Abstract
In the protozoan parasite Leishmania, drug resistance can be a complex phenomenon. Several metabolic pathways and membrane transporters are implicated in the resistance phenotype. To monitor the expression of these genes, we generated custom DNA microarrays with PCR fragments corresponding to 44 genes involved with drug resistance. Transcript profiling of arsenite and antimony resistant mutants with these arrays pinpointed a number of genes overexpressed in mutants, including the ABC transporter PGPA, the glutathione biosynthesis genes γ-glutamylcysteine synthetase (GSH1) and the glutathione synthetase (GSH2). Competitive hybridisations with total RNA derived from sensitive and methotrexate resistant cells revealed the overexpression of genes coding for dihydrofolate reductase (DHFR-TS), pteridine reductase (PTR1) and S-adenosylmethionine synthase (MAT2) and a down regulation of one gene of the folate transporter (FT) family. By labelling the DNA of sensitive and resistant parasites we could also detect several gene amplification events using DNA microarrays including the amplification of the S-adenosyl homocysteine hydrolase gene (SAHH). Alteration in gene expression detected by microarrays was validated by northern blot analysis, while Southern blots indicated that most genes overexpressed were also amplified, although other mechanisms were also present. The microarrays were useful in the study of resistant parasites to pinpoint several genes linked to drug resistance.
INTRODUCTION
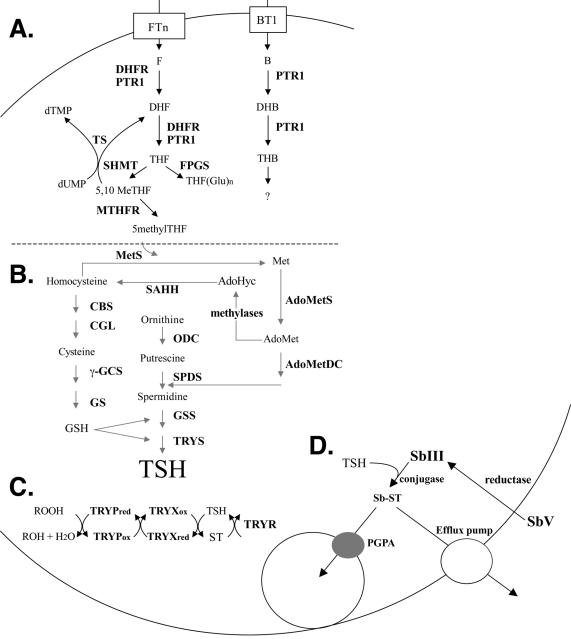
Leishmania is a protozoan parasite distributed worldwide for which we have limited means to control its spread. No vaccines are available and chemotherapy relies mainly on pentavalent antimony containing drugs, although in endemic regions with high antimony resistance the palliative miltefosine appears very useful (1,2). Unresponsiveness of parasites to antimonials is a serious health problem and resistance has now reached epidemic proportions in parts of India (3). Numerous resistance mechanisms have been uncovered from in vitro studies that were related to either transport defect, amplification of genes coding for drug targets, or altered metabolism (reviewed in 4). Antimony is given as pentavalent antimony (SbV) sugar conjugates but the metal is likely to be reduced in vivo to the trivalent form (SbIII). Therefore, a loss of this reduction activity could lead to resistance (5). The route of entry of SbV is not known, but reduced uptake could also lead to resistance and cells selected for SbIII resistance show a reduction in the steady-state accumulation of the metal (6). All antimonite and arsenite resistant Leishmania mutants share a metabolic signature: a significant increase in the intracellular trypanothione (TSH) level (7,8). TSH is a glutathione–spermidine conjugate (9), which is formed following several enzymatic steps (Fig. 1B). Overexpression of some of the genes in this pathway can lead to an increase in the intracellular level of TSH (10,11). It is proposed that an Sb–TSH conjugate is formed inside the parasite cell and the complex is either extruded outside the cell by an ATP-dependent efflux system (Fig. 1D) or sequestered within a vacuole by the intracellular ABC transporter PGPA (12). Other genes not related to the metabolic pathways shown in Figure 1D can also contribute to Sb resistance (13,14).
Figure 1.
Biochemical pathways of TSH and folate metabolism in Leishmania. (A) Folate and pterin metabolic pathways. (B) TSH biosynthetic pathway including selected aspects of cysteine and methionine pathways. (C) TSH-dependent peroxide reduction pathway. (D) Model for antimony resistance in Leishmania. All annotated genes involved in these pathways and their abbreviations are listed in Table 1 and were arrayed on DNA microarrays. F, DHF, THF, B, DHB and THB correspond to Folate and Biopterin and their Dihydro and Tetrahydro forms.
Similarly, resistance to the model antifolate drug methotrexate (MTX) in Leishmania can be explained by several mechanisms (Fig. 1A) including reduced uptake mediated by the modulation of the expression of a series of folate transporters (FT) (15,16). This is often compensated for by an overexpression of the biopterin transporter BT1 (17,18). Amplification of the target gene dihydrofolate-reductase-thymidylate synthase DHFR-TS (19) or of the pterin reductase PTR1 that has folate reductase activity (20,21) are two other frequent MTX resistance mechanisms. Modulation in the level of polyglutamylation of folates and MTX was also shown to contribute to MTX resistance in this parasite (22).
Several different mechanisms are responsible for resistance to a drug and multiple resistance mechanisms may co-exist in the same cell and act either additively or synergistically. The multiplicity of resistance mechanisms to a certain class of drug justifies an approach allowing the simultaneous analysis of the expression of several genes. DNA microarrays are well suited for that purpose. The utility of this technology has been demonstrated for drug responses and resistance mechanisms in microorganisms and cancer cells (23–29), and could allow the identification of novel pathways potentially involved in resistance. DNA microarrays have recently been used to investigate stage-specific gene expression in Leishmania (30–32). Since drug resistance in this parasite is often linked to alterations in gene expression due to either gene amplification (33,34), gene deletion (15) or other mechanisms (11), we chose DNA microarrays since it is well suited for analysing drug resistant mutants. The Leishmania genome-sequencing project is near completion (www.genedb.org) and most of the genes involved in the TSH biosynthetic pathway or in the folate metabolism pathway shown in Figure 1, are now part of the public domain. We have generated custom microarrays with DNA fragments corresponding to genes related to these two metabolic pathways as well as the linked pathways of cysteine and methionine metabolism (Fig. 1). We present a study of the expression of these genes in Leishmania mutants selected for metal or MTX resistance.
MATERIALS AND METHODS
Cell lines and cultures
The Leishmania tarentolae wild-type cell line TarIIWT, the arsenite resistant mutant TarIIAs50.1 and the antimonite resistant mutant TarIISbIII400.1 have been described previously (8,35). The Leishmania major LV39 strain was selected step by step for MTX resistance in M199 medium. This process generated two independent clones named MTX60.2 and MTX60.4, both resistant to 60 µM of MTX (EC50 of wild-type cell is 100 nM) (36). The growth properties of the different wild-type species as well as of the drug resistant mutants were carefully monitored to ensure that cells were collected in the same growth phase.
