Abstract
A mouse mutation (p100H/p100H) has been identified that is associated with cardioskeletal myopathy, heart block, delayed growth and early postnatal death. The gene that is disrupted in this mutation encodes the transcription factor Sox6. P19CL6 cells were used as an in vitro cardiomyocyte differentiation system and revealed that Sox6 is expressed exclusively when the cells are committed to differentiate to beating cardiac myocytes. We used the yeast two-hybrid system to identify the Prtb (Proline-rich transcript of the brain) protein as a Sox6 interactor, and subsequently confirmed the interaction by co-immunoprecipitation. Prtb expression in P19CL6 cells increased with differentiation to beating cardiomyocytes. Using the P19CL6 cells stably transfected with noggin, an antagonist of BMP (Bone Morphogenic Protein), we found that BMP expression is required for Sox6 expression in cardiomyocyte differentiation. Surprisingly, the expression of the α1c-subunit gene of the L-type Ca2+ channel decreased in P19CL6 cells as they differentiated to beating cardiac cells. Ectopic expression of Sox6 or Prtb alone in P19CL6 cells caused down-regulation of L-type Ca2+ α1c expression, but when Sox6 and Prtb were co-transfected to the cells, L-type Ca2+ α1c remained at basal levels. A similar relationship of Sox6 and L-type Ca2+ α1c expression was seen in vivo (comparing wild-type and p100H/p100H mutant mice). Thus, Sox6 is within the BMP pathway in cardiac differentiation, interacts with Prtb and may play a critical role in the regulation of a cardiac L-type Ca2+ channel.
INTRODUCTION
In distinct muscle cell lineages, individual muscle-specific genes exhibit unique temporal-spatial patterns of expression. Regulatory programs for myocyte transcription result from developmental cues and positional information that are mediated by specific transcription factors. The transcriptional expression pattern of muscle-specific genes is ultimately dependent on combinatorial interactions among the transcription factors that bind different regulatory proteins (1).
The Sox (Sry related HMG box) gene family encodes an important group of transcription factors that are key regulators of embryonic development and cell fate determination (2). Mutations of Sox6, a member of this family, are associated with neonatal lethality in the mouse (3,4). Sox6 was initially isolated from an adult mouse testis cDNA library (5), but its functional significance in this tissue is unknown. Sox6 has been suggested to play a role in the development of the central nervous system (5), chondrogenesis (4,6–8) and cardiac and skeletal muscle cell maintenance (3). Mice homozygous for a Sox6 null mutation, p100H, show delayed growth and die within 2 weeks after birth (3). Analysis of the p100H phenotype revealed that the p100H mutant develops myopathy and arterioventricular (AV) heart block, a cardiac conduction defect associated with lethality in human cardiac myopathies (9). Thus, among its diverse functions, the Sox6 protein is likely to be involved in maintaining the normal physiological function of muscle tissue, including the heart (3). We have recently cloned and sequenced the human SOX6 cDNA, isolated from a myoblast cDNA library (10). The human SOX6 protein shows 94.3% amino acid identity to mouse Sox6 throughout the gene, and 100% identity in the critical HMG box and coiled-coil domains. Northern blot analysis revealed that human SOX6, like mouse Sox6, is expressed in a wide variety of tissues, and is most abundantly expressed in skeletal muscle (3,10).
The muscle cell-specific effects of the Sox6 protein may be achieved by its interactions with other transcription factors. To identify Sox6 interactors in muscle cells and elucidate how the Sox6 protein achieves such remarkable cell type specificity, a yeast two-hybrid screening was performed. This identified the Prtb (Proline-rich transcript of the brain) protein as an interactor of the Sox6 protein.
To understand the role of Sox6 in cardiac muscle development, we utilized P19CL6, a cell line that differentiates exclusively to beating cardiomyocytes in the presence of DMSO (11). Sox6 is not expressed in untreated P19CL6 cells, but is up-regulated following DMSO exposure, reaching maximum levels when the cells have fully differentiated into beating cardiomyocytes. In addition, we found that Sox6 is downstream of the BMP (Bone Morphogenetic Protein) pathway in the cardiac system, using P19CL6noggin, a P19CL6 cell line stably transfected with noggin that antagonizes the BMP pathway (12).
To assess the effect of Sox6 in cardiac differentiation, we characterized the expression of the L-type Ca2+ channel α1c gene, encoding a subunit normally expressed in the heart (13). We found that the mRNA levels of the L-type Ca2+ channel α1c gene decreased when the P19L6 cells terminally differentiate or when they are transfected with Sox6. Moreover, in Sox6 null mutant mice (p100H/p100H) (3), the expression level of this calcium channel gene is ∼1.5-fold higher, correlating with the in vitro analysis. The Sox6 interactor, Prtb, also down-regulated the expression levels of the L-type Ca2+ channel α1c gene, whereas Prtb co-expressed with Sox6 did not.
MATERIALS AND METHODS
Plasmid construction
DNA fragments for plasmid constructs were generated by PCR or RT–PCR and confirmed by sequence analysis. To construct the plasmid pGBKT7 (Clontech) as bait for the yeast two-hybrid screen and for the co-immunoprecipitation (Co-IP), an amplified PCR fragment encoding amino acids 139–304 (including the coiled-coil domain) of mouse Sox6 (GenBank accession no. U32614) was cloned in-frame into EcoRI–SalI sites. To generate the plasmid pGADT7 (Clontech) for the Co-IP, the amplified PCR fragment encoding the first 108 amino acids of the Prtb gene (GenBank accession no. AF085348), and RT–PCR of the entire gene (168 amino acids), were cloned in-frame into EcoRI–XhoI sites. For transient transfection, complete cDNAs of the mouse Sox6 and Prtb genes were cloned into EcoRI, and EcoRI–XhoI, respectively, to pCDNA3.1/Zeo vector (Invitrogen) driven by the CMV promoter.
