Abstract
Auxin-response factors (ARFs) bind with specificity to TGTCTC auxin-response elements (AuxREs), which are found in promoters of primary/early auxin-response genes. Nine different ARFs have been analyzed for their capacity to activate or repress transcription in transient expression assays employing auxin-responsive GUS reporter genes. One ARF appears to act as a repressor. Four ARFs function as activators and contain glutamine-rich activation domains. To achieve transcriptional activation on TGTCTC AuxREs in transient expression assays, ARFs require a conserved dimerization domain found in both ARF and Aux/IAA proteins, but they do not absolutely require their DNA-binding domains. Our results suggest that ARFs can activate or repress transcription by binding to AuxREs directly and that selected ARFs, when overexpressed, may potentiate activation further by associating with an endogenous transcription factor(s) (e.g., an ARF) that is bound to AuxREs. Transfection experiments suggest that TGTCTC AuxREs are occupied regardless of the auxin status in cells and that these occupied AuxREs are activated when exogenous auxin is applied to cells or when ARF activators are overexpressed. The results provide new insight into mechanisms involved with auxin regulation of primary/early-response genes.
Keywords: transcriptional activators, dimerization domain, AuxRE
Indole-3-acetic acid or auxin is a plant hormone that plays key roles in regulating cell division, extension, and differentiation. One site of auxin action centers on the activation of primary/early-response genes (reviewed in ref. 1). Auxin-response elements (AuxREs) have been identified functionally in natural auxin-responsive promoters of primary/early genes, and these elements, at least in some cases, contain the core sequence TGTCTC (2–4). Highly active, synthetic AuxREs have been created as direct or palindromic repeats of the TGTCTC element (2, 5). A palindromic TGTCTC AuxRE was used as bait in a yeast one-hybrid system to isolate a transcription factor, referred to as auxin-response factor 1 (ARF1) (2). ARF1 was shown to bind with specificity to the TGTCTC element, and the sequence and spacing requirements for ARF1 binding to palindromic AuxREs in vitro correlated perfectly with those requirements for AuxRE activity in vivo (2).
The ARF1 protein contains an amino-terminal DNA-binding domain (DBD), which has some sequence similarity to a carboxyl-terminal B3 domain found in the maize transcription factor VIVIPAROUS 1 (VP1) and its relatives (2, 6–8). ARF1 contains a carboxyl-terminal domain (CTD) related to motifs III and IV found in the CTDs of Aux/IAA proteins, which are short-lived nuclear proteins encoded by a family of primary/early auxin-response genes (2). The CTDs in ARF1 and Aux/IAA proteins facilitate dimerization among the members of both ARF and Aux/IAA protein families (2, 5, 9). Transcriptional activation or repression domains in ARFs have not been identified in previous studies.
Here, we show that some ARFs contain middle regions (i.e., regions separating the DBD from the CTD) that function as activation domains. Our results suggest that TGTCTC AuxREs are occupied regardless of the auxin status in cells and that auxin is unlikely to directly affect the targeting of ARFs to their DNA-binding sites. These prebound transcription factors facilitate activated transcription when auxin levels are elevated or when ARF activators are overexpressed in transfected protoplasts. Our results also suggest that ARF CTDs facilitate interactions between ARF activators that are not bound to DNA and transcription factors (e.g., ARFs) that are bound to AuxREs to further enhance transcription.
MATERIALS AND METHODS
Isolation of ARF cDNA Clones from Arabidopsis Tissue Culture and Flower Libraries.
An Arabidopsis thaliana (cv. Columbia) λZAPII AC16H cell suspension culture cDNA library (10) was provided by M. Axelos (Laboratoire de Biologie Moleculaire des Relations Plantes-Microorganisms, Centre INRA d’Auzeville, Castanet-Tolosan, France), and an A. thaliana (cv. Landsberg erecta) λZAPII flower cDNA library was obtained from the Arabidopsis Biological Resource Center at Ohio State University, Columbus. Two million clones from each library were screened by using an ARF1 cDNA probe that encoded the DBD. Filters were screened by using low-stringency hybridization (6× standard saline phosphate/EDTA, 55°C). Of more than 100 positive clones obtained, those producing strong hybridization signals encoded ARF1, whereas several of those producing weak signals encoded other ARFs. Partial cDNAs of ARF4, ARF6, ARF7, and ARF8 were isolated from this screen, and their full-length cDNAs were retrieved either by rescreening phage libraries or by using PCR from a plasmid two-hybrid cDNA library.
Reporter and Effector Plasmids.
Reporter plasmids have been described previously (5, 11, 12). The 35S-CAT reporter gene was obtained from William Folk (University of Missouri, Columbia) and was added to equalize the amount of DNA in transfection experiments in which no effector plasmids or different amounts of effector plasmids were used. For effector plasmids, PCR was used to create constructs encoding an amino-terminally truncated version of ARFs. Constructs were sequenced to confirm that no mistakes were created by PCR. Restriction enzyme digests were used to make carboxyl-terminally truncated versions of different ARFs. All effector plasmids with ARFs or ARF truncations were placed under the control of the 35S cauliflower mosaic virus (CaMV) promoter followed by the translational enhancer from tobacco mosaic virus (TMV) 5′ leader and contained a nopaline synthase 3′ untranslated region as described by Ulmasov et al. (5).