Total RNA preparation and labelling
RNA was isolated from 108 Leishmania cells during the mid-log growth phase using Trizol reagent (Invitrogen) as described by the manufacturer. The RNA preparation was treated with DNase I (Ambion) to avoid any genomic contamination and purified further using the RNeasy kit (Qiagen). Purified total RNA was quantified spectrophotometrically at 260 and 280 nm, and ratiosA260 nm/A280 nm between 1.9 and 2.1 were considered acceptable. RNA integrity was assessed by visualising the ribosomal bands on a 1% TAE agarose gel. RNA was converted to fluorescent cDNA probes by direct incorporation of Cy3/Cy5-linked dUTP (Amersham Pharmacia) mediated by the Superscript II reverse transcriptase (RT) (Invitrogen) according to the supplier recommendations. For each labelling reaction, 15 µg of purified RNA were spiked with two exogenous RNAs (CAB1 at 2 pg/µl and NAC1 at 5 pg/µl from Arabidopsis thaliana; Stratagene) added as references to adjust for variations in the incorporation efficiency of the two dyes and for differences in first strand cDNA synthesis reactions. Briefly, total RNA was first denatured by heating at 95°C for 5 min and the reverse transcription was performed at 42°C for 3 h with the RT enzyme, random nonamers and anchored oligo(dT)25. The RNA was then degraded by an RNase treatment and the cDNA was purified using the Qiaquick PCR purification kit (Qiagen). The synthesised cDNA was quantified at 260 nm while the Cy3 and Cy5 fluorophor incorporations were monitored at 550 and 650 nm, respectively. Aliquots of cDNA containing 20 pmol of incorporated dye were lyophylised and stored at –80°C until use.
DNA labelling
Total DNA was isolated using the DNAzol technique (Invitrogen). Thirty micrograms were digested with Sau3AI, purified using Microcon 30 filter columns (Millipore) then quantified at 260 nm. Two micrograms of digested DNA were labelled by direct incorporation of Cy3/Cy5-linked dUTP using a random priming method with a highly concentrated Klenow enzyme (NEB). In addition to the genomic template DNA, each labelling reaction contained random hexamers, 120 µM of each of dATP, dGTP and dCTP, 60 µM dTTP, 60 µM Cy3 or Cy5-dUTP in a Tris-MgCl2 buffer. Reactions were performed at 37°C for 2 h. The DNA preparation was then purified with Microcon 30 filter columns and quantified using procedures similar than those described above for labelled cDNAs. Aliquots containing 20 pmol of incorporated dye were lyophylised and stored at –80°C until use. Hybridisations were performed with the same protocol used for labelled cDNA.
Generation of customised DNA microarrays
When this work was initiated, not all genes found in Table 1 were known and some were obtained by PCR homology cloning, but with the Leishmania genome-sequencing project advancing steadily (and now almost completed) it has been possible to obtain most genes from in silico screening. The remaining exception is the cystathionine γ-synthase (Table 1). Prior to genome data, several genes were cloned using degenerated oligonucleotides derived from the conserved sequences of aligned homologues of these proteins. For all 44 genes, depending on the size and sequence, one to three pairs of primers were synthesised and used to amplify 500 bp fragments from Leishmania genomic DNA. These PCR fragments (one to three fragments per gene, see Table 1) were cloned in the pGEMt-easy vector system (Promega), which contains the sequences necessary for amplification using T7 and SP6 primers. The identity of all clones was confirmed by sequencing. For layering on DNA arrays, each cloned fragment was amplified by PCR using T7 and SP6 primers, purified on a Qiaquick PCR column (Qiagen) and quantified using the Picogreen reagent (Molecular Probes). Aliquots containing 2 µg of each PCR fragment were transferred to a 384 well plate, dried and subsequently resuspended in 5 µl of a 50% DMSO solution. The DNA was denatured by heating at 95°C for 15 min and printed onto CMT-GAPS II slides (Corning) using an SDDC-2 Arrayer (Virtek) in a 70% humidity constant atmosphere. Slides were UV cross-linked at 200 mJ then baked at 80°C for 2 h. Printing quality controls of each slide batch include staining with ToTo-3 iodine (Molecular Probes) and hybridisation with Cy3-labelled random oligonucleotides (Spotcheck, Genetix).
Table 1. Genes represented in custom DNA microarrays.
| Genes | Protein | Access. # | Species used | # PCR Fragment |
|---|---|---|---|---|
| ABCTP1 | ABC transporter | AC005766 | L.major | 2 |
| Actin | Actina | L16961 | L.major | 1 |
| MAT2 | S-Adenosylmethionine synthase (AdoMetS) | AF179714 | L.donovani | 2 |
| AdoMetDC | S-Adenosylmethionine decarboxylase | U20091 | L.donovani | 2 |
| α-tubulin | Alpha-tubulina | U09612 | L.donovani | 2 |
| Amastine | Amastigote-specific gene | AF195531 | L.donovani | 1 |
| β-tubulin | Beta-tubulina | X93566 | L.major | 3 |
| BT1 | Biopterin transporter 1 | AF244919 | L.major | 3 |
| CBL | Cystathionine β-lyase | AL359773 | L.major | 3 |
| CBS | Cystathionine β-synthase | AF256080 | L.tarentolae/L.major | 2 |
| CGS | Cystathionine γ-synthase | PCR homology | L.major | 1 |
| DHFR-TS | Dihydrofolate reductase thymidylate synthase | X51733 | L.major | 3 |
| FPGS | Folylpolyglutamate synthase | AF284554 | L.tarentolae | 2 |
| FTn | Folate transporters | AF084469 | L.donovani | 3 |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenasea | AF047497 | L.major | 1 |
| GSH2 | Glutathione synthetase (GS) | AL356246 | L.major | 2 |
| GSH1 | γ-Glutamylcysteine synthetase (γ-GCS) | Y10049 | L.tarentolae/L.major | 3 |
| GSS | Glutathione spermidine synthase | AQ849191 | L.major | 1 |
| HSP70 | Heat shock protein 70 | X13441 | L.major | 2 |
| L4468.01 | Transporter, pot. ATP-dep. permease precursor | AL121864 | L.major | 3 |
| L673.01 | Transporter, MDR copy 1, Pgp | AL135898 | L.major | 3 |
| L673.02 | Transporter, MDR copy 2, Pgp | AL135898 | L.major | 1 |
| L8329.03 | Transporter, bacterial type ABC transporter | AL446004 | L.major | 3 |
| MDR | Transporter, multiple drug resistance related | U63320 | L.donovani | 2 |
| METS | Methionine synthase | AQ848249 | L.major | 1 |
| METH | Methionine synthase cobalamine dependant | Chr7-11_tmp.570b | L.major | 1 |
| METE | Methionine synthase cobalamine independent | AQ845725 | L.major | 1 |
| ODC | Ornithine decarboxylase | AF159564 | L.tarentolae/L.major | 3 |
| ORF SbV | Na stibogluconate resistance protein | AF047351 | L.tarentolae | 2 |
| ORF19 | Cysteine-leucine rich protein gene | AF262948 | L.tarentolae | 2 |
| PAH | Phenylalanine hydroxylase | CHR28_tmp.72b | L.tarentolae | 2 |
| PFK1 | Phosphofructokinase Ia | AY029213 | L.major | 2 |
| PGPA | p-Glycoprotein A (ABC transporter) | X17154 | L.tarentolae/L.major | 1 |
| PGPE | p-Glycoprotein E, (ABC transporter) | L29485 | L.tarentolae | 2 |
| PTR1 | Pterine reductase 1 | L01699 | L.major | 1 |
| SAHH | S-Adenosylhomocysteine hydrolase | AQ902547 | L.major | 2 |
| SHMT | Serine hydroxymethyltransferase | AL138973 | L.major | 2 |
| SOD A | Fe-dep. superoxide dismutase A | AF003964 | L.major | 2 |
| SOD B | Fe-dep. superoxide dismutase B | AF003963 | L.major | 1 |
| SPDS | Spermidine synthase | AF298195 | L.donovani | 1 |
| TRYR | Trypanothione reductase | Z23135 | L.donovani | 3 |
| TRYP | Tryparedoxin peroxidase | AF069386 | L.major | 1 |
| TRYX | Tryparedoxin | AQ849312 | L.major | 1 |
| TRYS | Trypanothione synthetase | AL133443 | L.major | 1 |
aHousekeeping and structural genes used as controls.