Yeast two-hybrid screening
The MATCHMAKER two-hybrid system 3 (Clontech) was used according to the supplier’s protocol with the Sox6 bait plasmid, detailed above. The screened library was mouse 11 day embryo MATCHMAKER cDNA (Clontech) that was pre-transformed to the Y187 yeast strain. The two-hybrid screening of the pre-transformed library was accomplished by yeast mating using the AH109 strain containing the bait plasmid. The screening was preformed for growth on minimal medium lacking histidine (His–) and in the presence of 2.5 mM 3-amino-1,2,4-triazole. Of 2.3 × 106 cDNAs, approximately 300 positive colonies grew on His–. To exclude false positive clones, the 300 colonies were replicated to high stringency plates, with simultaneous selection for three reporter genes (lacking histidine and adenine and containing X-α-Gal as substrate that employs blue/white screening directly on the plate). Approximately 200 of these clones grew under high stringency. These were sequenced and evaluated by BLAST search for being in frame with the activation domain, for the potential as a transcription factor and for expression in the developing heart. Of these, one, partially encoding Prtb, was selected for further analysis.
In vitro protein–protein interaction assay
To confirm true protein interactions, we used the MATCHMAKER Co-IP kit (Clontech) for in vitro Co-IP, according to the supplier’s protocol. The Co-IP contains two vectors: pGBKT7 (includes the mouse Sox6 coiled-coil domain), and pGADT7 (includes a library protein isolated from the yeast two-hybrid screening). These vectors contain a T7 RNA polymerase promoter and either a c-Myc or HA epitope tag, so they can be used directly in an in vitro transcription/translation reaction.
RNA isolation and northern blot analysis
At varying times and treatments, cells were harvested for total RNA isolation using the UltraSpec RNA Isolation kit (Biotecx). For northern blot hybridization, equal amounts of total RNA (10 µg/lane) were subjected to electrophoresis on a 1.2% agarose gel in the presence of 5.5% formaldehyde and transferred by the alkaline method to a nylon membrane (Hybond N+; Amersham). The membranes were hybridized with distinct cDNA probes obtained by PCR or RT–PCR and confirmed by sequencing. The Sox6 probe is a mouse cDNA fragment of 575 bp including nucleotides 1353–1927 (GenBank accession no. U32614). The Prtb probe is a mouse cDNA fragment of 562 bp including nucleotides 15–576 (GenBank accession no. AF085348). The Sox9 probe is a mouse cDNA fragment of 278 bp including nucleotides 1049–1326 (GenBank accession no. BC034264). The L-type Ca2+α1c probes are mouse cDNA fragments of 548 and 540 bp including nucleotides 6294–6841 and 3072–3611, respectively (GenBank accession no. NM_009781). Probes were labeled with [32P]dCTP, by random primer labeling (RediprimeII; Amersham Pharmacia Biotech). The hybridization was performed in phosphate buffered 7% SDS hybridization solution (14). Blots were washed with 0.2× SSC, 1% SDS at 60°C prior to exposure to X-ray film (Kodak) at –80°C from 3 h to 6 days. Human and mouse multiple tissue northern filters were purchased from Clontech. The filters were hybridized with mouse Prtb cDNA nucleotides 15–576 (GenBank accession no. AF085348) following the manufacturer’s protocols and exposed to X-ray film (Kodak) at –80°C for 19 h.
Cell culture and differentiation
P19CL6 and P19CL6noggin cell lines were cultured as described previously (11). To stimulate differentiation under adherent conditions, the P19CL6 and P19CL6noggin cells were plated at a density of 3.7 × 105 cells in a 60 mm tissue culture dish with medium including 1% DMSO. The medium was changed every 2 days. The P19CL6 cells started to beat after 11 days, and virtually all of the cells on the plate were beating after 14 days. Days of differentiation were numbered consecutively (i.e. day 0, no DMSO treatment; day 1, 24 h after DMSO added).
Transient transfection assay
Transient transfections were performed in a 6-well dish using FuGENE6 reagent (Roche), according to the manufacturer’s protocol. Bluescript II SK(-) plasmid was used as a control. To evaluate transfection efficiency, cells were co-transfected with a reporter vector containing the β-gal gene driven by the CMV promoter (pCMV SPORT–βgal; Invitrogen). In each well of a 6-well plate, 3.5 × 105 cells were transfected with 1 µg of pCDNA3.1/Zeo harboring Sox6 or Prtb or both plasmids, and 0.5 µg of pCMV SPORT–βgal. Cells were harvested after 24 h post-transfection for RNA for northern blot analysis (as described above). The band intensities were determined with Quantity One software (GS-700 densitometer; Bio-Rad, Hercules, CA). Also at this point protein extracts were used for the β-gal assay using the β galactosidase enzyme assay kit (Promega). The experiment was repeated three times, and the statistical analysis was done using t-test.
RT–PCR analysis of L-type Ca2+ α1c gene
Total RNA from embryonic day 18.5 heart was isolated from littermate wild-type and p100H/p100H mutant mice. One microgram of RNA, Oligo (dT)12–18 primer and reverse transcriptase were used for cDNA production. L-type Ca+2 α1c cDNA analysis was performed by PCR amplification of a fragment of 548 bp using the primers: MHB1401, 5′-TATCAGAGTGACAGCAGGGGCAAC-3′; and MHB1402, 5′-AGAGAGGCAGAGCGAAGGAAAC-3′. PCRs were carried out with several dilutions of cDNAs and/or over a range of amplifying cycles to insure a linear range of amplification. A fragment of the mouse GPDH gene was amplified as internal control. Amplified products were electrophoresed on a 2% agarose gel, visualized under UV light and analyzed using a densitometer (Gel Doc 1000; Bio-Rad).