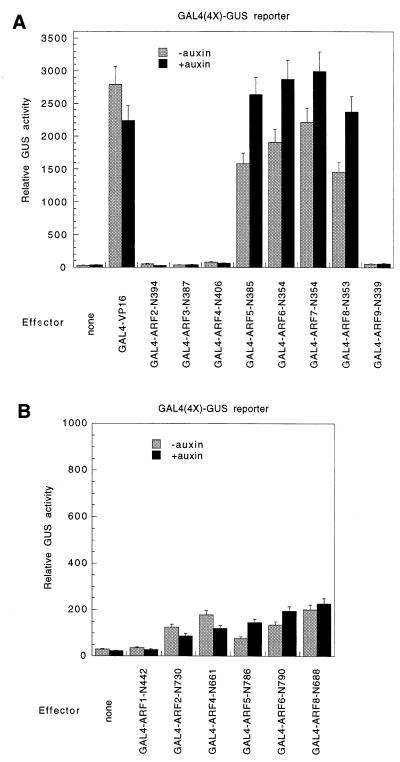
All GAL4 effector constructs contained the GAL4 DNA-binding domain (amino acids 1–147) with its nuclear localization signal fused in-frame with amino-terminally truncated forms of ARFs. The amino acid positions of the amino termini of the truncated ARF proteins are indicated in Fig. 3.
Figure 3.
The MRs of ARF5, ARF6, ARF7, and ARF8 contain ADs. (A) Transfections with truncated ARFs containing MRs plus the CTD. Effector plasmids consisted of the GAL4 DBD fused to the VP16 acidic AD from herpes virus or to ARF proteins that had their DBDs truncated. (B) Transfections with truncated ARFs containing only CTDs. Effector plasmids consisted of the GAL4 DBD fused to ARF proteins that had their DBDs plus MRs truncated. The reporter gene used in both A and B was the GAL4(4X) promoter-GUS gene (12). Effector genes contained the same promoter as those effectors tested in Fig. 2. The amino acid position of the amino termini in the truncated ARFs is indicated by N.
For ARF constructs with truncated DBDs in Fig. 4, the same primers used to create the GAL4 fusion constructs were employed. PCR products were cloned in-frame with the hemagglutinin (HA) epitope by using an NdeI site generated with the primers. The truncated ARFs were tagged on their amino termini with an MGYPYDVPDYAH peptide containing the HA epitope, which provided the initiator M. Constructs containing a VP16 activation domain were HA-tagged in-frame with the amino terminus of the VP16 fragment.
Figure 4.
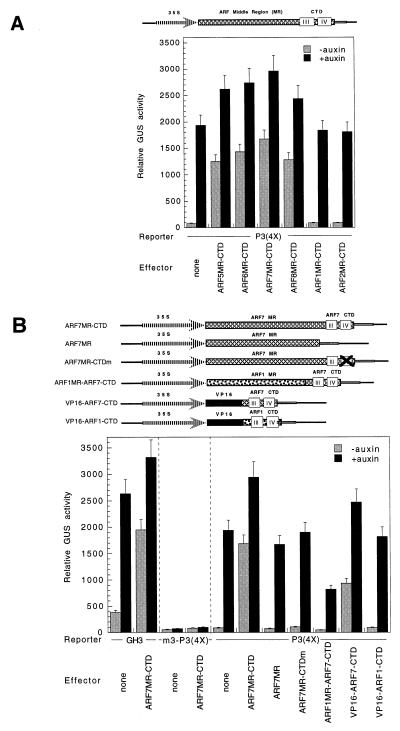
ARF5, ARF6, ARF7, and ARF8 that lack DBDs activate transcription on TGTCTC AuxREs. (A) ARFs with glutamine-rich MRs do not require a DBD to activate transcription of a P3(4X) promoter-GUS reporter gene (2). Diagrams of the effector plasmids are shown at the top. ARF1, ARF2, ARF5, ARF6, ARF7, and ARF8 contained their MRs and CTDs (MR-CTD). The amino termini of the truncated ARF proteins are identical to those shown in Fig. 3. (B) In the absence of a DBD, the CTD of ARF7 fused to homologous or heterologous ADs or RDs can modulate TGTCTC AuxRE promoter-GUS reporter gene expression. Diagrams of the effector plasmids are shown at the top. ARF7MR-CTD is ARF7 with its DBD truncated. ARF7MR is ARF7 with both its DBD and CTD truncated. ARF7MR-CTDm is identical to ARF7MR-CTD with the exception that point mutations were introduced into motif IV of the CTD, resulting in two conserved amino acid substitutions (PW → RS). ARF1MR-ARF7-CTD is derived from ARF7MR-CTD, with the ARF1 MR swapped for the ARF7 MR. VP16-ARF7-CTD has the VP16 AD swapped for the ARF7 MR. VP16-ARF1-CTD has the VP16 AD swapped for the ARF1 MR. The reporter plasmid was the GH3 promoter-GUS reporter gene, m3-P3(4X)promoter-GUS reporter gene, and P3(4X) promoter-GUS reporter gene, as indicated. The amino termini of the truncated ARF1 and ARF7 constructs is the same as that in Fig. 3. Details on the constructs are presented in Materials and Methods. Effector genes contained the same promoter as those effectors tested in Fig. 2.
