Abstract
It has been long recognized that in mammalian cells, DNA damage is preferentially repaired in the transcribed strand of transcriptionally active genes. However, recently, we found that in Chinese hamster ovary (CHO) cells, UV-induced cyclobutane pyrimidine dimers (CPDs) are preferentially repaired in both the transcribed and the non-transcribed strand of exon 1 of the dihydrofolate reductase (DHFR) gene. We mapped CPD repair at the nucleotide level in the transcriptionally active DHFR gene and the adjacent upstream OST gene, both of which have been translocated to two chromosomal positions that differ from their normal endogeneous positions. This allowed us to study the role of transcription, genomic context and chromatin structure on repair. We found that CPD repair in the transcribed strand is the same for endogenous and translocated DHFR genes, and the order of repair efficiency is exon 1 > exon 2 > exon 5. However, unlike the endogenous DHFR gene, efficient repair of CPDs in the non-transcribed strand of exon 1 is not observed in the translocated DHFR gene. CPDs are efficiently repaired in the transcribed strand in endogenous and translocated OST genes, which indicates that efficient repair in exon 1 of the non-transcribed strand of the endogenous DHFR gene is not due to the extension of transcription-coupled repair of the OST gene. Using micrococcal nuclease digestion, we probed the chromatin structure in the DHFR gene and found that chromatin structure in the exon 1 region of endogenous DHFR is much more open than at translocated loci. These results suggest that while transcription-coupled repair is transcription dependent, global genomic repair is greatly affected by chromatin structure.
INTRODUCTION
Nucleotide excision repair (NER) is a versatile repair system that repairs a wide range of bulky DNA lesions, including UV light-induced cyclobutane pyrimidine dimers (CPDs) (1–7). Two distinct NER subpathways—transcription-coupled repair (TCR) and global genomic repair (GGR)—have been found in mammalian cells (3–8). It is generally believed that the TCR pathway is responsible for the repair of DNA damage in the transcribed (T) strand of transcriptionally active genes, and that the GGR pathway is responsible for the repair of DNA damage occurring in locations other than the T strand of transcriptionally active genes in the genome. In general, TCR is more efficient than GGR. It has been found that human cells are proficient in both TCR and GGR of CPDs, while rodent cells lack GGR of CPDs (9).
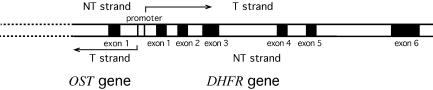
The basic enzymology of NER in mammalian cells has been well determined in vitro using naked DNA as substrates; more than 30 proteins are involved in NER in mammalian cells (10–13). However, the molecular mechanism of NER on its natural substrate, chromatin, remains largely unknown, especially the mechanism of TCR in mammalian cells. Increasing evidence suggests that NER in mammalian cells is much more complex than our current understanding of TCR and GGR would indicate (14–19). Recently, we found that in Chinese hamster ovary (CHO) AT3-2 cells, CPDs in the T strand of the DHFR gene are efficiently repaired and CPDs in the non-transcribed (NT) strand of the first exon of the DHFR gene are also efficiently repaired. In contrast, CPDs in exons 2 and 5 of the NT strand of the DHFR gene are poorly repaired (19). These results raise the possibility that the efficient repair in the first exon of the NT strand of the endogenous DHFR gene may result from two mechanisms. The first mechanism is the extension of TCR from the OST gene (20,21) to the DHFR gene. The OST gene is located immediately 5′ of the DHFR gene, shares the same promoter region with the DHFR gene and is transcribed in the direction opposite from the DHFR gene (20,21) (Fig. 1). The second possible mechanism is that the genomic context and/or chromatin structure of exon 1 is different from that found in exons 2 and 5. To test these possibilities, we mapped the CPD repair at nucleotide resolution in the DHFR and OST genes in three CHO cell lines, AT3-2, C26 and C38, using the T4 endonuclease V (T4 endo V) incision method in combination with ligation-mediated PCR (LMPCR) (18,19). We also probed the chromatin structure in the DHFR gene by determining the kinetics of micrococcal nuclease (MN) digestion. The DHFR and OST genes are located in different genomic contexts in these three cell lines: the AT3-2 cells contain DHFR and OST genes located in their normal endogenous positions, while in the C26 and C38 cells, the entire endogenous DHFR gene and at least 10 kb of the 5′ portion of the OST genes are deleted, and a fragment containing a single copy of the intact DHFR gene and 8 kb of its upstream region containing the OST gene has been introduced into different positions of the chromosome in these two cell lines (22). Since the DHFR genes in C26 and C38 are located at chromosome positions differing from the endogenous DHFR gene position in AT3-2 cells but are still transcriptionally active, these cells provide us with tools for studying the effects of genomic context, transcription and chromatin structure on TCR and GGR in this gene.
Figure 1.
Map of the endogenous DHFR and OST gene domains in CHO cells. The DHFR and OST genes share a common promoter region and are transcribed (arrow indicates the transcription direction) divergently from this common promoter region.
MATERIALS AND METHODS
Cell lines and culture conditions
AT3-2 cells contain diploid DHFR loci, and both loci are transcriptionally active. Both C26 and C38 cells contain a single copy of a transcriptionally active DHFR gene; they were constructed by transfection of CHO-DG44, a double deletion (>115 kb) mutant lacking both copies of the entire DHFR gene and at least 10 kb of the 5′ portion of the OST gene, with a cosmid containing a 41 kb DNA fragment from Chinese hamster genomic DNA containing the intact DHFR gene and flanking sequences (∼8 kb upstream containing the OST gene and 7 kb downstream) (22). As determined by fluorescence in situ hybridization, the chromosomal position of integration differed from the endogenous DHFR locus (22) and the site of integration was also different in these two transfectants. Northern blot and RT–PCR methods were used to confirm that the DHFR gene in these three cell lines is transcriptionally active. Cells were grown in α-minimum Eagle’s medium supplemented with 10% fetal calf serum.