RESULTS
Screening for a candidate protein that interacts with Sox6
To identify factors that interact with Sox6, we employed the yeast two-hybrid system. As bait, we used the coiled-coil domain including amino acids 139–304 of the mouse Sox6 protein. The leucine zipper motif and the Q-box create a coiled-coil domain (6) that is 100% conserved between the mouse and human SOX6, and has a 91% identity to L-Sox5 (10). This domain mediates homodimerization and heterodimerization. We screened a cDNA library from an 11-day mouse embryo, at a stage when the Sox6 gene is already expressed (3). One cDNA clone was selected for detailed analysis based on numerous criteria (see Materials and Methods). A BLAST search revealed that this clone contained two-thirds of the Prtb gene, including 108 amino acids (of 168 total) at the N-terminal (15). The full size cDNA of Prtb was subsequently isolated by RT–PCR. Although the two-hybrid system in yeast provides an in vivo analysis, it does not necessarily demonstrate direct interaction between these two proteins. To verify protein–protein interaction, we applied the Co-IP technique (Fig. 1). A c-Myc epitope tag was attached to the Sox6 bait, and an HA epitope tag was attached to both the isolated Prtb fragment (108 amino acids) and the full-length protein. The tagged proteins were expressed in vitro and non-denatured proteins and complexes were precipitated by anti-Myc or anti-HA. The proteins and complexes were then denatured and resolved by PAGE, verifying the interaction between the Sox6 and Prtb proteins. Both the 108 amino acid Prtb peptide and the full-length protein were co-precipitated with Sox6, confirming the yeast two-hybrid system results (data not shown).
Figure 1.
Protein–protein interaction assay. To verify interactions, we applied the Co-IP technique. The Sox6 bait was tagged with the a c-Myc epitope and each of the two Prtb proteins were tagged with an HA epitope. The first three lanes assay each protein by itself precipitated with an antibody to the corresponding tag. Lanes 4 and 5 assay a mix of two proteins, the Sox6 bait and one of two Prtb proteins (168 or 108 aa, lanes 4 and 5, respectively) allowed to associate prior to precipitation with the HA antibody. The antibody pulled down HA-tagged Prtb proteins and Sox6. Standard marker fragments in kDa are indicated at the left.
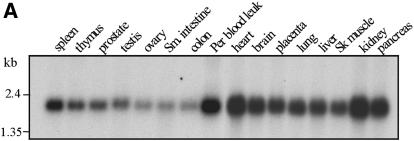
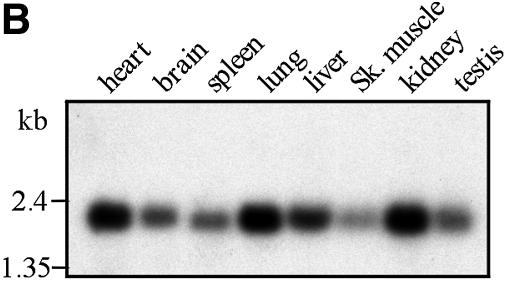
The Prtb protein has no known function. An analysis using the PROSITE motif search showed a possible ‘paired box’ domain (16) that may function in sequence-specific DNA recognition (17). There is a 92% sequence identity between the human and mouse Prtb genes. This high conservation between the two species may suggest a shared, and perhaps crucial, function of this protein. Prtb is expressed in various levels in a variety of tissues. The expression patterns of Prtb in human and in mouse are shown in Figure 2A and B, respectively. In both organisms Prtb is expressed at high levels in the heart, and at lower levels in skeletal muscle (when compared with Sox6) (3,10).
Figure 2.
Human (A) and mouse (B) multiple tissue northern blots. Each lane contains 2 µg of poly(A)+ RNA from the tissues indicated above. The filters were hybridized with a 32P-labeled 562 bp mouse Prtb cDNA fragment (nucleotides 15–576). Control hybridization with β-actin was performed to confirm equal loading of RNAs (data not shown). Abbreviations used: sm. intestine, small intestine; per. blood leuk., peripheral blood leukocyte; sk. muscle, skeletal muscle; colon (mucosal lining). Standard marker fragments in kb are indicated at the left.
Sox6 and Prtb expression during differentiation of P19CL6 cells and P19CL6noggin cells
Cardiomyocyte differentiation can be studied in vitro using P19CL6 cells (11), a clonal derivative of P19 murine embryonal carcinoma cells (18). Unlike the parental P19 cells, this subline efficiently differentiates into beating cardiomyocytes under adherent conditions when treated with DMSO. This cell line expresses BMPs that have been shown to play a pivotal role in the induction of the cardiac cell lineage (19,20). To further investigate the functions of BMP in cardiac development, Monzen et al. (12) established a variant P19CL6 cell line, P19CL6noggin, that constitutively over-expresses noggin, an antagonist of BMP. Because of the continual presence of noggin in P19CL6noggin cells, myogenic induction is suppressed and neither cardiac transcription factors nor contractile protein genes are expressed. To understand where in the cardiomyocyte developmental pathway Sox6 and Prtb may be acting, we used P19CL6 and P19CL6noggin to analyze the expression of these genes. The mRNA of Sox6 is only detected when the cells are beating (Fig. 3) or committed to differentiate (Fig. 4). Sox6 mRNA was not detected in untreated or DMSO treated P19CL6noggin cells that do not differentiate into cardiomyocytes (Fig. 3). These results indicate that the expression of Sox6 in P19CL6 is associated with the initiation of the cardiomyogenic program and not with the DMSO treatment itself. In contrast to the Sox6 expression in P19CL6, the expression of Prtb is less affected by the presence of noggin, since Prtb is expressed in both P19CL6 and P19CL6noggin cells (Fig. 3). The highest expression of Prtb was observed when the P19CL6 cells are beating. In P19CL6noggin cells, DMSO treatment increased the Prtb mRNA level, but not to the level observed in beating P19CL6 cells (Fig. 3). It is, therefore, possible that DMSO treatment itself may have a partial effect on the expression of the Prtb gene.