The MscI site in the ARF7 cDNA sequence was used to make chimeric ARF7/ARF1/VP16 truncation, replacement, or domain-swapping constructs. PCR was used to isolate corresponding ARF1 cDNA fragments. All fusion constructs were sequenced to confirm PCR and cloning fidelity. The constructs consisted of the following amino acid end points (i.e., the amino acid position number of the amino terminus and the carboxyl terminus; C indicates that the amino acid position is the natural carboxyl-terminal end): ARF7MR, 643-ARF7–1029; ARF1MR-ARF7-CTD, 325-ARF1–533 fused to 1030-ARF7–1164C; VP16-ARF1-CTD, 413-VP16–490 fused to 531-ARF1–665C; and VP16-ARF7-CTD, 413-VP16–490 fused to 1030-ARF7–1164C.
The ARF7 effector plasmid with site-directed mutations in motif IV of its CTD (ARF7MR-CTDm) construct was created by using the mutagenizing primer: CGGCGATGATCgatcGGAAGAATTCGTAAA. The corresponding mutation (shown in lowercase) replaces a conserved NcoI site in ARF7 with a PvuI site and changes amino acids PW in motif IV to RS. The Chameleon (Stratagene) kit was used to perform site-directed mutagenesis.
Transfection of Carrot Protoplasts.
Transfections of carrot cell suspension culture protoplasts were as described previously (5, 11, 12), with auxin treatments of 25 μM 1-naphthalene acetic acid. Transfections were performed in triplicate for both plus- and minus-auxin treatments, and at least two independent transfection experiments were performed with each construct. Effector plasmids were cotransfected with GUS reporter plasmids at a ratio of 1 effector to 3 reporter plasmids unless indicated otherwise.
RESULTS
A Family of ARFs in Arabidopsis.
We have described previously the cloning of ARF1, ARF3, and ARF1-binding protein (ARF1-BP) from A. thaliana (2). ARF1-BP contained a carboxyl-terminal region related to ARF1. Retrieval of additional cDNA clones revealed that ARF1-BP contains an amino-terminal region similar to the DBD found in ARF1. Based on analysis of the full-length cDNA clone, ARF1-BP henceforth is referred to as ARF2. A fourth ARF protein described by Ulmasov et al. (2) is identical to IAA24 (9) and MONOPTEROS (13), and we refer to this as ARF5.
Additional ARFs were identified by using low-stringency hybridization to screen Arabidopsis cDNA libraries with an ARF1 DBD cDNA probe. These ARFs are referred to as ARF4, ARF6, ARF7, and ARF8, with GenBank accession numbers AF013466, AF013467, AF042195, and AF042196, respectively. The ARF9 gene was identified in GenBank (accession number AC002343), and a cDNA clone for ARF9 was isolated by using PCR (GenBank accession number AF082176).
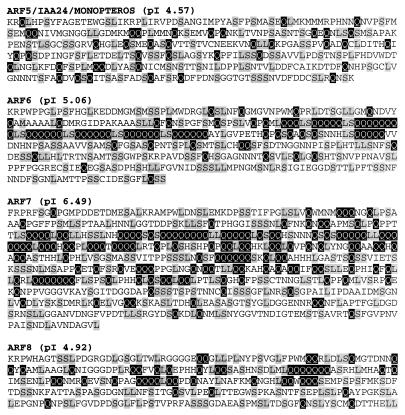
Each ARF contains a conserved amino-terminal region spanning about 300 aa (ref. 4; unpublished results). This region was shown to function as a DBD in ARF1 (2). With the exception of ARF3, ARFs contain a CTD with two motifs (referred to as motifs III and IV) related to motifs found in the CTD of Aux/IAA proteins (2). The middle region (MR) of ARF1 is rich in P, S, and T (2), whereas the MRs of ARF5 through ARF8 are rich in Q, L, and S (Fig. 1). ARF2, ARF4, and ARF9 contain MRs without any obvious biased amino acid sequences.
Figure 1.
Predicted sequences of Q-rich ARF MRs in Arabidopsis. Sequences are presented for MRs of ARF5, ARF6, ARF7, and ARF8. Glutamines are shaded with a black background and serines and leucines are shaded with a gray background. pI values for the middle regions are indicated for the sequences shown.
ARFs Function as Activators or Repressors in Transfected Carrot Protoplasts.