UV irradiation and genomic DNA isolation
For UV irradiation, AT3-2, C26 and C38 cells were grown to 50–70% confluence in 150 mm dishes. Prior to UV irradiation, the culture medium was removed, the cells were washed with phosphate-buffered saline (68 mM NaCl, 1.94 mM KCl, 1.07 mM KH2PO4, pH 7.4) and the cells were then UV irradiated at a fluence rate of 1 J/m2/s for 15 s using GE15118 germicidal lamps (predominant emission 254 nm) as the UV source. After irradiation, the cells were incubated in fresh medium containing 10 µM 5-bromo-2′-deoxyuridine and 1 µM 5-fluorodeoxyuridine for various periods of time to allow DNA repair for repair kinetic analysis. After incubation, cells were lysed with lysing buffer (0.5% SDS, 10 mM Tris, pH 7.8, 10 mM EDTA, 10 mM NaCl, 100 µg/ml proteinase K) at room temperature for 1 h. Protein was removed by repeated phenol extractions followed by diethyl ether extractions. DNA was then ethanol precipitated and resuspended in TE buffer (10 mM Tris, pH 7.5, 1 mM EDTA). RNA was removed by treatment with RNase A (50 µg/ml) for 1 h followed by repeated phenol and diethyl ether extractions. DNA was then ethanol precipitated and resuspended in TE buffer (pH 7.5). Replicated and non-replicated DNA were separated by CsCl gradient centrifugation in a Ti 50 rotor (3.7 × 104 r.p.m. for 72 h at 21°C). Only the unreplicated DNA was used for repair kinetics analysis (14,15,19).
Cleavage of CPDs with T4 endo V
A known quantity of purified genomic DNA (10 µg) was treated with T4 endo V (protein:DNA molar ratio 6:1, assuming the average DNA length was 14 kb) in a solution of 100 mM NaCl, 10 mM Tris, pH 7.5, and 0.5 mM EDTA at 37°C for 60 min to cleave CPDs. Escherichia coli photolyase (0.5 µg/µg DNA) was then added and the mixtures were irradiated with 366 nm UV light (Sylvania 15 watt F15T8) for 60 min at room temperature in the presence of 10 mM dithiothreitol for photoreactivation. The reactions were stopped by repeated phenol and diethyl ether extractions. The resultant genomic DNA was then precipitated by ethanol, resuspended in TE buffer (pH 7.5) and subjected to LMPCR (14,15,19).
Mapping the repair of CPDs at the nucleotide level with LMPCR
To investigate the repair of CPDs in the DHFR and OST genes, a known quantity (1 µg) of T4 endo V-treated genomic DNA was subjected to LMPCR to map the distribution of CPDs along exons 1, 2 and 5 of the DHFR gene and the T strand of exon 1 in the OST gene. Control genomic DNA was subjected to Maxam–Gilbert sequencing (23) followed by LMPCR, in order to serve as a sequencing ladder. The LMPCR method was the same as previously described (14,15,18,19). The oligonucleotide primers (Midland Certified Reagent Co., Midland, TX) used for LMPCR analysis of exons 1, 2 and 5 of the DHFR gene were the same as previously described (19), and the primers used for LMPCR analysis of the T strand of exon 1 in the OST gene are shown in Table 1. Oligonucleotide primer 1 was used in the first primer extension step of LMPCR, primer 2 was the PCR primer, and primer 3 was used to make the single-stranded hybridization probe. The template used for hybridization probe synthesis was prepared by PCR amplification from CHO genomic DNA with primers 3 and 4. The resultant LMPCR products were separated by electrophoresis in 8% denaturing polyacrylamide gels and electro-transferred to GeneScreen nylon membranes (NEN, Boston, MA). Blots were hybridized with 32P-labeled DNA probes specific for exons 1, 2 or 5 of the DHFR gene and exon 1 of the OST gene in hybridization buffer (0.25 M Na2HPO4, pH 7.2, 1 mM EDTA, 7% SDS, 1% bovine serum albumin) at 60°C for 12 h. The membranes were then exposed to a Cyclone Storage Phosphor screen (Packard, Meriden, CT), and the intensities of T4 endo V incision bands were quantified with the Cyclone Storage Phosphor System (Packard). Approximately 20 000 d.p.m. of 32P-labeled linearized pBR322 plasmid DNA was added to each genomic DNA sample at the beginning of the LMPCR as an internal standard to monitor sample recovery. After LMPCR, equivalent counts of 32P, as measured by a liquid scintillation counter (LKB-Wallac, Turku, Finland) and representing equivalent amounts of sample DNA, were loaded into each lane of the sequencing gel to separate DNA fragments of different sizes.
Table 1. Synthetic oligomer primers for LMPCR analysis of the exon 1 transcribed strand of the OST gene.
| Primer | Sequence | Tm (°C) |
|---|---|---|
| 1 | ACTCCGCCTCCACCAG | 48.1 |
| 2 | CGCCCAGTCCGGCGTGGC | 66.4 |
| 3 | CGCTCCAGGCGCGGGGTAGT | 65.0 |
| 4 | GGTAGACGCTGGGGGCGCTGAG | 65.6 |
Isolation of nuclei and digestion of chromatin by micrococcal nuclease
Methods for isolation of nuclei and subsequent MN digestion were the same as described previously (19,24). Briefly, AT3-2, C26 and C38 cells were harvested and nuclei were immediately isolated. The freshly isolated nuclei (1 × 107) of each cell line were immediately digested with MN (Amersham Pharmacia Biotech, Piscataway, NJ) (1 U) in 100 µl of digestion buffer (100 mM Tris, pH 8.0, 50 mM NaCl, 3 mM MgCl2, 1 mM CaCl2) for 1, 2, 5 or 10 min at 37°C. The digestion was stopped by adding an equal volume of stop solution (200 mM Tris, pH 8.0, 200 mM NaCl, 20 mM EDTA, 2% SDS, 200 µg/ml proteinase K). The control was an undigested, freshly lysed sample of nuclei. Genomic DNA was purified as described above and then separated by electrophoresis in 1.5% agarose gels. After staining with ethidium bromide, the separated DNA was transferred to nylon membranes and hybridized with a 32P-labeled probe specific for DHFR exon 1. After de-probing, the membranes were further hybridized with a 32P-labeled probe specific for DHFR exon 2.