Figure 3.
The expression of Sox6 and Prtb in P19CL6 and P19CL6noggin cells, with or without DMSO induction. Each lane contains 10 µg of total RNA from the cells treated as labeled (above). The same filter was serially hybridized with the probes indicated on the right. Hybridization with GPDH is presented to show that the same amount of intact RNA was loaded in each lane. Standard marker fragments in kb are indicated at the left.
Figure 4.
Northern blot analysis of Sox6, Prtb and Sox9 expression in P19CL6 cells treated with DMSO over a time course. Each lane contains 10 µg of total RNA. Numbers above refer to days of DMSO exposure: 0 (before adding DMSO), 2, 4, 6, 8, 10, 11, 12, 14 and 16 days. The cells started to beat on day 11. Hybridization with GPDH is presented to show that the same amount of intact RNA was loaded in each lane.
The temporal expression of Sox6 and Prtb in DMSO-treated P19CL6 cells was also analyzed by northern hybridization. The expression of Sox9 was also analyzed, because Sox9 has been shown to be required for the expression of Sox6 in chondrogenesis (21). The expression of Sox6 was faintly detectable at day 6, with highest expression on day 11 (Fig. 4), the first day that we could see beating cells. Prtb was faintly detected on day 0, with highest expression on day 6 (Fig. 4). Sox9 expression did not show any significant changes as DMSO-treated P19CL6 cells differentiated into cardiomyocytes (Fig. 4).
Regulation of L-type Ca2+ α1c in the P19CL6 cell line
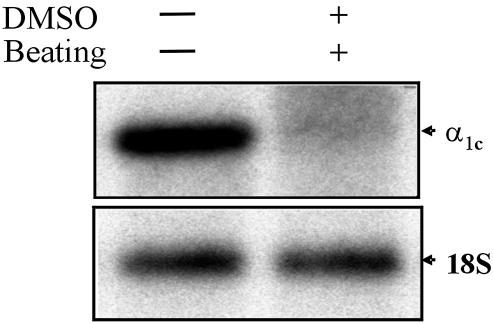
Ca2+ channels play a crucial role in maintaining muscle contraction in response to depolarization of the plasma membrane (22). At least five types of Ca2+ channels have been identified and characterized. The L-type channel is the predominant type in heart and vascular tissues. There are four subunits of the L-type Ca2+ channel present in the heart: α1, α2, δ and β, and there are also distinct isoforms of the α1 subunit, each with a unique gene product present in various tissues. The α1c subunit is expressed in cardiac and vascular smooth muscle, as well as in the brain (23). The L-type Ca2+ channel is critical for cardiomyocyte contraction (24). Expression of L-type Ca2+ α1c in P19CL6 cells is dramatically down-regulated following DMSO induction to beating cardiac cells (Fig. 5). However, expression of L-type Ca2+ α1c in P19CL6noggin cells was the same as in P19CL6 cells (with and without DMSO, data not shown).
Figure 5.
The L-type Ca2+ channel α1c (α1c) is down-regulated in P19CL6 cells treated with DMSO. The expression levels of the α1c subunit were determined by northern blot analysis. Each lane contains 10 µg of total RNA from the cells treated as labeled (above). 18S rRNA is presented to show that the same amount of intact RNA was loaded in each lane.
It is possible that the expression of Sox6 and Prtb may have a significant effect on the transcription of the L-type Ca2+ α1c gene, as it has been reported that the upstream sequence of the rat L-type Ca2+ α1c gene contains a repressor sequence between –1000 and –2000 bp relative to the transcription initiation site (25). This region contains seven consensus binding sites for Sox proteins (the potential binding sites of Prtb are unknown). We analyzed 4.1 kb of the regulatory region upstream of the L-type Ca2+ α1c cDNA in mouse (accession no. NM_009781), and detected eight perfect consensus binding sites for Sox proteins within a 1650 bp fragment located 1270 bp upstream of the published cDNA (data not shown).
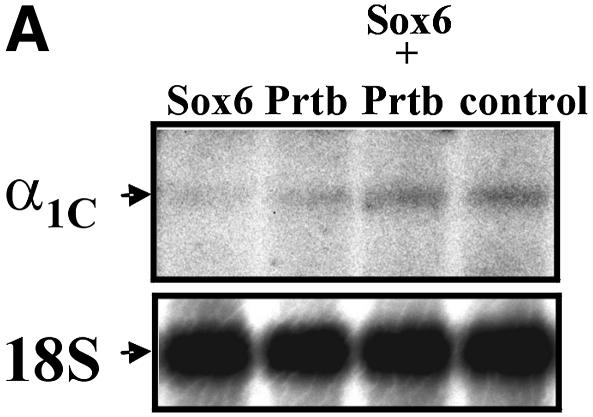
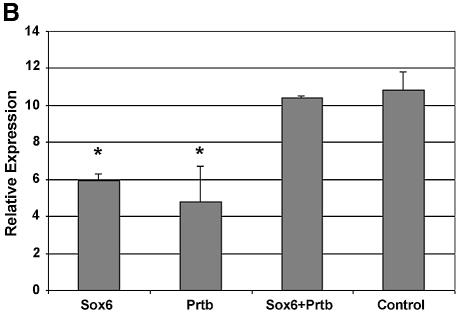
To examine the direct effect of Sox6 on the expression level of L-type Ca2+ α1c, we performed transient transfections using P19CL6 cells (Fig. 6A). We introduced Sox6 and Prtb cDNA individually, or together, under the control of a CMV promoter, and analyzed the mRNA levels of the L-type Ca2+ α1c gene by northern blot analysis. To determine the transfection efficiency, we co-transfected the cells with β-gal, driven by the CMV promoter (Fig. 6B). When transfected individually, both Sox6 and Prtb down-regulated the expression of L-type Ca2+ α1c (Fig. 6A and B). In contrast, when Prtb was co-transfected with Sox6, the expression of L-type Ca2+ α1c remained the same as the control level.