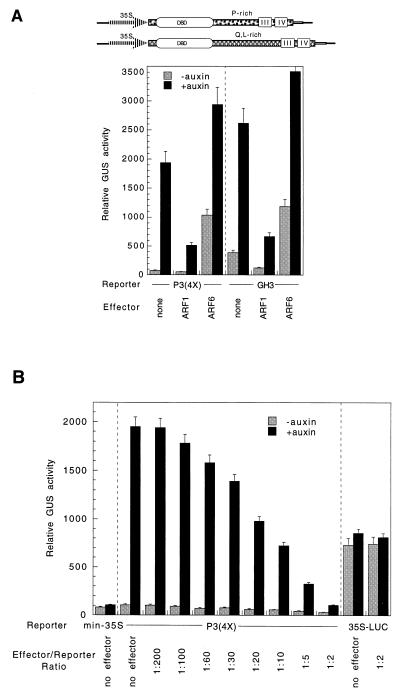
ARF1 and ARF6 contain MRs that are P/S/T-rich and Q/L/S-rich, respectively. To determine whether ARF1 and ARF6 had the potential to activate or repress transcription, effector plasmids that express full-length ARF1 and ARF6 were tested with auxin-responsive GUS reporter plasmids that were fused to either a P3(4X) promoter (2) or a soybean GH3 promoter (14). The P3(4X) reporter gene had low basal activity without auxin and was induced about 20-fold by auxin (Fig. 2A). The GH3 reporter gene had higher basal activity compared with the P3(4X) reporter gene but was still induced greater than 5-fold by auxin.
Figure 2.
ARF1 functions as a repressor, and ARF6 functions as an activator. (A) Diagrams of the ARF1 and ARF6 effector genes are shown at the top. The CaMV 35S promoter and TMV 5′ leader used to drive expression of the effector genes have been described previously (5). The DBD, MRs (i.e., P-rich in ARF1 and Q-rich in ARF6), and conserved motifs III and IV in the CTD are indicated in the diagrams. Transient assays were carried out in carrot protoplasts with a P3(4X) promoter-GUS reporter gene or a GH3 promoter-GUS reporter gene and the effector plasmid indicated or no effector plasmid (“none”). (B) Titration of the ARF1 effector plasmid with the P3(4X) promoter-GUS reporter plasmid. A CaMV 35S promoter-LUC reporter gene (11, 12) was used as a control to show that the effector was specific for TGTCTC auxin-responsive reporter genes. min−35S is a −46 minimal CaMV promoter-GUS reporter gene. (Bars = SE.)
Cotransfection of either reporter plasmid with the ARF1 effector plasmid resulted in repression of both basal and auxin-induced reporter gene expression compared with transfections carried out with no effector (Fig. 2A, “none”). Because of the low basal activity with the P3(4X) reporter gene, the level of repression for basal activity was difficult to assess, but auxin-induced activity was repressed by about 4-fold. With the GH3 reporter gene, basal and auxin-induced activities were repressed by about 3- and 4-fold, respectively. This repression is unlikely to result from squelching, because only repression and not activation was observed over a titration series from low to high ratios of effector to reporter plasmids (Fig. 2B). In contrast to results with TGTCTC AuxRE reporter genes, repression was not observed with a CaMV 35S promoter-LUC reporter gene.
Cotransfection of reporter plasmids with the ARF6 effector plasmid resulted in a severalfold increase in basal activity and a lesser increase in auxin-induced activity (Fig. 2A). With the P3(4X) reporter gene, there was a 10-fold increase in basal activity and a 30% increase in auxin-induced activity. With the GH3 reporter gene, there was a 9-fold increase in basal activity and a 25% increase in auxin-induced activity. We assume that the ARF6 effector plasmid had a greater effect on basal activity compared with auxin-induced activity because the reporter plasmids are already highly activated by auxin because of endogenous activators (2) and that the auxin response is overridden largely by the overexpression of the ARF6 activator.
Q-Rich MRs in ARF5, ARF6, ARF7, and ARF8 Function as Activation Domains (ADs) in Transfected Carrot Protoplasts.
To determine whether other ARFs might function as repressors or activators, effector plasmids encoding full-length ARF2, ARF3, ARF4, ARF5, ARF7, ARF8, and ARF9 were transfected into carrot protoplasts along with a P3(4X) promoter-GUS reporter gene. Effector plasmids encoding ARF2, ARF3, ARF4, and ARF9 had little, if any, influence on reporter gene activity, whereas effector plasmids encoding ARF5, ARF7, and ARF8, which contain Q-rich MRs, resulted in activation (i.e., 2-fold or more; data not shown).