bTemporary systematic name in Leishmania Gene databank.
Prehybridisation/hybridisation and statistical analysis
Slides were prehybridised at 42°C for 60 min in 5× SSC, 0.1% SDS, 1% BSA, 100 µg/ml ssDNA. For hybridisation, Cy3 and Cy5 labelled cDNAs were first resuspended and mixed together in the hybridisation solution (50% formamide, 5× SSC, 0.1% SDS, 25 µg/ml ssDNA, 460 µg/ml tRNA) pre-heated to 42°C. The mixture was heated at 95°C for 5 min, cooled slowly to 42°C and applied under a coverslip on the array. Hybridisation was performed at 42°C for at least 12 h under immersion (Corning chambers). Slides were first washed at 55°C for 5 min in 1× SSC, 0.03% SDS. Subsequent washes were at room temperature in 0.2× SSC and 0.05× SSC, each for 5 min under agitation. Slides were dipped in water and 99% ethanol then dried by centrifugation. Scanning was performed using a confocal 4000XL scanner (GSI Lumonics). Spots were manually examined and those of low intensity or presenting unusual shape were ignored. Data were analysed using QuantArray software. Four different RNA preparations of each mutant and their corresponding wild-type cell line were analysed, corresponding to two dye-swapping experiments. Data were corrected for local background, and ratios were adjusted using the two spiked RNAs as references. Statistical significance was calculated by the Student’s test using StatView software, and the two spiked RNAs as controls. A cut-off of two for significant differences was chosen although we do agree that smaller differences may also be of interest when statistically significant. Genes with consistent altered expression (at least 2-fold differences) were further studied by Southern and northern blot analysis, while following standard protocols (37) to confirm results obtained by DNA microarrays. Blots were hybridised to the same PCR DNA fragments layered on the arrays, and the hybridisation intensity was derived by densitometry analysis using an alphaImager 2000 (Alpha Innotech) with the software Alpha Ease version 4, and also a BioImage Visage 100S and compared with the signal intensity of blots hybridised with an α-tubulin probe.
RESULTS
Generation of DNA microarrays
The strategy to generate the DNA microarrays is laid out under Materials and Methods. Our goal was to build DNA microarrays containing most of the genes known to be implicated in antimony and MTX resistance and to complete these arrays with genes known to be involved in TSH and folate metabolic pathways including some involved in cysteine and methionine metabolic pathways (Fig. 1) as well as a number of ‘housekeeping’ genes. The 44 genes investigated, with their corresponding accession numbers, are listed in Table 1. Each PCR fragment was spotted in triplicate on the array. Arrays were hybridised to Cy3 and Cy5 labelled cDNA from the same sample preparation and hybridisation was found to be uniform (not shown). Once the spotting, hybridisation and washing conditions were optimised, and hybridisation signals could be consistently reproduced, the arrays were used for the parallel analysis of gene expression in Leishmania drug resistant mutants.
Analysis of metal resistant mutants by DNA microarrays
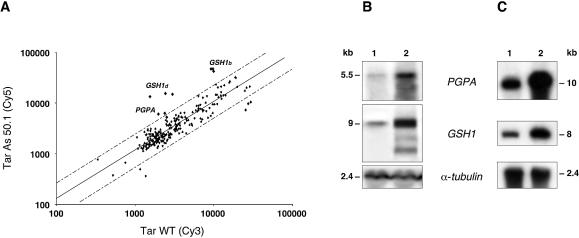
Our previous work on metal resistance in Leishmania has highlighted the importance of TSH biosynthesis and the increased expression of the ABC transporter PGPA (38). All the known genes of the TSH metabolic pathway were thus included on the array. To validate the use of our custom DNA microarrays, we first studied the arsenite resistant mutant TarIIAs50.1, a mutant in which PGPA is known to be overexpressed (35) and in which the level of TSH is known to be increased (7). Most genes were equally expressed between wild-type and TarIIAs50.1, except for γ-glutamylcysteine synthetase (GSH1) (P < 0.0001), the gene product of which catalyses the rate limiting step of glutathione biosynthesis, and PGPA (P < 0.0177), which according to this (Fig. 2) and other experiments were found to be overexpressed in the mutant. The same hybridisation profile was obtained whether the mutants were grown in the presence of drugs or in the absence of drugs for one passage (result not shown). It is also worth noting that arrays containing mostly genes derived from L.major (Table 1) are suitable for analysing the expression of genes of L.tarentolae. DNA microarray expression results were confirmed by northern blot analysis (Fig. 2B). While microarrays suggested a 2–3-fold overexpression, quantification of northern blots indicated that GSH1 was overexpressed by 11-fold and PGPA by 15-fold (Table 2). However, when the microarrays were scanned with a lower laser power, the fold difference values were higher and closer to northern blot values but weaker signals were lost. Thus, in some experiments, our custom microarrays became rapidly saturated for genes highly overexpressed.
Figure 2.
Gene expression analysis of the L.tarentolae As50.1 mutant as determined by DNA microarrays and northern blot analysis. (A) Scatter plot of hybridisation intensities between TarAs50.1 (Cy5) and wild-type L.tarentolae cells (Cy3). The expression of genes represented by dots within the dashed lines are considered as similar in the two tested strains. Dashed lines indicate 2-fold differences and genes whose expression differ significantly are indicated. (B) Confirmation of DNA microarray results by northern blot analysis showing that PGPA and GSH1 were overexpressed in the same mutant. Presumably, for genes grossly overexpressed, microarray results can only be qualitative as the signals are rapidly saturated. An α-tubulin hybridisation was performed to monitor RNA and DNA loading. For each figure we show one representative of several control gels used for the various genes. For quantification see Table 2. (C) Southern blot analysis to test whether increased RNA expression is mediated, at least in part, by gene amplification. The DNA of the parasites was digested with HindIII. 1, L.tarentolae wild-type cell; 2, TarII As50.1. Sizes were determined using the 1 kb Plus DNA ladder and the 0.24–9.5 kb RNA ladder from Invitrogen.