Figure 6.
(A) Northern blot analysis of L-type Ca2+ channel α1c (α1c) expression in P19CL6 cells transiently transfected with Sox6 and Prtb (as described in Materials and Methods). 18S rRNA is presented to show that the same amount of intact RNA was loaded in each lane. This panel shows one of three independent northern experiments. (B) The expression level values of α1c, normalized for transfection efficiency by the corresponding β-Gal value. Three independent transfection experiments were done. Asterisk denotes P ≤ 0.02 when compared with both control and co-transfection.
It has been shown previously that the L-type Ca2+ α1c transcript undergoes alternative splicing and produces three different transcripts in heart, ∼15.5, 8.9 and 20 kb (26). To confirm that we were assaying gene expression and not alternative splicing, we used two different probes for L-type Ca2+ α1c (see Materials and Methods). One probe encompassed the 5′ region of the cDNA spanning three exons and the other probe encompassed the 3′ region of the cDNA spanning four exons. We detected only the ∼15.5 kb transcript in the P19CL6 and P19CL6noggin cells using either the 5′ (data not shown) or 3′ (Figs 5 and 6) probes. Thus, the different expression levels are not due to alternative splicing.
Regulation of L-type Ca2+ α1c in p100H/p100H mutant mice
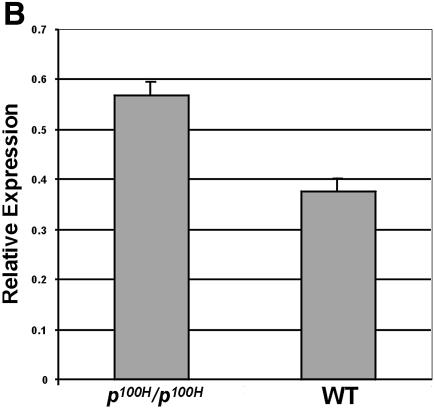
Next, we analyzed the expression of the L-type Ca2+α1c gene in the p100H/p100H mutant mouse, which is a Sox6 null mutation that develops significant changes in the ultrastructure of cardiac and skeletal muscle, AV heart block and a cardiac conduction defect (3). RT–PCR was performed to analyze mRNA levels of the L-type Ca2+α1c gene in wild type and p100H/p100H hearts (Fig. 7A). Primers were selected to amplify a 548-bp fragment corresponding to nucleotides 6294–6841 of the α1c subunit. Total RNA of the samples was calibrated to insure amplification in the linear range. Expression of the mouse GPDH gene was used as an internal control. In this in vivo assay, normal expression of Sox6 is associated with ∼1.5-fold down-regulation of the L-type Ca+2 α1c channel (Fig. 7B), which is consistent with our in vitro studies (Fig. 6).
Figure 7.
The expression of the L-type Ca2+ channel α1c (α1c) is up- regulated by ∼1.5-fold in Sox6 null mice (p100H/p100H). (A) RT–PCR products amplified from embryonic day 18.5 cardiac tissue of wild-type (WT) and p100H homozygous mice (p100H/p100H). The numbers on the bottom indicate cycle numbers of the PCR. Sizes of the marker standards (M) are: 622, 527 and 404 bp. Amplification of the GPDH fragment was used as an internal control. (B) The amplified L-type Ca2+ channel α1c fragment was quantified and normalized with the GPDH fragment using densitometry, and each bar represents the SD of three independent experiments.
DISCUSSION
Sox proteins distinguish their regulatory targets in a cell type-specific fashion via co-factor binding. They recognize a motif of only 6–7 bp of DNA with considerable degeneracy. However, they appear to regulate different sets of target genes, depending on the cell type in which they are expressed. Cooperative interactions between Sox proteins and their partner factors may allow stable associations with their specific target sequences (2). To identify an interactor of the Sox6 protein we used the yeast two-hybrid system. Screening a cDNA library prepared from an embryonic day-11 mouse, we identified the Prtb gene (15) as an interactor with the Sox6 coiled-coil domain. The interaction was confirmed by Co-IP, demonstrating a physical interaction between the two proteins. Sequence analysis of the Prtb gene indicated a possible ‘pair box’ domain (16), suggesting that Prtb may function as a transcription factor. Prtb is expressed in embryonic and adult stages in multiple tissues at different levels.
P19CL6 cells provide an in vitro model for cardiac myocyte differentiation (11). Sox6 is expressed exclusively when these cells are committed to differentiate to cardiac beating cells (Figs 3 and 4). Sox6 is expressed at day 6 following DMSO induction in P19CL6 cells (Fig. 4). A minimum exposure time of 4 days of DMSO is required for differentiation of P19CL6 cells to beating myocytes (27). These findings correlate with the fact that Sox6 is not expressed unless the cells are committed to beating, with the highest expression of Sox6 on day 11, at which point rhythmic contractions begin. The temporal expression of Sox6 in P19CL6 cells (and the mutant phenotype p100H/p100H mice) suggests that Sox6 plays a significant role in cardiogenesis.
It has been shown that Sox6, together with L-Sox5 and Sox9, undergoes up-regulation by BMP-2 during murine fracture healing (28). On the other hand, it has been shown that Sox6 is an important downstream mediator of BMP-2 signaling in chondrogenesis, whereas other signals control the expression and function of Sox9 as a chondrogenic transcription factor (8). BMPs are members of the transforming growth factor-β super family and play a crucial role in chondrogenesis as well as in cardiomyocyte differentiation (reviewed in 29). In the present study, we have shown that Sox6 is regulated through the BMP pathway in the differentiation of cardiomyocytes (Fig. 3) using P19CL6 cells stably transfected with noggin, an antagonist of BMP (12). In addition, we have shown that Sox9 expression was high and equal through the DMSO time course assay (Fig. 4). Thus, we suggest that Sox6 is a downstream mediator of BMP signaling in cardiogenesis as well as in chondrogenesis, and Sox6 and Sox9 are probably regulated differently in the cardiac system. Another study showed that Sox9 is required for the expression of Sox6 in chondrocyte differentiation (21). In cardiomycyte differentiation, the expression of Sox9 may be required for the expression of Sox6, but it is clearly not sufficient. Hence, the inducible P19CL6 cells may make it possible to define additional factors that are upstream of Sox6.