RESULTS
CPD repair in the transcribed strand of the DHFR gene in different genomic contexts is the same
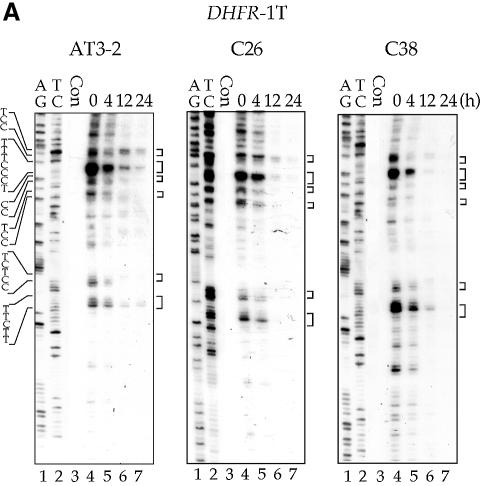
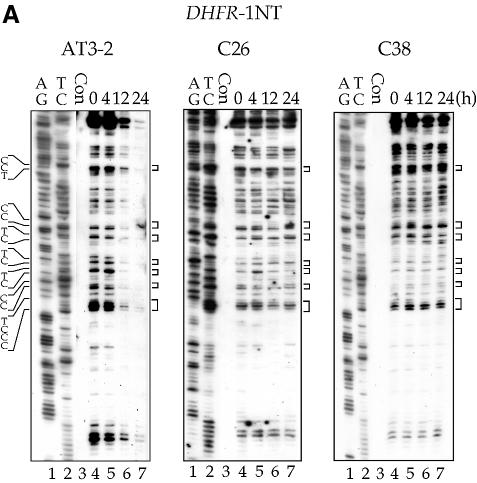
The repair of CPDs was mapped at the sequence level using the T4 endo V incision method in combination with LMPCR. CPD repair in the T strand of the DHFR gene in these three cell lines is shown in Figures 2 and 3. The kinetics of CPD repair in the T strand of exons 1, 2 and 5 in the translocated DHFR gene in C26 and C38 cells are very similar, if not identical, to the kinetics of repair in the endogenous DHFR gene in AT3-2 cells. The initial repair rate of CPDs along the T strand of the translocated DHFR genes showed a 5′ to 3′ polarity effect similar to that observed in the endogenous DHFR gene in AT3-2 cells: the initial repair rate was faster in exon 1 than in exon 2, and much faster in exons 1 and 2 than in exon 5. In addition, there were no significant differences in the initial repair rates along the T strand between these three cell lines. The time required for 50% CPD removal (T1/2) in exons 1, 2 and 5 of the DHFR gene in these three cell lines was ∼4, 6 and 12 h, respectively. However, 24 h post-irradiation, the CPDs were almost completely removed in these three exons of the DHFR gene in all three cell lines.
Figure 2.
The time course of CPD repair in the transcribed strand of exon 1 (A), exon 2 (B) and exon 5 (C) of the DHFR gene in CHO AT3-2, C26, and C38 cells. Cultured cells were UV irradiated (15 J/m2) and then incubated for various periods of time. Genomic DNA was isolated, treated with T4 endo V followed by photoreactivation, and then subjected to LMPCR. The LMPCR products were separated by electrophoresis in 8% denaturing polyacrylamide gels, transferred to nylon membranes, and hybridized with 32P-labeled probes specific for the transcribed strand of DHFR exons 1, 2 or 5. A + G and T + C represent Maxam–Gilbert sequencing reactions. Sequences of contiguous pyrimidines with the potential to form CPDs are indicated on the left, and T4 endo V incision sites are indicated on the right (bracketed). Lanes 4–7 show the relative frequency of T4 endo V cutting at dipyrimidine sites along each sequence at different post-UV repair time points (0, 4, 12 and 24 h). Lane 3 (Con) represents DNA isolated from unirradiated control cells and treated with T4 endo V. Very similar results were obtained from three independent experiments.
Figure 3.
The kinetics of CPD repair in the transcribed strand of the DHFR gene in AT3-2, C26 and C38 cells. The relative amount of CPD formed at the dipyrimidine sites (bracketed) along the T strand of exons 1 (1T), 2 (2T) and 5 (5T) of the DHFR gene for each time point shown in Figure 2 was quantified with a Cyclone Storage Phosphor System. The percentage of CPDs remaining in the T strand of each exon was plotted as a function of repair time. The results represent three independent experiments.
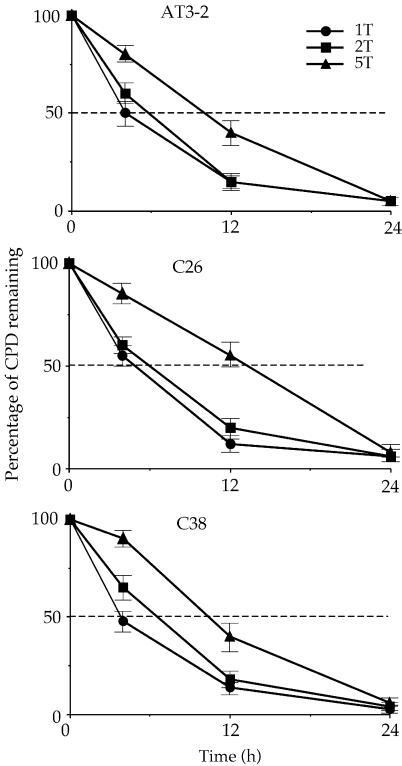
CPD repair in the non-transcribed strand of the DHFR gene in different genomic contexts is not the same
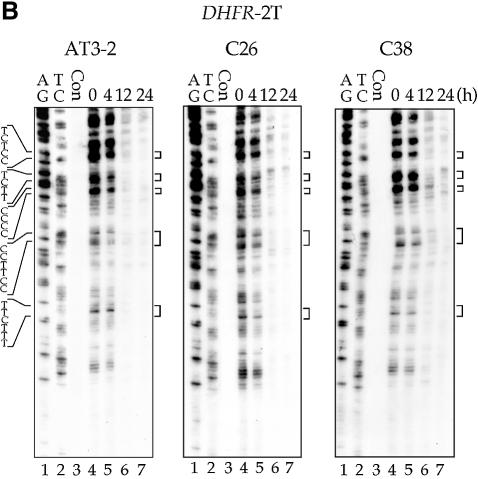
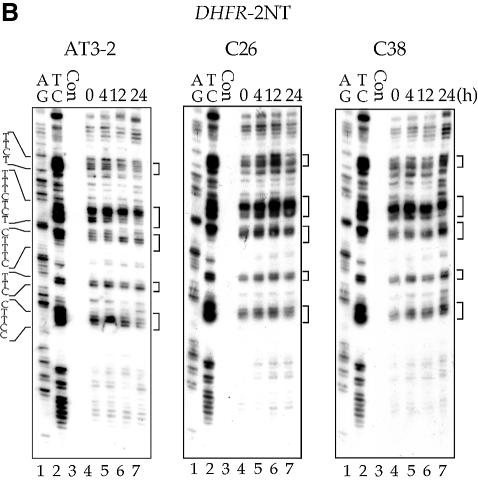
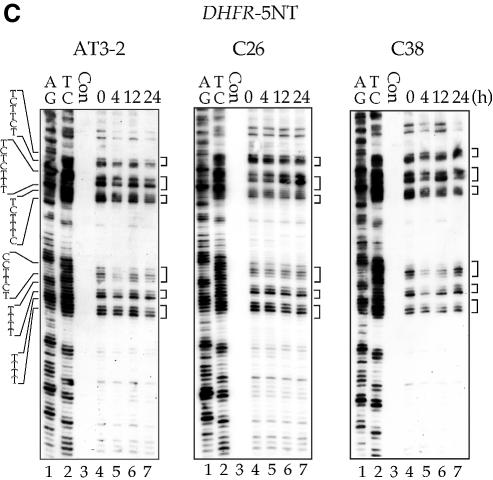
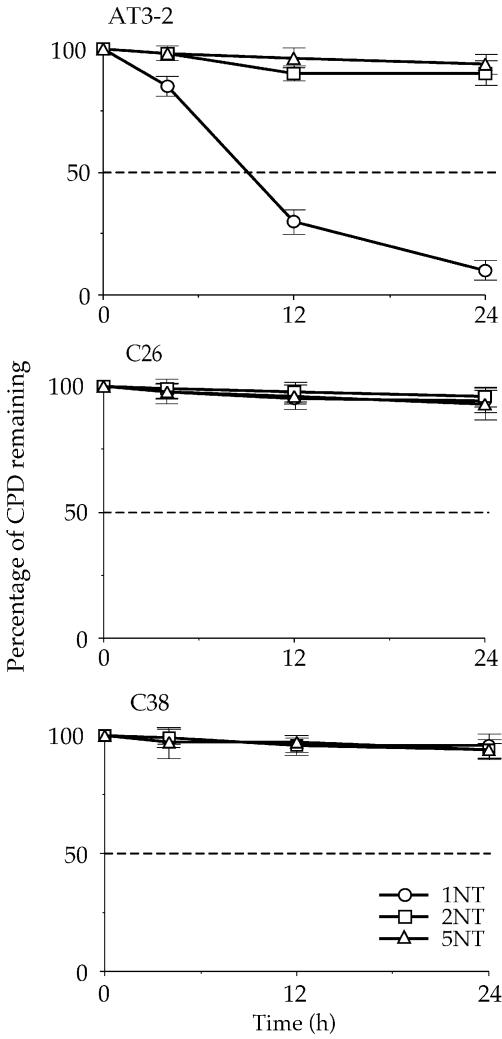