Table 2. Correlation between microarray results, RNA expression and DNA copy number.
| Average fold increase compared to wild-type cells | ||||
|---|---|---|---|---|
| Strains | Genes | Microarrays | Northern | Southern |
| TarAs50.1 | GSH1 | 3.0 ± 0.2*a | 10.9 | 12.9 |
| PGPA | 2.0 ± 0.3**b | 15 | 3.2 | |
| TarIISbIII400.1 | GSH1 | 11.5 ± 0.9* | 44.7 | 3.9 (1.9 ± 0.1*)c |
| PGPA | 2.2 ± 0.3** | 2.9 | 12.7 (6.2 ± 0.5*)c | |
| GSH2 | 2.8 ± 0.3* | 3.6 | 0.8 (1.0)c | |
| SAHH | 0.7 ± 0.1** | 1.1 | 2.2(2.8 ± 0.1*)c | |
| LV39 MTX60.2 | PTR1 | 25.3 ± 4.0* | 20 | 17.2 |
| MAT2 | 4.1 ± 0.2* | 4 | 1.1 | |
| LV39 MTX60.4 | FT | 0.6* | 0.12d | 0e |
| DHFR | 2.8 ± 0.1* | 11.5 | 7.1 | |
a*P value <0.0001.
b**P value <0.02.
cValues within parenthesis correspond to gene amplification as determined using DNA microarrays.
dThis signal is probably coming from one of the several other FT members.
eSome of the copy number of the FT family remains unchanged but other genes, indicated by asterisks, were deleted.
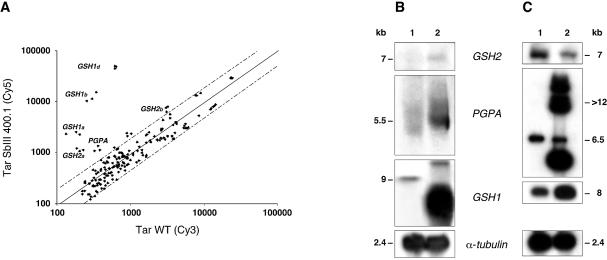
The antimonite resistant mutant L.tarentolae TarIISbIII400.1 has been described previously. It is already known that GSH1 and PGPA are overexpressed (8) in this strain, but revertants of these cells having lost the GSH1 containing amplicon nonetheless had higher TSH levels than wild-type cells (8), suggesting that another gene of the TSH biosynthetic pathway might be increased. Analysis of the competitive hybridisation between labelled wild-type and TarIISbIII400.1 cDNAs on our custom DNA microarrays indeed confirmed that PGPA and GSH1 (P < 0.0079 and P < 0.0001) were overexpressed, but we also found that the glutathione synthase (GSH2) gene, the second step in glutathione biosynthesis (Fig. 1), was also overexpressed (P < 0.0001) (Fig. 3). The Leishmania GSH2 gene has not yet been described but was revealed by the ongoing Leishmania genome project (accession number AL356246). The gene product shows 35 and 38% identity with the GSH synthase proteins of yeast and human (accession numbers Y138804 and BC007927), respectively. These array results were confirmed by northern blot analysis (Fig. 3B). In this mutant a better correlation was observed between microarrays and northern blots to estimate the fold increase in gene expression, although saturation was evident for the highly overexpressed genes (Table 2).
Figure 3.
Gene expression analysis of the L.tarentolae TarIISbIII400.1 mutant as determined by DNA microarrays and northern blot analysis. (A) Scatter plot of hybridisation intensities between Tar SbIII400.1 (Cy5) and wild-type L.tarentolae cells (Cy3). Dashed lines indicate 2-fold differences. (B) Confirmation of DNA microarray results by northern blot analysis showing that PGPA, GSH1 and GSH2 are overexpressed. An α-tubulin hybridisation was performed to monitor RNA and DNA loading. For each figure we show one representative of several control gels used for the various genes. For quantification see Table 2. (C) Southern blot analysis to test whether increased RNA expression is mediated by gene amplification. The DNA of the parasites was digested with HindIII (blots hybridised to GSH2 and tubulin probes); EcoRI (blot hybridised to a GSH1 probe); and BamHI (blot hybridised to a PGPA probe). 1, Leishmania tarentolae wild-type cell; 2, TarIISbIII400.1. Sizes were determined using the 1 kb Plus DNA ladder and the 0.24–9.5 kb RNA ladder from Invitrogen.
Analysis of L.major mutants selected for MTX resistance by DNA microarrays
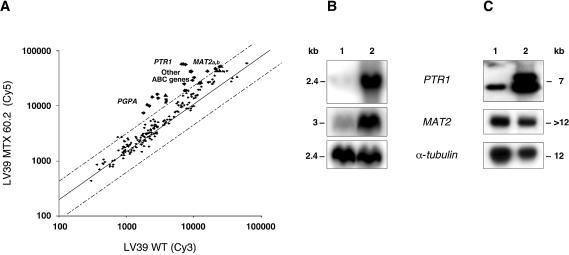
Our success in analysing metal resistance using DNA microarrays has prompted us to investigate MTX resistant mutants of L.major that had not been systematically characterised previously. The first mutant analysed was L.major MTX60.2 and DNA microarrays have revealed that PTR1 (P < 0.0001) as well as PGPA (P < 0.0001) (Fig. 4A) were overexpressed. PTR1 is a known MTX resistance gene (39,40) and its overexpression was confirmed by northern blots (Fig. 4B). In addition to PTR1, the microarrays suggested that the S-adenosylmethionine synthase gene (MAT2), recently described in L.infantum (41) and involved in a critical step in methionine metabolism (Fig. 1), was also overexpressed (P < 0.0001). This microarray result was validated by northern blot analysis (Fig. 4B). As for mutant TarIISbIII400.1 there was a good correlation between the fold expression determined by DNA microarrays and northern blots (Table 2).
Figure 4.
Gene expression analysis of the L.major LV39 MTX60.2 mutant as determined by DNA microarrays and northern blot analysis. (A) Scatter plot of hybridisation intensities between LV39 MTX60.2 (Cy5) and wild-type L.major LV39 cells (Cy3). Dashed lines indicate 2-fold differences. A number of fragments spanning ABC transporter genes cross-hybridised to the overexpressed PGPA. (B) Confirmation of DNA microarray results by northern blot analysis showing overexpression of PTR1 and MAT2. An α-tubulin hybridisation was performed to monitor RNA and DNA loading. For quantification see Table 2. (C) Southern blot analysis to test whether increased RNA expression is mediated by gene amplification. The DNA of the parasites was digested with HindIII except for the blot hybridised to the PTR1 probe where the DNA was digested with SacI. 1, Leishmania major wild-type cell; 2, L.major LV39 MTX60.2. Sizes were determined using the 1 kb Plus DNA ladder and the 0.24–9.5 kb RNA ladder from Invitrogen.