To better assess the activation potency of MRs in ARF2–ARF9, the DBD was deleted from each ARF, and the MR plus CTD of each ARF (with the exception of ARF3, which lacks a CTD) was fused to the yeast GAL4 DBD. The GAL4-ARF MR-CTD constructs were tested as effector plasmids. These effector plasmids were cotransfected into carrot cells along with a GAL4(4X) promoter-GUS reporter gene containing four tandem GAL4 DNA-binding sites in the promoter. Fig. 3A shows that effector plasmids encoding ARF2, ARF3, ARF4, and ARF9 resulted in little, if any, activation or repression of the GUS reporter gene in the presence or absence of auxin when compared with the control (i.e., no effector plasmid). On the other hand, effector plasmids encoding ARF5, ARF6, ARF7, and ARF8, which contain Q-rich MRs (see Fig. 1), resulted in strong activation of 40- to 100-fold, depending on the construct and the auxin treatment. Activation with these GAL4-ARF effectors was significantly greater in the presence of auxin, and the activation potency in the presence of auxin was about equal to that observed with the strong VP16 AD (i.e., GAL4-VP16). Furthermore, the overexpression of GAL4-VP16 did not show the increased auxin response observed with the overexpression of GAL4-ARF5 through GAL4-ARF8. Control experiments showed that transfection with ARF5–ARF8 effector plasmids did not result in nonspecific transcription, because there was no activation or repression with a constitutive Arabidopsis RNA polymerase II 19.5 subunit (RPB7) promoter-GUS reporter gene (5) or 35S promoter-LUC reporter gene (data not shown). In contrast to other ARF effectors, preliminary experiments indicate that GAL4-ARF1MR-CTD effectors repress GAL4(4X) promoter-GUS reporter gene (T.U., unpublished results).
These results suggest that the Q-rich MRs in ARF5 through ARF8 function as ADs and that activation potency is enhanced in the presence of auxin. The experiments do not rule out the possibility that MRs in ARF2, ARF4, and ARF9 (and the region carboxyl-terminal to the DBD in ARF3) function as activation domains in certain cells and tissues, but not in carrot suspension cells. It is also possible that full-length ARF2, ARF3, ARF4, and ARF9 as well as truncated forms fused to GAL4 DBD are unstable in carrot cells or fail to reach their nuclear target site.
The results in Fig. 3A do not rule out the possibility that the conserved CTDs containing motifs III and IV might be responsible for or contribute to the activation potential of ARF5–ARF8. To test this possibility, amino-terminal truncations of selected ARF proteins were constructed that deleted both the DBDs and the MRs, and these truncations were fused to the GAL4 DBD. These truncated proteins contained conserved motifs III and IV, but little sequence amino-terminal to the CTD. The effector constructs were tested in carrot protoplasts by using the GAL4(4X) promoter-GUS reporter gene. The ARF1 effector had activity equivalent to that observed with no effector construct (Fig. 3B). ARF2, ARF4, ARF5, ARF6, and ARF8 effectors activated transcription 3- to 6-fold, which is substantially less than the 40- to 100-fold activation observed with ARF5– ARF8 effectors that contained the Q-rich MRs (Fig. 3A). The auxin response was inconsistent, resulting in a small decrease in activity with the ARF2 and ARF4 effectors and a small increase in activity with the ARF5, ARF6, and ARF8 effectors. With ARF5, ARF6, and ARF8, it is possible that the low amount of activation observed results from protein–protein interactions between the GAL4-ARF that is bound to the GAL4 DNA-binding sites and endogenous ARF activators that exist in carrot cells. These types of interactions might be facilitated by conserved motifs III and IV present in GAL4-ARF proteins and endogenous ARF proteins. In this case, the activation potential might be conferred by an endogenous ARF activator that associates with DNA-bound truncated ARF, similar to what occurs in a two-hybrid system.
ARFs with Q-Rich MRs Can Function as Activators in the Absence of a DBD with TGTCTC AuxRE Promoter-GUS Reporter Genes.
If transcriptional activation can be achieved by a two-hybrid-like system, where ARF CTDs facilitate interactions between a DNA-bound ARF and an unbound ARF activator, the overexpression of ARFs containing Q-rich ADs should potentiate transcription on TGTCTC AuxREs [e.g., P3(4X) promoter-GUS reporter genes] even if their DBDs are deleted. To test this prediction, truncated ARF1, ARF2, ARF5, ARF6, ARF7, and ARF8 proteins lacking their DBDs were expressed from effector plasmids. These truncated ARFs also were tested for in vitro DNA binding by using gel mobility-shift assays and were found to be incapable of binding P3(4X) probes (T.U., unpublished results). Effector plasmids were cotransfected into carrot protoplasts with a P3(4X) promoter-GUS reporter gene, and reporter activity was tested in the absence and presence of auxin.
Fig. 4A shows that overexpression of truncated ARFs that contain a Q-rich MR and the CTD (i.e., ARF5–ARF8) results in strong activation (i.e., 15- to 20-fold) of the GUS reporter construct in protoplasts not treated with auxin. Overexpression of these truncated ARFs also leads to increased activity in the presence of auxin compared with the control with no effector plasmid. ARF1 and ARF2 do not have Q-rich MRs, and when tested without their DBDs, overexpression of these truncated ARFs (ARF1MR-CTD and ARF2MR-CTD effectors) failed to activate or repress reporter activity compared with the control with no effector plasmid. This suggests that those truncated ARFs with Q-rich MRs are able to activate transcription when the DBD is deleted, but the truncated ARF1 and ARF2 do not activate, similar to untruncated forms of ARF1 and ARF2. The results also show that for ARF1 to repress transcription with a P3(4X) promoter-GUS reporter gene, the DBD of ARF1 is required. Furthermore, the results suggest the endogenous ARFs (or some other factors that can interact with ARFs lacking DBDs) must be bound to TGTCTC AuxREs before auxin stimulation, because high levels of activation are observed with effector plasmids that encode ARF activators that are unable to bind TGTCTC AuxREs.