The efficient CPD repair in the NT strand of exon 1 in the endogenous DHFR gene in AT3-2 cells could be due to the DNA sequence effects; it is possible that the sequence of exon 1 of DHFR may intrinsically allow more efficient CPD repair. It is also possible that the same mechanism facilitating efficient repair in the T strand of exon 1, such as the transcription process of the DHFR gene, may also facilitate efficient repair in the NT strand. In either case, our findings that CPD repair in the T strand is the same in translocated and endogenous DHFR genes led us to expect that CPD repair in the NT strand of exons 1, 2 and 5 of the translocated DHFR gene in C26 and C38 cells should be the same as that observed in the endogenous DHFR gene in AT3-2 cells. To test this possibility, we mapped the CPD repair in the NT strand of exons 1, 2 and 5 of the translocated and endogenous DHFR genes in these three cell lines. The results in Figures 4 and 5 show that efficient repair was not observed in the NT strand of exon 1 of the translocated DHFR gene in either transfectant cell line. CPDs along the NT strand were poorly repaired in exon 1 as well as in exons 2 and 5 in the translocated DHFR genes, and >90% of CPDs remained unrepaired 24 h after UV irradiation. In contrast, CPDs along the NT strand of exon 1 in the endogenous DHFR gene were efficiently repaired and were almost completely removed 24 h after UV irradiation. The CPD repair in the NT strand of exons 2 and 5 was the same in both the endogenous and translocated DHFR genes. These results exclude the possibility that DNA sequence causes efficient CPD repair in the NT strand of exon 1 and strongly suggest that the mechanism facilitating the efficient CPD repair in the T strand of exon 1 in the DHFR gene in different genomic contexts differs from the mechanism facilitating the efficient repair in the NT strand of the endogenous DHFR gene. Since the DHFR genes in these three cell lines are all transcriptionally active, these results also exclude the possibility that the efficient repair in the NT strand of exon 1 in the endogenous DHFR gene results from the transcription process of the DHFR gene.
Figure 4.
The time course of CPD repair in the non-transcribed strand of exon 1 (A), exon 2 (B) and exon 5 (C) of the DHFR gene in AT3-2, C26 and C38 cells. Cultured cells were UV irradiated (15 J/m2) and then incubated for various periods of time. Genomic DNA was isolated, treated with T4 endo V followed by photoreactivation, and then subjected to LMPCR. A + G and T + C represent Maxam–Gilbert sequencing reactions. Sequences of contiguous pyrimidines with the potential to form CPD are indicated on the left, and T4 endo V incision sites are indicated on the right (bracketed). Lanes 4–7 show the relative frequency of T4 endo V cutting at dipyrimidine sites along each sequence at different post-UV repair time points (0, 4, 12 and 24 h). Lane 3 (Con) represents DNA isolated from unirradiated control cells treated with T4 endo V. Very similar results were obtained from three independent experiments.
Figure 5.
The kinetics of CPD repair in the non-transcribed strand of the DHFR gene in AT3-2, C26 and C38 cells. The relative amount of CPD formed at the dipyrimidine sites (bracketed) along the non-transcribed strand of exons 1 (1NT), 2 (2NT) and 5 (5NT) of the DHFR gene for each time point shown in Figure 4 was quantified with a Cyclone Storage Phosphor System. The percentage of CPD remaining in the non-transcribed strand of each exon was plotted as a function of repair time. The results represent three independent experiments.