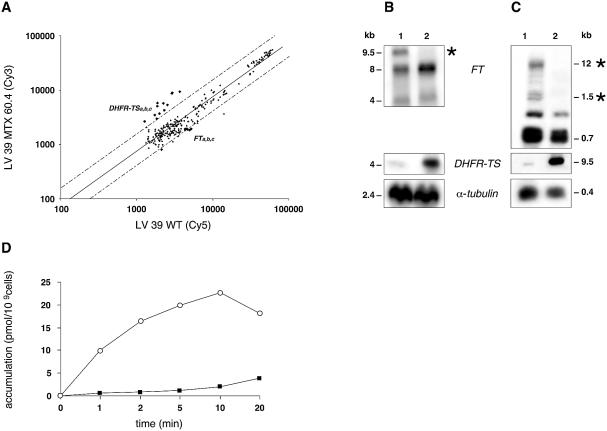
The analysis of the mutant L.major MTX60.4 using DNA microarrays has revealed a different set of genes whose expression was altered compared with the L.major MTX60.2 mutant. Indeed, DHFR-TS was overexpressed (P < 0.0001) while the expression of one of the FT genes was down regulated (P < 0.0001) (Fig. 5A). These expression results were confirmed by northern hybridisations where DHFR-TS RNA was increased (Fig. 5B). Several messengers hybridised to an FT probe, but the expression of at least one message was significantly down-regulated (Fig. 5B, marked with an asterisk). The down-regulation of FT expression is translated into a marked decrease of folate/MTX transport in mutant cells (Fig. 5D). There was a lack of correlation in the estimation of expression levels of DHFR-TS between microarrays and northern blots (Table 2).
Figure 5.
Gene expression analysis of the L.major LV39 MTX60.4 mutant as determined by DNA microarrays and northern blot analysis. (A) Scatter plot of hybridisation intensities between LV39 MTX 60.4 (Cy3) and wild-type L.major LV39 cells (Cy5). Dashed lines indicate 2-fold limits. (B) Confirmation of DNA microarray results by northern blot analysis showing overexpression of DHFR-TS, while the expression of one member of the FT family labelled with an asterisk is repressed. An α-tubulin hybridisation was performed for the determination of equal RNA and DNA loading. For quantification see Table 2. (C) Southern blot analysis to test whether variation in RNA expression is mediated by variation in gene’s copy number. The DNA of the parasites was digested with SalI for the FT and α-tubulin blots and with SacI for the DHFR-TS blot. 1, Leishmania major wild-type cell; 2, L.major LV39 MTX60.4. Sizes were determined using the 1 kb Plus DNA ladder and the 0.24–9.5 kb RNA ladder from Invitrogen. (D) Transport experiment showing a decrease in MTX accumulation in the mutant LV39 MTX60.4 cells (filled squares) compared with wild-type L.major LV39 cells (open circles).
Gene overexpression is mediated by gene amplification and other mechanisms
Gene overexpression in Leishmania is often due to gene amplification (33,34). We have tested whether DNA microarrays could be useful to detect gene amplification events by carrying out competitive hybridisations with digested total DNAs derived from sensitive cells and resistant mutants labelled with Cy3 and Cy5, respectively. Results are presented for the mutant TarIISbIII400.1 in which three genes, PGPA, GSH1 and GSH2 were found to be overexpressed (Fig. 3). While PGPA was amplified, and the copy number of GSH1 was also slightly increased, GSH2 was not amplified (Fig. 6). These array results were confirmed by Southern blot analysis (Fig. 3C) (Table 2). While GSH1 is amplified, the increase in copy number is not commensurate with the increase in RNA overexpression, suggesting that another mechanism than gene amplification also contributes to GSH1 overexpression. Moreover, since GSH2 is overexpressed without gene amplification (Fig. 3), we tested whether all the other genes found to be overexpressed in the other resistant mutant studied were amplified or not using Southern blots. In mutant TarIIAs50.1, overexpression of both GSH1 and PGPA is correlated to amplification of their respective genes (Fig. 2C). In the MTX resistant mutant L.major MTX60.2, the overexpression of PTR1 is due to gene amplification (Fig. 4C). PGPA is closely linked to PTR1 on the same locus (42,43) and thus the overexpression of PGPA observed by microarray (Fig. 4) is likely due to the co-amplification of its gene. In contrast to PTR1, the MAT2 gene in L.major MTX60.2 is overexpressed (Fig. 4B) but its gene is not amplified (Fig. 4C). In mutant L.major MTX60.4, the overexpression of DHFR-TS is due to the amplification of its gene (Fig. 5C). In the same mutant, one of the FT was down-regulated. We probed the DNA with a FT probe recognising several gene members (15) and observed a deletion event in the FT locus (Fig. 5C, marked with an asterisk). In general, a higher copy number of a gene will lead to more RNA but the correlation is only qualitative (Table 2). Copy number and RNA expression correlated best for PTR1 in LV39MTX60.2 and for GSH1 in TarIIAs50.1 (Table 2).
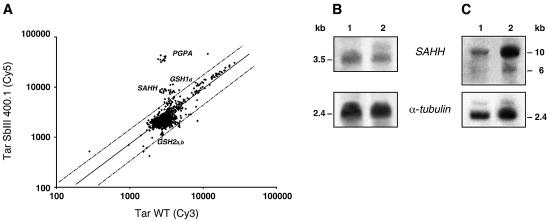
Figure 6.
Gene amplification events in L.tarentolae TarIISbIII400.1 as determined by DNA microarrays. (A) Scatter plot of hybridisation intensities between TarIISbIII400.1 and wild-type digested total DNA and labelled, respectively, with Cy5 and Cy3. This corresponds to a second generation of array with 50 genes each spotted 12 times. Dots outside the dotted lines representing 2-fold differences suggest changes in copy number in the mutant. (B) Northern blot analysis of the SAHH gene. 1, Leishmania tarentolae wild-type cell; 2, TarIISbIII400.1. Sizes were determined using the 1 kb Plus DNA ladder and the 0.24–9.5 kb RNA ladder from Invitrogen. (C) The amplification of the SAHH gene in this mutant was confirmed by Southern blot of DNA digested with HindIII, while the other amplification events are shown in Figure 3.
In our analysis of DNA amplification events using microarrays, we found in the mutant TarIISbIII400.1 (Fig. 6A), a new gene corresponding to S-adenosyl homocysteine hydrolase (SAHH) (Fig. 1), a putative drug target in Leishmania (44). This array result was validated by Southern blot analysis (Fig. 6C). This result was in contrast to the microarray results, however, which did not show that the SAHH gene was overexpressed. A northern blot using a SAHH probe revealed that in this mutant there were two RNA species, as opposed to one in the wild-type cell (Fig. 6B). The fold increase in expression was low, however, and explains why this was missed by the microarray analysis. Further detailed analysis has confirmed that there is no statistical difference in the expression array data for this gene between wild-type and TarIISbIII400.1 (Table 2).
DISCUSSION
DNA microarrays are powerful tools now being employed in the field of parasitology. They have already showed their usefulness in malaria research (45–48) and also for looking at gene expression in Trypanosoma brucei (49). Both cDNAs and random 1 kb genomic DNAs of L.major spotted on microarrays appear to have provided useful results to study differential gene expression between L.major promastigotes and metacyclics (30–32). We took advantage of the ongoing Leishmania genome-sequencing project and analysed four diverse resistant mutants. These studies have validated the use of microarrays in studying drug resistance in Leishmania and have pinpointed new genes overexpressed either by gene amplification or by other mechanisms.