Expression analysis of the Prtb gene in P19CL6 and P19CL6noggin cell lines showed that Prtb is up-regulated when the cells differentiate to beating cardiac cells. Although DMSO, by itself, has a moderate effect on the expression of this gene, its maximum expression is observed 11 days after DMSO treatment when the cardiomyocytes are beating (Figs 3 and 4). Thus, Prtb function might be specified by the cooperation of other genes such as Sox6.
In addition to playing a role in cardiac myocyte development, Prtb and Sox6 may also be involved in bone formation. It has been shown that Prtb is a serum-responsive gene in osteoblasts and is up-regulated during adhesion and possibly involved in processes such as cell cycle control and proliferation as a participant in the extracellular matrix (16). L-Sox5 and Sox6 are essential for cartilage formation (4) and are required for notochord extracellular matrix (30). Thus, it is possible that Sox6 and Prtb cooperate in bone formation.
Elucidation of the target genes of Sox6 and Prtb will be important to understand the process of cardiac myocyte differentiation. The correlation between beating cardiomyocytes and Sox6 expression allows us to evaluate proteins important in contraction as potential targets of Sox6 regulation. For example, cardiac contraction is highly dependent on the gating function of the L-type calcium channel (31). The α1c subunit of the calcium channel provides the pore structure for Ca2+ ion entry. It has clinical relevance, since it contains the binding sites for multiple classes of drugs, collectively known as the calcium antagonists (13). To gain insights into the regulation of this channel, we analyzed its expression in P19CL6 cells. L-type Ca2+ α1c expression in these cells is dramatically down-regulated following DMSO induction to beating cardiac cells (Fig. 5), opposite the expectation of a direct correlation between beating cells and calcium channel expression. The reduced expression of this gene might result from aberrant regulation by DMSO, as expression of L-type Ca2+ α1c was also down-regulated in P19CL6noggin cells treated with DMSO. Another possible explanation for this reduction is that the proliferation of the cells has an impact on expression of the L-type Ca2+ channel. The cells beat when they grow in multilayers. In smooth muscle cells of the rat aorta it was shown that transcription of the L-type Ca2+ channel α1c is down-regulated in the proliferative state and it is closely linked to cell growth (32). Our in vitro transfection data (Fig. 6A and B) suggest that the Sox6 or Prtb proteins might at least partially down-regulate the α1c channel in the P19CL6 cells treated with DMSO.
The regulatory region of the L-type Ca2+ α1c gene contains eight consensus binding sites for Sox proteins. Multiple Sox binding sites have been reported previously in several other genes regulated by Sox factors (33–35), suggesting that the L-type Ca2+ α1c gene might be similarly regulated. Sox6 heterodimerized with Sox5, and in the presence of Sox9, cooperatively activates the expression of col2a (a chondrocyte differentiation marker) (6). On the other hand, Sox6 interacts with CtBP2 (C-terminal binding protein) and causes repression of Fgf-3 expression (36). Thus, Sox6 can function as activator or as a repressor. The transient transfection analysis showed that Sox6 and Prtb (each one by itself) down-regulated the expression level of the L-type Ca2+ α1c channel gene (Fig. 6A and B). However, when Sox6 and Prtb are both over-expressed, they appear to antagonize the effects of each. Prtb might antagonize Sox6 repression (or vice versa) by interacting with Sox6 and creating a complex that can no longer interact with its DNA target (37), or a Prtb-associated complex might change the chromatin structure and abrogate Sox6 function (38). It is known that Sox transcription factors bind to the minor groove of DNA causing a 70–85° bend of the DNA, consequently introducing local conformational changes (39,40). Therefore, Sox proteins may perform part of their function as architectural proteins by organizing local chromatin structure and assembling other DNA-bound transcription factors into biologically active, sterically defined, multiprotein complexes (41,42).
The p100H/p100H mutant mouse is a Sox6 null mutant characterized by early postnatal lethality, associated with progressive atrioventricular heart block and myopathy (3). The cardiac expression of L-type Ca2+ α1c is up-regulated by ∼1.5-fold in the p100H/p100H mutant mouse (Fig. 7). This in vivo result correlates with our cell line observations (Fig. 6). Interestingly, knockout mice for the L-type Ca2+ α1c die before embryonic day 14.5 (43,44). Deletion of this channel leads to a selective perturbation of cardiac morphogenesis and function in early embryonic development. Further investigation will be required to understand the role of L-type Ca2+ α1c in the arrhythmia of the Sox6 mutant homozygote.
In conclusion, Sox6 is within the BMP pathway in the cardiac system, interacts with Prtb, and may regulate downstream genes in the heart such as the α1c-subunit of the L-type Ca2+ channel. The inducible P19CL6 cardiac cell lines and the p100H/p100H mutant mice represent useful models for studying cardiac development, with important implications for understanding congenital heart disease and therapeutic intervention.
Acknowledgments
ACKNOWLEDGEMENTS
We thank our colleagues, Drs Ray Runyan, John Gardner, Ricardo Samson and Drew T. Erickson for their helpful comments on the manuscript. This work was supported by NIH grant GM43840.