Genomic context does not affect TCR in the OST gene
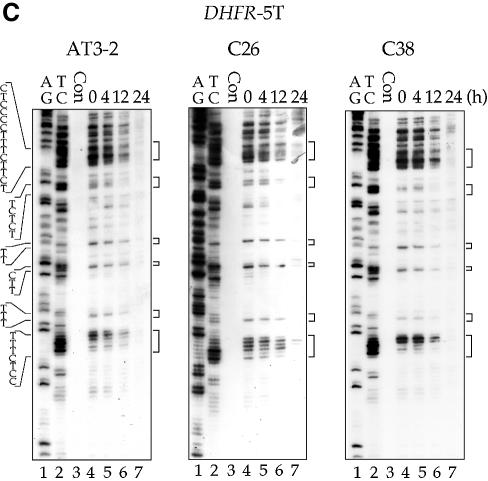
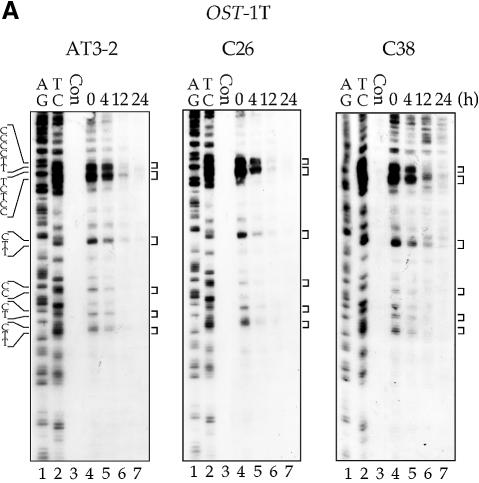
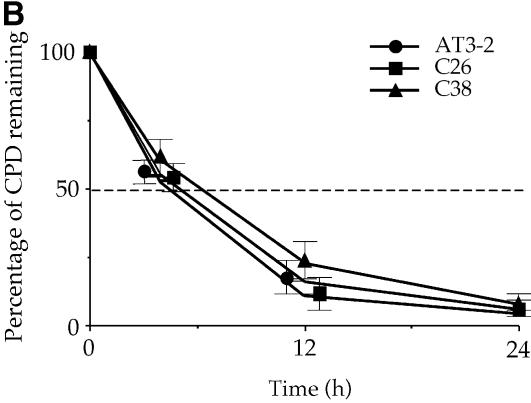
It has been reported that in some transcriptionally active gene domains, such as the p53 gene in human cells and the MFA2 gene in yeast cells, TCR appears to extend beyond the transcription start and termination sites of these active genes, i.e. the genomic regions affected by TCR are larger than the actual transcription unit (25–27). It is known that at the endogenous DHFR gene locus in CHO cells, there is an unidentified gene, the OST gene (also named Rep3), located immediately 5′ upstream of the DHFR gene, that shares the same promoter region with the DHFR gene (20,21). It encodes a homolog of the mismatch repair protein MSH3 in human cells (28). The OST gene is transcribed in the direction opposite to that of the DHFR gene; the DNA strand used as the NT strand of the DHFR gene is the T strand of the OST gene (Fig. 1) (20,21,29). It has been shown that in the OST gene, CPDs are preferentially repaired in the T strand and poorly repaired in the NT strand (30). These findings raise the possibility that the preferential repair of CPDs in the NT strand of exon 1 in the endogenous DHFR gene in AT3-2 cells comes from the extension of TCR in the T strand of the OST gene. If this is the case, the disappearance of the preferential repair of CPDs in exon 1 of the NT strand in the translocated DHFR gene in C26 and C38 cells should be accompanied by the disappearance of TCR from the T strand of the OST gene, which could be caused by the translocation of the fragment containing the intact DHFR gene and ∼8 kb of the OST gene 5′ end, including the promoter region. To test this possibility, CPD repair in exon 1 of the OST gene was mapped at the nucleotide level in the AT3-2, C26 and C38 cell lines. The results in Figure 6 show that in both endogenous and translocated OST genes, the CPDs in the T strand of exon 1 are efficiently repaired, and CPDs are almost completely removed 12 h after UV irradiation. Furthermore, the repair efficiency of CPDs in the T strand of the OST gene in these three cell lines is very similar. These results exclude the possibility that the efficient repair of CPDs in the NT strand of exon 1 of the endogenous DHFR gene in AT3-2 cells comes from the extension of TCR from the upstream OST gene.
Figure 6.
The time course of CPD repair in the transcribed strand of exon 1 of the OST gene in AT3-2, C26 and C38 cells. Cultured cells were UV irradiated (15 J/m2) and then incubated for various periods of time. Genomic DNA was isolated, treated with T4 endo V followed by photoreactivation, and then subjected to LMPCR. (A) Typical autoradiographs. A + G and T + C represent Maxam–Gilbert sequencing reactions. Sequences of contiguous pyrimidines with the potential to form CPDs are indicated on the left, and T4 endo V incision sites are indicated on the right (bracketed). Lanes 4–7 show the relative frequency of T4 endo V cutting at dipyrimidine sites along each sequence at different post-UV repair time points (0, 4, 12 and 24 h). Lane 3 (Con) represents DNA isolated from unirradiated control cells and treated with T4 endo V. Very similar results were obtained from three independent experiments. (B) The kinetics of CPD repair in the transcribed strand of exon 1 of the OST gene. The relative amount of CPD formed at the dipyrimidine sites (bracketed) along the transcribed strand of exons 1 of the OST gene for each time point shown in (A) was quantified with a Cyclone Storage Phosphor System. The percentage of CPD remaining in the transcribed strand was plotted as a function of repair time. The results represent three independent experiments.