A number of studies in bacterial or mammalian cells have dealt with changes of expression in the presence of drugs (24–28) but few have dealt with the analysis of resistance per se. One example is the use of microarrays for studying the genesis of resistance to fluconazole in Candida albicans (29). While it was known that TSH levels were increased in TarIIAs50.1 (7), the molecular mutation responsible for this increase was unknown. In other mutants, we have shown that overexpression of GSH1 and/or of the ornithine decarboxylase gene ODC were associated with increased TSH biosynthesis (11). This is due to GSH1 amplification in TarIIAs50.1. The work presented here has also pinpointed a novel gene corresponding to GSH2, whose overexpression is possibly linked to increased TSH in metal resistant mutants. Although GSH1 has been shown to be overexpressed in a number of circumstances in several organisms including Leishmania (10), this appears to be the first report of GSH2 overexpression in any drug resistant cells. While GS is not rate limiting under standard conditions, this situation could change following the amplification of GSH1 in Leishmania, as observed in TarIISbIII400.1. In rats, treatment known to increase the expression of GSH1 also increase the expression of GSH2 (50). While overexpression of GSH2 in yeast does not appear to increase GSH levels (51), induction of GS increased glutathione levels in a human liver cell line (50).
The custom DNA microarrays have also been used for the analysis of MTX resistant mutants where several genes known to be implicated in resistance, such as PTR1, DHFR-TS and FT were pinpointed. The analysis of LV39 MTX60.2 has highlighted the overexpression of MAT2. Overexpression of this gene is novel and has not been described in any other drug resistant organism. Ongoing proteomic studies have shown that overexpression of the MAT2 protein is a frequent event in MTX resistant mutants or in cells put in contact with MTX, but transfection of MAT2 alone does not seem to directly confer MTX resistance (J.Drummelsmith and M.Ouellette, unpublished). Since the MAT2 gene is not amplified, its overexpression cannot be explained by co-amplification of another nearby resistance gene. Genes in Leishmania are transcribed as large polycistronic RNAs and only one region on the entire chromosome 1 appears to correspond to a pol II-like promoter (52). Thus, we think it is unlikely that the overexpression of MAT2 is due to a mutation in a promoter-like element of an upstream gene. Mutations in either the 5′ or 3′ untranslated regions of MAT2 or mutations in a trans acting factor may explain the increase in RNA level of this gene or of the other genes revealed here that are overexpressed without an increase in copy number. Because MAT2 is often overexpressed following MTX selection, we believe that it is linked to MTX selection. Since transfection excludes a direct role (unpublished observation), we must assume a more indirect one. AdoMet is not only an intermediate metabolite in methionine catabolism but also a key intracellular molecule involved in several processes (53). One of these may be perturbed upon alteration of folate metabolism. Since folate and methionine pathways are linked (Fig. 1), a modulation in folate metabolism may indeed have consequences on the expression of genes involved in downstream pathways.
Upon drug selection, Leishmania often uses DNA amplification to overexpress its genes (33,34). In the mutants studied, we found that the GSH1, PGPA, PTR1 and DHFR-TS genes were amplified (Figs 2–5) and that some members of the FT family were deleted. The ability of DNA microarrays to detect gene amplification events is less exploited than for expression profiling, but it was used, for example, in the study of human breast tumours (54). We have demonstrated the suitability of DNA microarrays to detect gene amplification events in Leishmania (Fig. 6 and results not shown). While simple techniques such as comparison of digested DNA on ethidium bromide stained gels can lead to the detection of gene amplification events (19,55), this does not identify the number of unique amplicons nor their identity. These arrays have allowed the detection of the amplification of the SAHH gene in a SbIII resistant mutant (Fig. 6). The amplification of this gene does not seem to lead to an increase in RNA levels, as this was not detected using microarray. Northern blot analysis (Fig. 6) indicated that while the main message was at an estimated size of 3.5 kb, another band at 3.8 kb seems to be unique to the mutant. It is possible that this second SAHH RNA species is more easily transported or translated and thus could lead to increase gene product expression. It is intriguing that the expression of another gene of the methionine pathway is changed in drug resistant mutants, although in this case a more simple explanation can be put forward. An increase in SAHH will lead to increase homocysteine, the precursor of de novo cysteine biosynthesis, and cysteine is the main building block of TSH which is increased in mutants.
The overexpression of most genes detected in our custom arrays were linked to gene amplification events. Nonetheless, two genes GSH2 and MAT2 were overexpressed without gene amplification (Figs 3 and 4) and often, for other genes, the fold increase in expression is not commensurate with the level of gene amplification (Table 2). Increased expression without amplification was found previously for ODC overexpression in arsenite resistant mutants (11) and for overexpression of BT1 or PTR1 in cells grown in biopterin poor medium for prolonged periods (56). As there seems to be no control at the level of transcription initiation in Leishmania, it is possible that an increased in RNA stability is the reason for the augmented levels of RNA, as uncovered using microarrays and confirmed by northern blot analysis. This could either be due to changes in untranslated regions or to mutations in trans acting factors.
Overall, we have shown the utility of DNA microarrays to study drug resistance in Leishmania. Indeed, we could find several genes in the various mutants that were overexpressed that correspond to known resistance gene, hence validating the technique. Our small targeted arrays have not only allowed the efficient analysis of several resistant mutants but have also permitted the discovery of three new genes GSH2, SAHH and MAT2, whose expression was modulated in drug resistant mutants. Once the whole genome of Leishmania become available and is put on arrays, many more genes could be found overexpressed (or repressed) and/or amplified (or deleted) in the same mutants. These arrays will be a valuable tool to study the mechanisms of resistance in field isolates.