To confirm that this two-hybrid-like activation depended on an AuxRE reporter gene, the ARF7 truncation (ARF7MR-CTD effector) was tested with the auxin-responsive GH3 promoter-GUS reporter gene and the m3-P3(4X) promoter-GUS reporter gene (2). The m3-P3(4X) promoter contains a T → G mutation at position 3 within each copy of TGTCTC element. The mutant promoter is not auxin-responsive in vivo nor does it bind ARF1 in vitro (2). Overexpression of truncated ARF7 did not activate the control m3-P3(4X) reporter gene, but did activate the GH3 reporter gene in a manner similar to that observed with the P3(4X) reporter gene (Fig. 4B). Additional control experiments indicated that the truncated ARF7 had no effect on promoter activity with GAL4(4X) or the constitutive Arabidopsis RPB7 promoter-GUS reporter genes (ref. 5; data not shown). These results are consistent with truncated ARF7 activation being limited to promoters containing functional TGTCTC AuxREs.
To achieve activation on AuxREs with the Q-rich ARFs lacking their DBDs, it is likely that the CTD (i.e., motifs III and IV, which facilitate protein–protein interactions among ARFs; refs. 2 and 5) would be required for the unbound truncated ARF to associate with the DNA-bound protein (i.e., presumably an endogenous ARF). To test this prediction, ARF7 with its DBD truncated (ARF7MR-CTD effector) was truncated further to remove the CTD (ARF7MR effector). Overexpression of the ARF7MR protein failed to activate transcription in the absence or presence of auxin with the P3(4X) promoter-GUS reporter gene (Fig. 4B). Another effector derived from ARF7MR-CTD (i.e., ARF7MR-CTDm effector) contained site-directed mutations within two conserved amino acids found in motif IV (i.e., PW → RS in GDDPW at positions 1103 and 1104). Overexpression of the ARF7MR-CTDm protein also failed to activate transcription, suggesting that these amino acids in motif IV are critical for the association between CTDs in ARFs.
To test further the role of the MR and CTD in conferring activation in the absence of DNA binding, domain-swap experiments were performed. When the MR of ARF1 was fused to the CTD of ARF7 (ARF1MR-ARF7-CTD effector), overexpression of ARF1MR-ARF7-CTD resulted in slight repression, not activation. These results suggest that the MR in ARF7 is necessary and sufficient to confer activation when combined with its CTD protein–protein interaction domain. We tested two other constructs in which the herpes virus VP16 AD was fused to ARF7 or ARF1 CTD (VP16-ARF7-CTD or VP16-ARF1-CTD effectors). Overexpression of VP16-ARF7-CTD resulted in activation, but not as great as that achieved with the ARF7MR-CTD construct. On the other hand, overexpression of a chimeric protein containing the VP16 AD and the ARF1 CTD (VP16-ARF1-CTD effector) had no effect compared with the reporter activity observed with the reporter plasmid alone. Taken together, these results suggest that the CTD in ARF7 is able to interact with a protein (i.e., presumably an ARF) that is bound to a TGTCTC AuxRE in carrot cells, but that the CTD in ARF1 apparently does not interact with the DNA-bound protein. The implications of these results are that CTDs facilitate associations between ARFs that are bound to DNA as well as ARFs that are not bound to DNA (2, 5, 9), that an endogenous ARF(s) is bound to DNA target sites whether auxin levels are high or low, and that there is at least some degree of specificity for these interactions among different ARFs. Furthermore, the results suggest that Q-rich ARFs can activate transcription on TGTCTC AuxREs without directly binding to their DNA target sites.
DISCUSSION
ARFs bind to TGTCTC AuxREs, examples of which are found in a variety of primary/early auxin-response genes (3, 4). ARF1–ARF9 contain a conserved amino-terminal domain of about 300 aa (ref. 3; unpublished results), and this domain has been shown to be required and sufficient for binding of ARF1 to TGTCTC AuxREs (2). The DBD is unique to plants, and related motifs spanning about 120 aa are found in maize VP1 and Arabidopsis ABI3 transcription factors, which function in abscisic acid hormone responses. In VP1, a carboxyl-terminal B3 domain, which is highly conserved in ABI3 and related to the ARF1 DBD, has been shown to bind DNA in a sequence-specific manner (8). In addition, a number of uncharacterized proteins are found in Arabidopsis databases that contain a sequence related to a portion of the DBD in ARF1 and the B3 domain in VP1 (2–4). Each ARF, with the exception of ARF3, contains a CTD related to motifs III and IV in the Aux/IAA proteins (2, 4). The Aux/IAA protein family contains more than 15 members in Arabidopsis, and several Aux/IAA mRNAs are induced within minutes after auxin application (9, 15). The CTDs in ARFs and Aux/IAA proteins have been shown to be important for associations (i.e., dimerization) between members of both protein families (2, 5, 9). Four of the ARFs (i.e., ARF5, ARF6, ARF7, and ARF8) described here contain Q-rich MRs that are also enriched in L and S. ARF1 contains a P-rich MR, whereas ARF2, ARF4, and ARF9 have MRs with no striking amino acid sequence bias.