The kinetics of MN digestion in the exon 1 region are slower in the translocated DHFR genes than in the endogenous DHFR gene
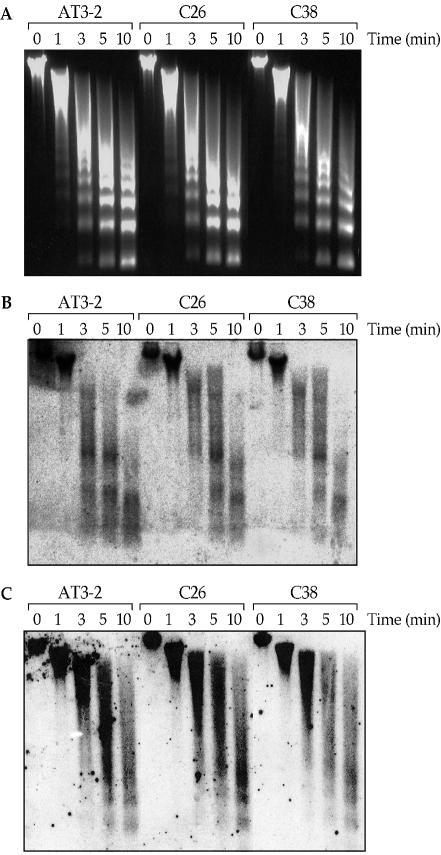
It is known that packaging of eukaryotic DNA into chromatin affects all aspects of DNA processing, including DNA repair, because it modulates access of proteins to DNA (31–33). Several recent studies have demonstrated that nucleosomal structure can inhibit repair of CPDs and <6–4> photoproducts in in vitro NER assays using mononucleosome or dinucleosome systems, suggesting that the assembly of nucleosomes may restrict the access of DNA repair proteins to the damaged DNA bases (34–36). We previously found that the exon 1 region of the endogenous DHFR gene is more sensitive to MN digestion than the exon 2 and 5 regions, which indicates that the exon 1 region has a more open chromatin structure than the exon 2 and 5 regions (19). These results raise the possibility that the more open chromatin structure in exon 1 may contribute to the preferential CPD repair in the NT strand of exon 1 in the endogenous DHFR gene in AT3-2 cells. If this is the case, then we would expect the chromatin structure of exon 1 in the translocated DHFR gene to be different from that found in exon 1 of the endogenous DHFR gene, since we have found that CPD repair in the NT strand of exon 1 is much more efficient in the endogenous than in the translocated DHFR gene. To test this possibility, we probed the chromatin structure of exons 1 and 2 in both the endogenous and translocated DHFR genes by determining their sensitivity to MN digestion. Nuclei were isolated from AT3-2, C26 and C38 cells, and subjected to digestion with MN for different time periods. The results in Figure 7 show that the chromatin structure of the exon 1 region of the endogenous DHFR gene in AT3-2 cells was much more sensitive to MN digestion than in the translocated DHFR gene in C26 and C38 cells. After 3 min of MN digestion, a significant amount of the endogenous DHFR gene exon 1 was in tri-, di- and mononucleosome structures. In contrast, most of exon 1 in the translocated DHFR genes remained in trinucleosome and higher nucleosome structures. After 5 min of MN digestion, there were more di- and mononucleosomes in exon 1 of the endogenous DHFR gene than that in exon 1 of the translocated DHFR genes. However, the kinetics of MN digestion for exon 2 in the endogenous and translocated DHFR genes are very similar if not identical. These results indicate that the exon 1 region of the endogenous DHFR gene in AT3-2 cells has a much more open chromatin structure than that of the translocated DHFR genes in C26 and C38, which strongly suggests that the more efficient repair of CPDs in the NT strand of the exon 1 region of the endogenous DHFR gene is due to its more open chromatin structure.
Figure 7.
The MN digestion sensitivity of exons 1 and 2 of the DHFR gene in AT3-2, C26 and C38 cells. Nuclei were isolated from the three CHO cell lines and digested with MN (1 U/100 µl) for different times (0, 1, 3, 5 and 10 min). Genomic DNA was isolated, separated by electrophoresis in a 1.5% agarose gel, stained with ethidium bromide (A) and then transferred to a nitrocellulose membrane and hybridized with 32P-labeled probes specific for exon 1 (B) or exon 2 (C) of the DHFR gene.
DISCUSSION
It is generally accepted that NER consists of two pathways: TCR and GGR. While the TCR pathway repairs DNA damage in the T strand of actively transcribed genes, the GGR pathway repairs DNA damage in the NT strand of actively transcribed genes and non-coding DNA (3–8). Most of our knowledge about TCR and GGR is derived from the results of mapping CPD repair in defined regions of the DHFR gene in rodent and human cells (8,30,37–40). Ample evidence has demonstrated that in both rodent and human cells, CPD repair is much faster in the coding region of this gene than in the 3′ downstream non-coding region, and is also much faster in the T strand than in the NT strand (8,30,37–40). Based on these results, the concept of two subpathways for the repair of bulky DNA damage by NER has emerged during the past decade. Hereditary defects and somatic mutations that lead to defects in either subpathway have also been found (41–45). Human cells are proficient in both pathways. Cultured rodent cells, however, are proficient in TCR but are deficient in GGR of CPDs (9). It has been found that human xeroderma pigmentosum complementation group C (XPC) cells are deficient in GGR, while Cockayne syndrome (CS) cells and rodent ERCC6 cells are deficient in TCR (39,40,44–47). Interestingly, despite great efforts by many laboratories, the mechanism of TCR in mammalian cells remains unclear. In E.coli cells, not only has the mfd gene, which regulates TCR, been identified, but also the role of the mfd protein in TCR has been elucidated in a cell-free system (48,49). In contrast, even though all the major NER factors, including CS proteins, have been purified and efficient NER can be reconstituted in vitro (13), the TCR phenomenon has not yet been seen in a cell-free cell lysate system.
The current model proposed in the literature suggests that DNA damage that serves as a substrate for TCR should block transcription; this blockage gives rise to a special signal that attracts NER factors to the damaged site and subsequently allows NER to proceed (50). However, it has been found that in some cell-free cell lysate systems, blockage of the transcription process by DNA damage actually hinders NER (51,52). This finding led to the proposition that the blocked transcription machinery may retreat from the damaged site to allow the attraction of NER factors to the site (4), which raises the possibility that TCR could occur more often at the 5′ end of the gene if significant amounts of abortive transcription take place. It is also possible that the transcriptionally active genes, because of transcription factor binding, exist in a ‘state’ of chromatin structure that is more susceptible to NER, thereby allowing bulky DNA damage along the transcriptionally active genes to be evenly repaired in the T strand. It is well established that the 5′ end of the DHFR gene is transcribed more frequently than the 3′ end of the gene because of abortive transcription (53). Controversial results, however, have been reported with regard to the repair pattern of CPDs along the T strand of the DHFR gene. Using a Southern blot-based DNA repair assay, both uniform repair and a 5′ to 3′ polarity effect on CPD repair along the T strand of the DHFR gene have been reported (30,37). In this study, a much more sensitive method—T4 endo V incision in combination with LMPCR—was used to measure CPD repair in exons 1, 2 and 5 of the DHFR gene at nucleotide resolution. Our finding clearly demonstrates that CPD repair in the T strand of the DHFR gene is subject to a polarity effect; CPDs are repaired more quickly at the 5′ than at the 3′ end of the gene in different chromosomal positions that exhibit different chromatin structures, which suggests that blockage of transcription triggers the process of TCR. It is likely that the transcription process per se opens up a limited area, thus allowing the repair process to take place. Our results, however, are unable to determine whether the blocked transcription machinery retreats from the DNA damage site.