REFERENCES
- 1.Herwaldt B.L. (1999) Leishmaniasis. Lancet, 354, 1191–1199. [DOI] [PubMed] [Google Scholar]
- 2.Guerin P.J., Olliaro,P., Sundar,S., Boelaert,M., Croft,S.L., Desjeux,P., Wasunna,M.K. and Bryceson,A.D. (2002) Visceral leishmaniasis: current status of control, diagnosis and treatment and a proposed research and development agenda. Lancet Infect. Dis., 2, 494–501. [DOI] [PubMed] [Google Scholar]
- 3.Sundar S., More,D.K., Singh,M.K., Singh,V.P., Sharma,S., Makharia,A., Kumar,P.C. and Murray,H.W. (2000) Failure of pentavalent antimony in visceral leishmaniasis in India: report from the Center of the Indian Epidemic. Clin. Infect. Dis., 31, 1104–1107. [DOI] [PubMed] [Google Scholar]
- 4.Ouellette M. and Ward,S. (2002) Drug resistance in parasites. In Marr,J., Nielsen,T. and Komuniecki,R. (eds), Molecular Medical Parasitology. Academic Press, San Diego, pp. 395–430. [Google Scholar]
- 5.Shaked-Mishan P., Ulrich,N., Ephros,M. and Zilberstein,D. (2001) Novel intracellular SbV reducing activity correlates with antimony susceptibility in Leishmania donovani. J. Biol. Chem., 276, 3971–3976. [DOI] [PubMed] [Google Scholar]
- 6.Brochu C., Wang,J., Roy,G., Messier,N., Wang,X.Y., Saravia,N.G. and Ouellette,M. (2003) Antimony uptake systems in the protozoan parasite Leishmania and accumulation alteration in antimony resistant parasites. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukhopadhyay R., Dey,S., Xu,N., Gage,D., Lightbody,J., Ouellette,M. and Rosen,B.P. (1996) Trypanothione overproduction and resistance to antimonials and arsenicals in Leishmania. Proc. Natl Acad. Sci. USA, 93, 10383–10387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haimeur A., Brochu,C., Genest,P., Papadopoulou,B. and Ouellette,M. (2000) Amplification of the ABC transporter gene PGPA and increased trypanothione levels in potassium antimonyl tartrate (SbIII) resistant Leishmania tarentolae. Mol. Biochem. Parasitol., 108, 131–135. [DOI] [PubMed] [Google Scholar]
- 9.Fairlamb A.H. and Cerami,A. (1992) Metabolism and functions of trypanothione in the Kinetoplastida. Annu. Rev. Microbiol., 46, 695–729. [DOI] [PubMed] [Google Scholar]
- 10.Grondin K., Haimeur,A., Mukhopadhyay,R., Rosen,B.P. and Ouellette,M. (1997) Co-amplification of the gamma-glutamylcysteine synthetase gene gsh1 and of the ABC transporter gene PGPA in arsenite-resistant Leishmania tarentolae. EMBO J., 16, 3057–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haimeur A., Guimond,C., Pilote,S., Mukhopadhyay,R., Rosen,B.P., Poulin,R. and Ouellette,M. (1999) Elevated levels of polyamines and trypanothione resulting from overexpression of the ornithine decarboxylase gene in arsenite-resistant Leishmania. Mol. Microbiol., 34, 726–735. [DOI] [PubMed] [Google Scholar]
- 12.Légaré D., Richard,D., Mukhopadhyay,R., Stierhof,Y.D., Rosen,B.P., Haimeur,A., Papadopoulou,B. and Ouellette,M. (2001) The Leishmania ABC protein PGPA is an intracellular metal-thiol transporter ATPase. J. Biol. Chem., 276, 26301–26307. [DOI] [PubMed] [Google Scholar]
- 13.Haimeur A. and Ouellette,M. (1998) Gene amplification in Leishmania tarentolae selected for resistance to sodium stibogluconate. Antimicrob.Agents Chemother., 42, 1689–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh N., Singh,R.T. and Sundar,S. (2002) Identification of a gene linked to drug resistance in field isolates of Leishmania donovani. Ann. Trop. Med. Parasitol., 96, 839–841. [DOI] [PubMed] [Google Scholar]
- 15.Richard D., Kundig,C. and Ouellette,M. (2002) A new type of high affinity folic acid transporter in the protozoan parasite Leishmania and deletion of its gene in methotrexate-resistant cells. J. Biol. Chem., 277, 29460–29467. [DOI] [PubMed] [Google Scholar]
- 16.Cunningham M.L. and Beverley,S.M. (2001) Pteridine salvage throughout the Leishmania infectious cycle: implications for antifolate chemotherapy. Mol. Biochem. Parasitol., 113, 199–213. [DOI] [PubMed] [Google Scholar]
- 17.Kündig C., Haimeur,A., Légaré,D., Papadopoulou,B. and Ouellette,M. (1999) Increased transport of pteridines compensates for mutations in the high affinity folate transporter and contributes to methotrexate resistance in the protozoan parasite Leishmania tarentolae. EMBO J., 18, 2342–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemley C., Yan,S., Dole,V.S., Madhubala,R., Cunningham,M.L., Beverley,S.M., Myler,P.J. and Stuart,K.D. (1999) The Leishmania donovani LD1 locus gene ORFG encodes a biopterin transporter (BT1). Mol. Biochem. Parasitol., 104, 93–105. [DOI] [PubMed] [Google Scholar]
- 19.Coderre J.A., Beverley,S.M., Schimke,R.T. and Santi,D.V. (1983) Overproduction of a bifunctional thymidylate synthetase-dihydrofolate reductase and DNA amplification in methotrexate-resistant Leishmania tropica. Proc. Natl Acad. Sci. USA, 80, 2132–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nare B., Hardy,L.W. and Beverley,S.M. (1997) The roles of pteridine reductase 1 and dihydrofolate reductase-thymidylate synthase in pteridine metabolism in the protozoan parasite Leishmania major. J. Biol. Chem., 272, 13883–13891. [DOI] [PubMed] [Google Scholar]
- 21.Wang J., Leblanc,E., Chang,C.F., Papadopoulou,B., Bray,T., Whiteley,J.M., Lin,S.X. and Ouellette,M. (1997) Pterin and folate reduction by the Leishmania tarentolae H locus short-chain dehydrogenase/reductase PTR1. Arch. Biochem. Biophys., 342, 197–202. [DOI] [PubMed] [Google Scholar]
- 22.El Fadili A., Richard,D., Kundig,C. and Ouellette,M. (2003) Effect of polyglutamylation of methotrexate on its accumulation and the development of resistance in the protozoan parasite Leishmania. Biochem. Pharmacol., 66, 999–1008. [DOI] [PubMed] [Google Scholar]
- 23.Cheok M.H., Yang,W., Pui,C.H., Downing,J.R., Cheng,C., Naeve,C.W., Relling,M.V. and Evans,W.E. (2003) Treatment-specific changes in gene expression discriminate in vivo drug response in human leukemia cells. Nature Genet., 34, 85–90. [DOI] [PubMed] [Google Scholar]
- 24.Wilson M., DeRisi,J., Kristensen,H.H., Imboden,P., Rane,S., Brown,P.O. and Schoolnik,G.K. (1999) Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl Acad. Sci. USA, 96, 12833–12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., Zhang,Y., Zhou,Y., An,S. and Cheng,J. (2002) Response of gene expression in Saccharomyces cerevisiae to amphotericin B and nystatin measured by microarrays. J. Antimicrob. Chemother., 49, 905–915. [DOI] [PubMed] [Google Scholar]