ADs in ARFs.
The Q-rich stretches in ARF6, ARF7, and ARF8 are reminiscent of Q-rich stretches that function as ADs in a number of metazoan transcription factors (16). Our results show that full-length ARF6 functions as a transcriptional activator, and our unpublished results have shown that full-length ARF5, ARF7, and ARF8 function as activators when expressed from effector plasmids that are cotransfected into carrot cells with P3(4X) promoter-GUS reporter or GH3 promoter-GUS reporter genes. Furthermore, when the Q-rich MRs plus CTDs are fused to the yeast GAL4 DBD and expressed from effector plasmids along with GUS reporter genes that contain promoters with GAL4 DNA-binding sites, the chimeric transcription factors function as strong activators in carrot cells.
Although ARF5 is enriched for Q in its MR, it differs from ARF6, ARF7, and ARF8 in having no homopolymeric stretches of Q. Nevertheless, the ARF5 MR has activation potency nearly equivalent to that found in ARFs with homopolymeric Q stretches. These results suggest that Q-rich motifs can function as ADs in plant cells; however, because these motifs are also somewhat rich in L, S, and acidic amino acids, it remains to be demonstrated that the Qs are responsible for activation. Although we are unaware of any previously characterized Q-rich ADs in plant transcription factors, it has been shown recently with a synthetic promoter that insertion of homopolymeric Q stretches can increase the activation potency of a VP16 acidic AD in plant cells (17).
In contrast to ARFs with Q-rich MRs, ARF1 contains a P-rich MR, which is also characteristic of some metazoan transcriptional ADs and RDs (18). P-rich domains in bZIP transcription factors from plants also have been shown to function as ADs or RDs (19, 20). Our results show that in transfected carrot protoplasts, overexpression of ARF1 represses transcription on TGTCTC AuxRE reporter genes. The mechanism for this repression remains to be assessed. Overexpression of full-length ARF2, ARF3, ARF4, and ARF9 does not activate or repress transcription with P3(4X) or GH3 reporter genes (T.U., unpublished results), nor do their MRs appear to function as ADs or RDs in carrot protoplasts. It is possible, however, that these apparently inert ARFs might activate or repress transcription in selected cell types that contain a different set of coactivators or corepressors than those found in carrot suspension culture cells. It is also possible that these apparently inert ARFs function by binding DNA target sites and serve as scaffolds for ARF and/or Aux/IAA activators and repressors.
ARFs May Activate Transcription by Binding Directly to AuxREs or by Associating with Factors That Occupy AuxREs.
Protoplasts prepared from transfected carrot suspension culture cells presumably contain endogenous ARFs that function as transactivators, because TGTCTC AuxRE promoter-GUS reporter genes are active and responsive to auxin in these cells (2). When a given ARF is overexpressed from an effector plasmid (i.e., an exogenous ARF), it would be expected to compete with endogenous ARFs for binding to AuxREs and result in activation (e.g., ARF5–ARF8) or repression (e.g., ARF1). Surprisingly, however, overexpression of Q-rich ARFs that lack DBDs results in activation of the reporter genes. Thus, the ability of an exogenous Q-rich ARF to activate transcription on transfected reporter genes depends on a functional TGTCTC AuxRE-reporter gene and the exogenous ARF’s MR and CTD, but is not strictly dependent on the exogenous ARF’s DBD. These results suggest that untruncated (i.e., full-length) forms of exogenous Q-rich ARFs might activate transcription by (i) competing with endogenous ARFs for DNA target sites on reporter genes and/or (ii) interacting with some endogenous factor that is bound to an AuxRE. The most likely candidate for this endogenous DNA-bound factor is an ARF that can both recognize the AuxRE through its DBD and associate with exogenous Q-rich ARFs through its CTD. The dependence on the CTD for activation by exogenous Q-rich ARFs lacking a DBD is consistent with the DNA-bound factor being an endogenous ARF. Thus, ARFs may have the capacity to activate or repress transcription on AuxREs by binding directly to TGTCTC target sites and further potentiate activation or repression by interacting with ARFs and possibly Aux/IAA proteins that are not in direct contact with DNA.
ARFs are not unique in their abilities to activate or repress transcription by binding directly to DNA target sites or by acting off the DNA through contacts with proteins that occupy the DNA target sites. It is well documented that glucocorticoid receptors and other members of the steroid hormone receptor superfamily can activate or repress transcription as DNA-binding proteins or as proteins that interact with other transcription factors that are bound to DNA (21, 22). Likewise, the maize transcription factor VP1, which has a DBD with some limited similarity to the ARF DBD (2), appears to function by binding DNA directly in some cases and acting off the DNA in other cases (8, 23). In general, the transcription factor that binds the DNA target interacts with a different type of transcription factor (e.g., a factor that cannot recognize the target). ARFs, on the other hand, can recognize the DNA target through an amino-terminal domain and interact with one another through a carboxyl-terminal domain, allowing one of the partners to participate without binding the DNA target.