We have found that 90% of CPDs of genomic DNA in UV (15 J/m2)-irradiated CHO cells remained unrepaired after 24 h of post-irradiation incubation. These results are consistent with the notion that cultured rodent cells are deficient in GGR of CPDs (9). The finding that CPDs in the NT strand of exon 1 of the endogenous DHFR gene are repaired as efficiently as in the T strand is intriguing. We have previously demonstrated that the NT strand of exon 1 of the DHFR gene is not transcribed; therefore, the repair in this region is not due to TCR of an immediately upstream transcription unit (19). In this study, we demonstrate that neither the DNA sequence nor the TCR of the upstream OST gene contributes to the efficient repair of CPDs in the NT strand of exon 1 of the endogenous DHFR gene. Our results also suggest that the efficient repair is not due to the transcription of the DHFR gene since the DHFR genes in these three cell lines are all transcriptionally active. However, we have found that chromatin in the exon 1 region of the endogenous DHFR gene is much more sensitive to MN digestion than chromatin in exons 2 and 5 (19). We have also found that in the translocated DHFR gene, the disappearance of efficient CPD repair in the NT strand of exon 1 is accompanied by the disappearance of sensitivity to MN digestion in this region. Together, these results suggest that certain open chromatin structures in rodent cells allow efficient repair to occur in the NT strand without the help of the transcription process. The effect of this open chromatin structure on NER may be similar to the effect of blocked transcription on NER and most probably is able to attract NER factors to the damaged sites and allows NER to proceed. Recently, Sancar and colleagues have shown that the chromatin-remodeling factor SWI/SNF can greatly enhance NER on mononucleosome core substrates, but has no effect on NER on a naked DNA substrate (54). Perhaps the chromatin at exon 1 of the endogenous DHFR gene is favored to associate with this type of remodeling factor and/or is remodeled by histone acetylation or phosphorylation, which consequently allows NER to take place.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Steven Lloyd for the generous gift of T4 endo V, Dr Aziz Sancar for the generous gift of plasmid pMS969 containing the photolyase gene, and Dr Yen-Yee Tang for critical review. This research was supported by ES03124 and ES08389.
REFERENCES
- 1.Friedberg E.C. (1985) DNA damage. In Friedberg,E.C. (ed.), DNA Repair. W.H.Freeman and Company, New York, pp. 1–78. [Google Scholar]
- 2.Friedberg E.C., Walker,G.C. and Siede,W. (1995) Nucleotide excision repair in mammalian cells: general considerations and chromatin dynamics. In Friedberg,E.C., Walker,G.C. and Siede,W. (eds), DNA Repair and Mutagenesis. ASM Press, Washington, DC, pp. 283–310. [Google Scholar]
- 3.Balajee A.S. and Bohr,V.A. (2000) Genomic heterogeneity of nucleotide excision repair. Gene, 250, 15–30. [DOI] [PubMed] [Google Scholar]
- 4.Hanawalt P.C. (2001) Controlling the efficiency of excision repair. Mutat. Res., 485, 3–13. [DOI] [PubMed] [Google Scholar]
- 5.Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- 6.Wood R.D. (1997) Nucleotide excision repair in mammalian cells. J. Biol. Chem., 272, 23465–23468. [DOI] [PubMed] [Google Scholar]
- 7.de Laat W.L., Jaspers,N.G. and Hoeijmakers,J.H. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- 8.Mellon I., Spivak,G. and Hanawalt,P.C. (1987) Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell, 51, 241–249. [DOI] [PubMed] [Google Scholar]
- 9.Hanawalt P.C. (2001) Revisiting the rodent repairadox. Environ. Mol. Mutagen., 38, 89–96. [DOI] [PubMed] [Google Scholar]
- 10.Evans E., Moggs,J.G., Hwang,J.R., Egly,J.M. and Wood,R.D. (1997) Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J., 16, 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzder S.N., Habraken,Y., Sung,P., Prakash,L. and Prakash,S. (1995) Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A and transcription factor TFIIH. J. Biol. Chem., 270, 12973–12976. [DOI] [PubMed] [Google Scholar]
- 12.Mu D., Hsu,D.S. and Sancar,A. (1996) Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem., 271, 8285–8294. [DOI] [PubMed] [Google Scholar]
- 13.Mu D., Park,C.H., Matsunaga,T., Hsu,D.S., Reardon,J.T. and Sancar,A. (1995) Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem., 270, 2415–2418. [DOI] [PubMed] [Google Scholar]
- 14.Zheng Y., Pao,A., Adair,G.M. and Tang,M.-s. (2001) Cyclobutane pyrimidine dimers and bulky chemical DNA adducts are efficiently repaired in both strands of either a transcriptionally active or promoter-deleted APRT gene. J. Biol. Chem., 276, 16786–16796. [DOI] [PubMed] [Google Scholar]
- 15.Feng Z., Hu,W., Komissarova,E., Pao,A., Hung,M., Adair,G.M. and Tang,M.-s. (2002) Transcription-coupled DNA repair is genomic context-dependent. J. Biol. Chem., 277, 12777–12783. [DOI] [PubMed] [Google Scholar]
- 16.Vreeswijk M.P., van Hoffen,A., Westland,A., Vrieling,H., van Zeeland,A.A. and Mullenders,L.H. (1994) Analysis of repair of cyclobutane pyrimidine dimers and pyrimidine 6–4 pyrimidone photoproducts in transcriptionally active and inactive genes in Chinese hamster cells. J. Biol. Chem., 269, 31858–31863. [PubMed] [Google Scholar]
- 17.Venema J., Bartosova,Z., Natarajan,A.T., van Zeeland,A.A. and Mullenders,L.H. (1992) Transcription affects the rate but not the extent of repair of cyclobutane pyrimidine dimers in the human adenosine deaminase gene. J. Biol. Chem., 267, 8852–8856. [PubMed] [Google Scholar]
- 18.Tu Y., Tornaletti,S. and Pfeifer,G.P. (1996) DNA repair domains within a human gene: selective repair of sequences near the transcription initiation site. EMBO J., 15, 675–683. [PMC free article] [PubMed] [Google Scholar]
