on the fact, established here for MOT3 and earlier for ROX1 (3,5,6), that expression of the repressors is de-repressed from an inhibited state by heme. This occurs, at least in part, through a regulatory system which controls both genes, with repression in anaerobic cells being fully dependent on Tup1–Ssn6, and partly dependent on the heme-unbound form of Hap1 (5,6). A second factor which appears to play a role in repression of ROX1 and MOT3 has not been identified, nor have factors responsible for activation under aerobic conditions.
We have concluded that the Rox1 repression mechanism requires the participation of the Tup1–Ssn6 co-repression complex as originally suggested (23), and that Mot3 functions in a different way. This is based on the observation that repression of ANB1 caused by over-expression of ROX1 in anaerobic cells is lost in a tup1Δssn6Δ strain, whereas repression resulting from over-expression of MOT3 is still in effect. However, the role of Tup1–Ssn6 is not clear. For example, though expression of ANB1 under anaerobic and aerobic conditions is equal in the absence of the heterodimer (see Fig. 1A), the mRNA level is consistently lower than in anaerobic wild-type cells or in aerobic rox1Δ cells. The de-repression of ROX1 in tup1Δssn6Δ cells may cause reduced expression of ANB1, though this seems unlikely in view of the dependence of Rox1 on Tup1–Ssn6 (Fig. 5). More likely, reduced expression of ANB1 results from de-repressed expression of Mot3 (in glucose cultures) (Fig. 4). In addition, the absence of Tup1–Ssn6 may lead to reduced expression through some other perturbation of the regulatory circuits in this system. The genetic interaction of Rox1 and Tup1–Ssn6 suggests that Rox1 recruits Tup1–Ssn6 to the promoter during aerobic growth. However, recent evidence of constitutive binding of Tup1–Ssn6 to the ANB1 promoter (28) suggests that the functional relationship between Rox1 and Tup1–Ssn6 may not be one of recruitment.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Fred Winston for yeast strains. This work was supported by a grant from the National Science Foundation.
REFERENCES
- 1.Lowry C.V., Cerdan,M.E. and Zitomer,R.S. (1990) A hypoxic consensus operator and a constitutive activation region regulate the ANB1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol., 10, 5921–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zitomer R.S. and Lowry,C.V. (1992) Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev., 56, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowry C.V. and Zitomer,R.S. (1988) ROX1 encodes a heme-induced repression factor regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol. Cell. Biol., 8, 4651–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasubramanian B., Lowry,C.V. and Zitomer,R.S. (1993) The Rox1 repressor of the Saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility-group motif. Mol. Cell. Biol., 13, 6071–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keng T. (1992) HAP1 and ROX1 form a regulatory pathway in the repression of HEM13 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 2616–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deckert J., Perini,R., Balasubramanian,B. and Zitomer,R.S. (1995) Multiple elements and auto-repression regulate Rox1, a repressor of hypoxic genes in Saccharomyces cerevisiae. Genetics, 139, 1149–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen B.D., Sertil,O., Abramova,N.E., Davies,K.J. and Lowry,C.V. (2001) Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res., 29, 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crowley J.H., Leak,F.W.,Jr, Shianna,K.V., Tove,S. and Parks,L.W. (1998) A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol., 180, 4177–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abramova N.E., Cohen,B.D., Sertil,O., Kapoor,R., Davies,K.J.D. and Lowry,C.V. (2001) Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics, 157, 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sertil O., Cohen,B.D., Davies,K.J.D. and Lowry,C.V. (1997) The DAN1 gene of S.cerevisiae is regulated in parallel with the hypoxic genes, but by a different mechanism. Gene, 192, 199–205. [DOI] [PubMed] [Google Scholar]
- 11.Donzeau M., Bourdineaud,J.P. and Lauquin,G.J. (1996) Regulation by low temperature and anaerobiosis of a yeast gene specifying a putative GPI-anchored plasma membrane protein. Mol. Microbiol., 20, 449–459. [DOI] [PubMed] [Google Scholar]
- 12.Abramova N., Sertil,O., Mehta,S. and Lowry,C.V. (2001) Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol., 183, 2881–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vik A. and Rine,J. (2001) Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 6395–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grishin A.V., Rothenberg,M., Downs,M.A. and Blumer,K.J. (1998) Mot3, a Zn finger transcription factor that modulates gene expression and attenuates mating pheromone signaling in Saccharomyces cerevisiae. Genetics, 149, 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madison J.M., Dudley,A.M. and Winston,F. (1998) Identification and analysis of Mot3, a zinc finger protein that binds to the retrotransposon Ty long terminal repeat (delta) in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 1879–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hongay C., Jia,N., Bard,M. and Winston,F. (2002) Mot3 is a transcriptional repressor of ergosterol biosynthetic genes and is required for normal vacuolar function in Saccharomyces cerevisiae. EMBO J., 21, 4114–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kastaniotis A.J., Mennella,T.A., Konrad,C., Torres,A.M. and Zitomer,R.S. (2000) Roles of transcription factor Mot3 and chromatin in repression of the hypoxic gene ANB1 in yeast. Mol. Cell. Biol., 20, 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van der Vaart J.M., Caro,L.P.H., Chapman,J.W., Klis,F.M. and Verrips,C.T. (1995) Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J. Bacteriol., 177, 3104–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winston F., Dollard,C. and Ricupero-Hovasse,S.L. (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast, 11, 53–55. [DOI] [PubMed] [Google Scholar]
- 20.Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- 21.Lowry C.V., Weiss,J.L., Walthall,D.A. and Zitomer,R.S. (1983) Modulator sequences mediate the oxygen regulation of CYC1 and a neighboring gene in yeast. Proc. Natl Acad. Sci. USA, 80, 151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zitomer R.S., Sellers,J.W., McCarter,D.W., Hastings,G.A., Wick,P. and Lowry,C.V. (1987) Elements involved in oxygen regulation of the Saccharomyces cerevisiae CYC7 gene. Mol. Cell. Biol., 7, 2212–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang M., Rosenblum-Vos,L.S., Lowry,C.V., Boakye,K.A. and Zitomer,R.S. (1991) A yeast protein with homology to the beta-subunit of G proteins is involved in control of heme-regulated and catabolite-repressed genes. Gene, 97, 153–161. [DOI] [PubMed] [Google Scholar]
- 24.Tzamarias D. and Struhl,K. (1994) Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature, 369, 758–761. [DOI] [PubMed] [Google Scholar]
- 25.Carrico P. and Zitomer,R.S. (1998) Mutational analysis of the Tup1 general repressor of yeast. Genetics, 148, 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keleher C.A., Redd,M.J., Schultz,J., Carlson,M. and Johnson,A.D. (1992) Ssn6–Tup1 is a general repressor of transcription in yeast. Cell, 68, 709–719. [DOI] [PubMed] [Google Scholar]
- 27.Redd M.J., Arnaud,M.B. and Johnson,A.D. (1997) A complex composed of tup1 and ssn6 represses transcription in vitro. J. Biol. Chem., 272, 11193–11197. [DOI] [PubMed] [Google Scholar]
- 28.Papamichos-Chronakis M., Petrakis,T., Ktistaki,E., Topalidou,I. and Tzamarias,D. (2002) Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol. Cell, 9, 1297–1305. [DOI] [PubMed] [Google Scholar]