Roles of ARFs and Aux/IAA Proteins in Auxin-Responsive Transcription.
Our results suggest that ARFs may associate with one another through their CTDs to regulate transcription on TGTCTC AuxREs and that there is some specificity to these CTD interactions. There is also evidence that Aux/IAA proteins can associate with themselves and with ARFs and that these interactions are facilitated by motifs III and IV in their CTDs (2, 5, 9). Furthermore, it has been shown that overexpression of at least some Aux/IAA proteins from effector plasmids represses transcription on TGTCTC AuxRE-reporter genes in transfected carrot protoplasts (5). Our interpretation of these latter results is that these exogenous Aux/IAA proteins might bind directly to endogenous DNA-bound ARFs and interfere with their ability to activate AuxREs or might bind to unbound ARFs and prevent them from interacting with ARFs that are bound to AuxREs. It remains possible, however, that some of the more than 15 Aux/IAA protein family members also could function as activators by interacting with ARFs that are bound to DNA.
ARF transcription factors presumably are present in cells and bound to TGTCTC AuxREs regardless of the auxin status, and auxin may somehow influence ARF protein–protein interactions required for transcriptional activation/repression. It is possible that when auxin levels are low, ARFs that are bound to TGTCTC AuxREs are associated with transcriptional repressors that are not bound to DNA. Aux/IAA proteins or, possibly, nonactivator ARF proteins might be responsible for such repression because both types of proteins contain CTDs that facilitate interactions among these proteins. When auxin levels are elevated, the repressors may dissociate, allowing a DNA-bound ARF to activate transcription, and this activation might be potentiated further by association with unbound ARF or, possibly, Aux/IAA activators.
Acknowledgments
We thank X. Feng for excellent technical assistance. This research was supported by National Science Foundation Grant MCB 9603678 and University of Missouri Food for the 21st Century Program.
ABBREVIATIONS
- AuxRE
auxin-response element
- ARF
auxin-response factor
- DBD
DNA-binding domain
- CTD
carboxyl-terminal domain
- CaMV
cauliflower mosaic virus
- MR
middle region between the DBD and CTD
- AD
activation domain
Footnotes
References
- 1.Abel S, Theologis A. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ulmasov T, Hagen G, Guilfoyle T J. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 3.Guilfoyle T J, Hagen G, Ulmasov T, Murfett J. Plant Physiol. 1998;118:341–347. doi: 10.1104/pp.118.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guilfoyle T J, Ulmasov T, Hagen G. Cell Mol Life Sci. 1998;54:619–627. doi: 10.1007/s000180050190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulmasov T, Murfett J, Hagen G, Guilfoyle T J. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarty D R, Hattori T, Carson C B, Vasil V, Lazar M, Vasil I K. Cell. 1991;66:895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- 7.Giraudat J, Hauge B M, Valon C, Smalle J, Parcy F, Goodman H M. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki M, Kao C Y, McCarty D R. Plant Cell. 1997;9:799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J, Harter K, Theologis A. Proc Natl Acad Sci USA. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Regad F, Bardet C, Tremousaygue D, Moisan A, Lescure B, Axelos M. FEBS Lett. 1993;316:133–136. doi: 10.1016/0014-5793(93)81201-a. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z-B, Ulmasov T, Shi X, Hagen G, Guilfoyle T J. Plant Cell. 1994;6:645–657. doi: 10.1105/tpc.6.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulmasov T, Liu Z-B, Hagen G, Guilfoyle T J. Plant Cell. 1995;7:1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardtke C S, Berleth T. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagen G, Martin G, Li Y, Guilfoyle T J. Plant Mol Biol. 1991;17:567–579. doi: 10.1007/BF00040658. [DOI] [PubMed] [Google Scholar]
- 15.Abel S, Nguyen M D, Theologis A. J Mol Biol. 1995;251:533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- 16.Courey A J, Tjian R. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 17.Schwechheimer C, Smith C, Bevan M W. Plant Mol Biol. 1998;36:195–204. doi: 10.1023/a:1005990321918. [DOI] [PubMed] [Google Scholar]
- 18.Williams T, Tjian R. Genes Dev. 1991;5:670–682. doi: 10.1101/gad.5.4.670. [DOI] [PubMed] [Google Scholar]
- 19.Schindler U, Terzaghi W, Beckmann H, Kadesch T, Cashmore A R. EMBO J. 1992;11:1275–1289. doi: 10.1002/j.1460-2075.1992.tb05171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z-B, Hagen G, Guilfoyle T J. Plant Physiol. 1997;115:397–407. doi: 10.1104/pp.115.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McEwan I J, Wright A P H, Gustafsson J-A. BioEssays. 1997;19:153–160. doi: 10.1002/bies.950190210. [DOI] [PubMed] [Google Scholar]
- 22.Lefstin J A, Yamamoto K R. Nature (London) 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 23.Carson C B, Hattori T, Rosenkrans L, Vasil V, Vasil I K, Peterson P A, McCarty D R. Plant J. 1997;12:1231–1240. doi: 10.1046/j.1365-313x.1997.12061231.x. [DOI] [PubMed] [Google Scholar]