REFERENCES
- 1.Firulli A.B. and Olson,E.N. (1997) Modular regulation of muscle gene transcription: a mechanism for muscle cell diversity. Trends Genet., 13, 364–369. [DOI] [PubMed] [Google Scholar]
- 2.Kamachi Y., Uchikawa,M. and Kondoh,H. (2000) Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet., 16, 182–187. [DOI] [PubMed] [Google Scholar]
- 3.Hagiwara N., Klewer,S.E., Samson,R.A., Erickson,D.T., Lyon,M.F. and Brilliant,M.H. (2000) Sox6 is a candidate gene for p100H myopathy, heart block and sudden neonatal death. Proc. Natl Acad. Sci. USA, 97, 4180–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smits P., Li,P., Mandel,J., Zhang,Z., Deng,J.M., Behringer,R.R., de Crombrugghe,B. and Lefebvre,V. (2001) The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell, 1, 277–290. [DOI] [PubMed] [Google Scholar]
- 5.Connor F., Wright,E., Denny,P., Koopman,P. and Ashworth,A. (1995) The Sry-related HMG box-containing gene Sox6 is expressed in the adult testis and developing nervous system of the mouse. Nucleic Acids Res., 323, 3365–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lefebvre V., Li,P. and de Crombrugghe,B. (1998) A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J., 17, 5718–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stokes D.G., Liu,G., Dharmavaram,R., Hawkins,D., Piera-Velazquez,S. and Jimenez,S.A. (2001) Regulation of type-II collagen gene expression during human chondrocyte de-differentiation and recovery of chondrocyte-specific phenotype in culture involves Sry-type high-mobility-group box (SOX) transcription factors. Biochem. J., 360, 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Lloris R., Vinals,F., Lopez-Rovira,T., Harley,V., Bartrons,R., Rosa,J.L. and Ventura,F. (2003) Induction of the Sry-related factor SOX6 contributes to bone morphogenetic protein-2-induced chondroblastic differentiation of C3H10T1/2 cells. Mol. Endocrinol., 17, 1332–1343. [DOI] [PubMed] [Google Scholar]
- 9.Morris G.E. and Manilal,S. (1999) Heart to heart: from nuclearproteins to Emery–Dreifuss muscular dystrophy. Hum. Mol. Genet., 8, 1847–1851. [DOI] [PubMed] [Google Scholar]
- 10.Cohen-Barak O., Hagiwara,N., Arlt,M.F., Horton,J.P. and Briliant,M.H. (2001) Cloning, characterization and chromosome mapping of the human SOX6 gene. Gene, 265, 157–164. [DOI] [PubMed] [Google Scholar]
- 11.Habara-Ohkubo A. (1996) Differentiation of beating cardiac muscle cells from a derivative of P19 embryonal carcinoma cells. Cell Struct. Funct., 21, 101–110. [DOI] [PubMed] [Google Scholar]
- 12.Monzen K., Shiojima,I., Hiroi,Y., Kudoh,S., Oka,T., Takimoto,E., Hayashi,D., Hosoda,T., Habara-Ohkubo,A., Nakaoka,T. et al. (1999) Bone morphogenetic proteins induce cardiomyocyte differentiation through the mitogen-activated protein kinase kinase kinase TAK1 and cardiac transcription factors Csx/Nkx-2.5 and GATA-4. Mol. Cell. Biol., 19, 7096–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L., O’Hara,D.S., Cala,S.E., Poornima,I., Hines,R.N. and Marsh,J.D. (2000) Developmental regulation of the L-type calcium channel α1C subunit expression in heart. Mol. Cell. Biochem., 205, 101–109. [DOI] [PubMed] [Google Scholar]
- 14.Durham-Pierre D., Gardner,J.M., Nakatsu,Y., Kink,R.A., Francke,U., Ching,A., Aquaron,R., del Marmol,V. and Brilliant,M.H. (1994) African origin of an intragenic deletion of the human P gene in tyrosinase positive oculocutaneous albinism. Nature Genet., 7, 176–179. [DOI] [PubMed] [Google Scholar]
- 15.Yang W. and Mansour,S.L. (1999) Expression and genetic analysis of prtb, a gene that encodes a highly conserved proline-rich protein expressed in the brain. Dev. Dyn., 215, 108–116. [DOI] [PubMed] [Google Scholar]
- 16.Sommerfeldt D.W., Zhi,J., Rubin,C.T. and Hadjiargyrou,M. (2002) Proline-rich transcript of the brain (prtb) is a serum-responsive gene in osteoblasts and upregulated during adhesion. J. Cell. Biochem., 84, 301–308. [DOI] [PubMed] [Google Scholar]
- 17.Tell G., Pellizzari,L. and Damante G. (1997) Transcription factor and cancer. The example of pax genes. Adv. Clin. Path., 1, 243–255. [PubMed] [Google Scholar]
- 18.Martin G.R. (1980) Teratocarcinomas and mammalian embryogenesis. Science, 209, 768–776. [DOI] [PubMed] [Google Scholar]
- 19.Frasch M. (1995) Induction of visceral and cardiac mesoderm by ectodermal Dpp in the early Drosophila embryo. Nature, 374, 464–467. [DOI] [PubMed] [Google Scholar]
- 20.Schultheiss T.M., Burch,J.B.E. and Lassar,A.B. (1997) A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev., 11, 451–462. [DOI] [PubMed] [Google Scholar]
- 21.Akiyama H., Chaboissier,M.C., Martin,J.F., Schedl,A. and de Crombrugghe,B. (2002) The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev., 16, 2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akaike N., Kanaide,H., Kuga,T., Nakamura,M., Sadoshima,J. and Tomoike,H. (1989) Low-voltage-activated calcium current in rat aorta smooth muscle cells in primary culture. J. Physiol., 416, 141–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez-Reyes E., Wei,X.Y., Castellano,A. and Birnbaumer,L. (1990) Molecular diversity of L-type calcium channels. Evidence for alternative splicing of the transcripts of three non-allelic genes. J. Biol. Chem., 265, 20430–20436. [PubMed] [Google Scholar]