- 26.Staunton J.E., Slonim,D.K., Coller,H.A., Tamayo,P., Angelo,M.J., Park,J., Scherf,U., Lee,J.K., Reinhold,W.O., Weinstein,J.N. et al. (2001) Chemosensitivity prediction by transcriptional profiling. Proc. Natl Acad. Sci. USA, 98, 10787–10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng W.L., Kazmierczak,K.M., Robertson,G.T., Gilmour,R. and Winkler,M.E. (2003) Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol., 185, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gmuender H., Kuratli,K., Di Padova,K., Gray,C.P., Keck,W. and Evers,S. (2001) Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res., 11, 28–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cowen L.E., Nantel,A., Whiteway,M.S., Thomas,D.Y., Tessier,D.C., Kohn,L.M. and Anderson,J.B. (2002) Population genomics of drug resistance in Candida albicans. Proc. Natl Acad. Sci. USA, 99, 9284–9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almeida R., Norrish,A., Levick,M., Vetrie,D., Freeman,T., Vilo,J., Ivens,A., Lange,U., Stober,C., McCann,S. et al. (2002) From genomes to vaccines: Leishmania as a model. Philos. Trans. R. Soc. Lond. B. Biol. Sci., 357, 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beverley S.M., Akopyants,N.S., Goyard,S., Matlib,R.S., Gordon,J.L., Brownstein,B.H., Stormo,G.D., Bukanova,E.N., Hott,C.T., Li,F. et al. (2002) Putting the Leishmania genome to work: functional genomics by transposon trapping and expression profiling. Philos. Trans. R. Soc. Lond. B. Biol. Sci., 357, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saxena A., Worthey,E.A., Yan,S., Leland,A., Stuart,K.D. and Myler,P.J. (2003) Evaluation of differential gene expression in Leishmania major Friedlin procyclics and metacyclics using DNA microarray analysis. Mol. Biochem. Parasitol., 129, 103–114. [DOI] [PubMed] [Google Scholar]
- 33.Beverley S.M. (1991) Gene amplification in Leishmania. Annu. Rev. Microbiol., 45, 417–444. [DOI] [PubMed] [Google Scholar]
- 34.Borst P. and Ouellette,M. (1995) New mechanisms of drug resistance in parasitic protozoa. Annu. Rev. Microbiol., 49, 427–460. [DOI] [PubMed] [Google Scholar]
- 35.Ouellette M., Hettema,E., Wust,D., Fase-Fowler,F. and Borst,P. (1991) Direct and inverted DNA repeats associated with P-glycoprotein gene amplification in drug resistant Leishmania. EMBO J., 10, 1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drummelsmith J., Brochu,V., Girard,I., Messier,N. and Ouellette,M. (2003) Proteome mapping of the protozoan parasite Leishmania and application to the study of drug targets and resistance mechanisms. Mol. Cell. Proteomics, 2, 146–155. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 38.Légaré D., Cayer,S., Singh,A.K., Richard,D., Papadopoulou,B. and Ouellette,M. (2001) ABC proteins of Leishmania. J. Bioenerg. Biomembr., 33, 469–474. [DOI] [PubMed] [Google Scholar]
- 39.Papadopoulou B., Roy,G. and Ouellette,M. (1992) A novel antifolate resistance gene on the amplified H circle of Leishmania. EMBO J., 11, 3601–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Callahan H.L. and Beverley,S.M. (1992) A member of the aldoketo reductase family confers methotrexate resistance in Leishmania. J. Biol. Chem., 267, 24165–24168. [PubMed] [Google Scholar]
- 41.Reguera R.M., Balana-Fouce,R., Perez-Pertejo,Y., Fernandez,F.J., Garcia-Estrada,C., Cubria,J.C., Ordonez,C. and Ordonez,D. (2002) Cloning expression and characterization of methionine adenosyltransferase in Leishmania infantum promastigotes. J. Biol. Chem., 277, 3158–3167. [DOI] [PubMed] [Google Scholar]
- 42.Ouellette M. and Papadopoulou,B. (1993) Mechanisms of drug resistance in Leishmania. Parasitol. Today, 9, 150–153. [DOI] [PubMed] [Google Scholar]
- 43.Nare B., Luba,J., Hardy,L.W. and Beverley,S. (1997) New approaches to Leishmania chemotherapy: pteridine reductase 1 (PTR1) as a target and modulator of antifolate sensitivity. Parasitology, 114 (Suppl), S101–110. [PubMed] [Google Scholar]
- 44.Henderson D.M., Hanson,S., Allen,T., Wilson,K., Coulter-Karis,D.E., Greenberg,M.L., Hershfield,M.S. and Ullman,B. (1992) Cloning of the gene encoding Leishmania donovani S-adenosylhomocysteine hydrolase, a potential target for antiparasitic chemotherapy. Mol. Biochem. Parasitol., 53, 169–183. [DOI] [PubMed] [Google Scholar]
- 45.Rathod P.K., Ganesan,K., Hayward,R.E., Bozdech,Z. and DeRisi,J.L. (2002) DNA microarrays for malaria. Trends Parasitol., 18, 39–45. [DOI] [PubMed] [Google Scholar]
- 46.Ben Mamoun C., Gluzman,I.Y., Hott,C., MacMillan,S.K., Amarakone,A.S., Anderson,D.L., Carlton,J.M., Dame,J.B., Chakrabarti,D., Martin,R.K. et al. (2001) Co-ordinated programme of gene expression during asexual intraerythrocytic development of the human malaria parasite Plasmodium falciparum revealed by microarray analysis. Mol. Microbiol., 39, 26–36. [DOI] [PubMed] [Google Scholar]
- 47.Hayward R.E., Derisi,J.L., Alfadhli,S., Kaslow,D.C., Brown,P.O. and Rathod,P.K. (2000) Shotgun DNA microarrays and stage-specific gene expression in Plasmodium falciparum malaria. Mol. Microbiol., 35, 6–14. [DOI] [PubMed] [Google Scholar]
- 48.Bozdech Z., Zhu,J., Joachimiak,M.P., Cohen,F.E., Pulliam,B. and DeRisi,J.L. (2003) Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol., 4, R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diehl S., Diehl,F., El-Sayed,N., Clayton,C. and Hoheisel,J. (2002) Analysis of stage-specific gene expression in the bloodstream and the procyclic form of Trypanosoma brucei using a genomic DNA-microarray. Mol. Biochem. Parasitol., 123, 115. [DOI] [PubMed] [Google Scholar]
- 50.Huang Z.A., Yang,H., Chen,C., Zeng,Z. and Lu,S.C. (2000) Inducers of gamma-glutamylcysteine synthetase and their effects on glutathione synthetase expression. Biochim. Biophys. Acta, 1493, 48–55. [DOI] [PubMed] [Google Scholar]
- 51.Inoue Y., Sugiyama,K., Izawa,S. and Kimura,A. (1998) Molecular identification of glutathione synthetase (GSH2) gene from Saccharomyces cerevisiae. Biochim. Biophys. Acta, 1395, 315–320. [DOI] [PubMed] [Google Scholar]
- 52.Martinez-Calvillo S., Yan,S., Nguyen,D., Fox,M., Stuart,K. and Myler,P.J. (2003) Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell, 11, 1291–1299. [DOI] [PubMed] [Google Scholar]
- 53.Mato J.M., Corrales,F.J., Lu,S.C. and Avila,M.A. (2002) S-Adenosylmethionine: a control switch that regulates liver function. FASEB J., 16, 15–26. [DOI] [PubMed] [Google Scholar]
- 54.Pollack J.R., Perou,C.M., Alizadeh,A.A., Eisen,M.B., Pergamenschikov,A., Williams,C.F., Jeffrey,S.S., Botstein,D. and Brown,P.O. (1999) Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nature Genet., 23, 41–46. [DOI] [PubMed] [Google Scholar]
- 55.Ouellette M., Haimeur,A., Grondin,K., Legare,D. and Papadopoulou,B. (1998) Amplification of ABC transporter gene PGPA and of other heavy metal resistance genes in Leishmania tarentolae and their study by gene transfection and gene disruption. Methods Enzymol., 292, 182–193. [DOI] [PubMed] [Google Scholar]
- 56.Roy G., Kundig,C., Olivier,M., Papadopoulou,B. and Ouellette,M. (2001) Adaptation of Leishmania cells to in vitro culture results in a more efficient reduction and transport of biopterin. Exp. Parasitol., 97, 161–168. [DOI] [PubMed] [Google Scholar]