- 24.Mikami A., Imoto,K., Tanabe,T., Niidome,T., Mori,Y., Takeshima,H., Narumiya,S. and Numa,S. (1989) Primary structure and functional expression of the cardiac dihydropyridine-sensitive calcium channel. Nature, 340, 230–233. [DOI] [PubMed] [Google Scholar]
- 25.Liu L., Fan,Q.I., El-Zaru,M.R., Vanderpool,K., Hines,R.N. and Marsh,J.D. (2000) Regulation of DHP receptor expression by elements in the 5′-flanking sequence. Am. J. Physiol. Heart Circ. Physiol., 278, H1153–H1162. [DOI] [PubMed] [Google Scholar]
- 26.Ma Y., Kobrinsky,E. and Marks,A.R. (1995) Cloning and expression of a novel truncated calcium channel from non-excitable cells. J. Biol. Chem., 270, 483–493. [DOI] [PubMed] [Google Scholar]
- 27.Peng C.F., Wei,Y., Levsky,J.M., McDonald,T.V., Childs,G. and Kitsis,R.N. (2002) Microarray analysis of global changes in gene expression during cardiac myocyte differentiation. Physiol. Genom., 9, 145–155. [DOI] [PubMed] [Google Scholar]
- 28.Uusitalo H., Hiltunen,A., Ahonen,M., Gao,T.G., Lefebvre,V., Harley,V., Kahari,V.M. and Vuorio,E. (2001) Accelerated up-regulation of L-Sox5, Sox6 and Sox9 by BMP-2 gene transfer during murine fracture healing. J. Bone Miner. Res., 16, 1837–1845. [DOI] [PubMed] [Google Scholar]
- 29.Monzen K., Nagai,R. and Komuro,I. (2002) A role for bone morphogenetic protein signaling in cardiomyocyte differentiation. Trends Cardiovasc Med., 12, 263–269. [DOI] [PubMed] [Google Scholar]
- 30.Smits P., and Lefebvre,V. (2003) Sox5 and Sox6 are required for notochord extracellular matrix sheath formation, notochord cell survival and development of the nucleus pulposus of intervertebral discs. Development, 130, 1135–1148. [DOI] [PubMed] [Google Scholar]
- 31.Marsh J.D. and Allen,P.D. (1989) Developmental regulation of cardiac calcium channels and contractile sensitivity to [Ca]o. Am. J. Physiol., 256, H179–H185. [DOI] [PubMed] [Google Scholar]
- 32.Ihara E., Hirano,K., Hirano,M., Nishimura,J., Nawata,H. and Kanaide,H. (2002) Mechanism of down-regulation of L-type Ca2+ channel in the proliferating smooth muscle cells of rat aorta. J. Cell. Biochem., 87, 242–251. [DOI] [PubMed] [Google Scholar]
- 33.Peirano R.I., Goerich,D.E., Riethmacher,D. and Wegner,M. (2000) Protein zero gene expression is regulated by the glial transcription factor Sox10. Mol. Cell. Biol., 20, 3198–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Callard G.V., Tchoudakova,A.V., Kishida,M. and Wood,E. (2001) Differential tissue distribution, developmental programming, estrogen regulation and promoter characteristics of cyp19 genes in teleost fish. J. Steroid Biochem. Mol. Biol., 79, 305–314. [DOI] [PubMed] [Google Scholar]
- 35.Hwang C.K., Wu,X., Wang,G., Kim,C.S. and Loh,H.H. (2003) Mouse µ opioid receptor distal promoter transcriptional regulation by SOX proteins J. Biol. Chem., 278, 3742–3750. [DOI] [PubMed] [Google Scholar]
- 36.Murakami A., Ishida,S., Thurlow,J., Revest,J.M. and Dickson,C. (2001) SOX6 binds CtBP2 to repress transcription from the Fgf-3 promoter. Nucleic Acids Res., 29, 3347–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latchman D.S. (1996) Inhibitory transcription factors. Int. J. Biochem. Cell Biol., 28, 965–974. [DOI] [PubMed] [Google Scholar]
- 38.Frank A., de-Camillis,M., Zirk,D., Chang,N., Brock,H.W. and Paro,R. (1992) Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J., 11, 2941–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferrari S., Harley,V., Pontiggia,A., Goodfellow,P.N., Lovell-Badge,R. and Bianchi,M.E. (1992) SRY, like HMG1, recognizes sharp angles in DNA. EMBO J., 11, 4497–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Connor F., Cary,P.D., Read,C.M., Preston,N.S., Driscoll,P.C., Denny,P., Crane-Robinson,C. and Ashworth,A. (1994) DNA binding and bending properties of the post-meiotically expressed Sry-related protein Sox-5. Nucleic Acids Res., 22, 3339–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Werner M.H. and Burley,S.K. (1997) Architectural transcription factors: proteins that-remodel DNA. Cell, 88, 733–736. [DOI] [PubMed] [Google Scholar]
- 42.Wolffe A.P. (1994) Architectural transcription factors. Science, 264, 1100–1101. [DOI] [PubMed] [Google Scholar]
- 43.Seisenberger C., Specht,V., Welling,A., Platzer,J., Pfeifer,A., Kuhbandner,S., Striessnig,J., Klugbauer,N., Feil,R. and Hofmann,F. (2000) Functional embryonic cardiomyocytes after disruption of the L-type α1C (Cav1.2) calcium channel gene in the mouse. J. Biol. Chem., 275, 39193–39199. [DOI] [PubMed] [Google Scholar]
- 44.Klugbauer N., Welling,A., Specht,V., Seisenberger,C. and Hofmann,F. (2002) L-type Ca2+ channels of the embryonic mouse heart. Eur. J. Pharmacol., 447, 279–284. [DOI] [PubMed] [Google Scholar]