- 19.Hu W., Feng,Z., Chasin L.A. and Tang,M.-s. (2002) Transcription-coupled and transcription-independent repair of cyclobutane pyrimidine dimers in the dihydrofolate reductase gene. J. Biol. Chem., 277, 38305–38310. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell P.J., Carothers,A.M., Han,J.H., Harding,J.D., Kas,E., Venolia,L. and Chaisn,L.A. (1986) Multiple transcription start sites, DNase I-hypersensitive sites and an opposite-strand exon in the 5′ region of the CHO dhfr gene. Mol. Cell. Biol., 6, 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wells J.M., Held,P., Illenve,S. and Heintz,N.H. (1996) Protein–DNA interactions at the major and minor promoters of the divergently transcribed dhfr and rep3 genes during the Chinese hamster ovary cell cycle. Mol. Cell. Biol., 16, 634–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C. and Chasin,L.A. (1998) Cointegration of DNA molecules introduced into mammalian cells by electroporation. Somat. Cell Mol. Genet., 24, 249–256. [DOI] [PubMed] [Google Scholar]
- 23.Maxam A.M. and Gilbert,W. (1980) Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol., 65, 499–560. [DOI] [PubMed] [Google Scholar]
- 24.Kuhnert P., Peterhans,E. and Pauli,U. (1992) Chromatin structure and DNase I hypersensitivity in the transcriptionally active and inactive porcine tumor necrosis factor gene locus. Nucleic Acids Res., 20, 1943–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng Y. and Waters,R. (2000) Excision repair at the level of the nucleotide in the upstream control region, the coding sequence and in the region where transcription terminates of the Saccharomyces cerevisiae MFA2 gene and the role of RAD26. Nucleic Acids Res., 28, 1114–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teng Y., Li,S., Waters,R. and Reed,S.H. (1997) Excision repair at the level of the nucleotide in the Saccharomyces cerevisiae MFA2 gene: mapping of where enhanced repair in the transcribed strand begins or ends and identification of only a partial rad16 requisite for repairing upstream control sequences. J. Mol. Biol., 267, 324–337. [DOI] [PubMed] [Google Scholar]
- 27.Tolbert D.M. and Kantor,G.J. (1996) Definition of a DNA repair domain in the genomic region containing the human p53 gene. Cancer Res., 56, 3324–3330. [PubMed] [Google Scholar]
- 28.Watanabe A., Ikejima,M., Suzuki,N. and Shimada,T. (1996) Genomic organization and expression of the human MSH3 gene. Genomics, 31, 311–318. [DOI] [PubMed] [Google Scholar]
- 29.Dijkwel P.A. and Hamlin,J.L. (1988) Matrix attachment regions are positioned near replication initiation sites, genes and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol. Cell. Biol., 8, 5398–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spivak G. and Hanawalt,P.C. (1996) Fine structure mapping of DNA repair within a 100 kb genomic region in Chinese hamster ovary cells. Mutat. Res., 350, 207–216. [DOI] [PubMed] [Google Scholar]
- 31.Morales V., Giamarchi,C., Chailleux,C., Moro,F., Marsaud,V., Le Ricousse,S. and Richard-Foy,H. (2001) Chromatin structure and dynamics: functional implications. Biochimie, 83, 1029–1039. [DOI] [PubMed] [Google Scholar]
- 32.Li S. and Smerdon,M.J. (2002) Nucleosome structure and repair of N-methylpurines in the GAL1-10 genes of Saccharomyces cerevisiae. J. Biol. Chem., 277, 44651–44659. [DOI] [PubMed] [Google Scholar]
- 33.Meijer M. and Smerdon,M.J. (1999) Accessing DNA damage in chromatin: insights from transcription. Bioessays, 21, 596–603. [DOI] [PubMed] [Google Scholar]
- 34.Liu X. and Smerdon,M.J. (2000) Nucleotide excision repair of the 5S ribosomal RNA gene assembled into a nucleosome. J. Biol. Chem., 275, 23729–23735. [DOI] [PubMed] [Google Scholar]
- 35.Hara R., Mo,J. and Sancar,A. (2000) DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol., 20, 9173–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ura K., Araki,M., Saeki,H., Masutani,C., Ito,T., Iwai,S., Mizukoshi,T., Kaneda,Y. and Hanaoka,F. (2001) ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J., 20, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bohr V.A., Okumoto,D.S., Ho, L. and Hanawalt, P.C. (1986) Characterization of a DNA repair domain containing the dihydrofolate reductase gene in Chinese hamster ovary cells. J. Biol. Chem., 261, 16666–16672. [PubMed] [Google Scholar]
- 38.Bohr V.A., Smith,C.A., Okumoto,D.S. and Hanawalt,P.C. (1985) DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell, 40, 359–369. [DOI] [PubMed] [Google Scholar]
- 39.Venema J., Mullenders,L.H.F., Natarajan,A.T., van Zeeland,A.A. and Mayne,L.V. (1990) The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc. Natl Acad. Sci. USA, 87, 4707–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Hoffen A., Natarajan,A.T., Mayne,L.V., van Zeeland,A.A., Mullenders,L.H. and Venema,J. (1993) Deficient repair of the transcribed strand of active genes in Cockayne′s syndrome cells. Nucleic Acids Res., 21, 5890–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Boer J. and Hoeijmakers,J.H. (2000) Nucleotide excision repair and human syndromes. Carcinogenesis, 21, 453–560. [DOI] [PubMed] [Google Scholar]
- 42.de Gruijl F.R., van Kranen,H.J. and Mullenders,L.H. (2001) UV-induced DNA damage, repair, mutations and oncogenic pathways in skin cancer. J. Photochem. Photobiol. B, 63, 19–27. [DOI] [PubMed] [Google Scholar]
- 43.Hanawalt P.C. (1996) Role of transcription-coupled DNA repair in susceptibility to environmental carcinogenesis. Environ. Health Perspect., 104 (Suppl.), 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Hoffen A., Venema,J., Meschini,R., van Zeeland,A.A. and Mullenders,L.H.F. (1995) Transcription-coupled repair removes both cyclobutane pyrimidine dimers and 6–4 photoproducts with equal efficiency and in a sequential way from transcribed DNA in xeroderma pigmentosum group C fibroblasts. EMBO J., 14, 360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venema J., van Hoffen,A., Karcagi,V., Natarajan,A.T., van Zeeland,A.A. and Mullenders,L.H. (1991) Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol. Cell. Biol., 11, 4128–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cooper P.K., Nouspikel,T., Clarkson,S.G. and Leadon,S.A. (1997) Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science, 275, 990–993. [DOI] [PubMed] [Google Scholar]
- 47.O'Donovan A. and Wood,R.D. (1993) Identical defects in DNA repair in xeroderma pigmentosum group G and rodent ERCC group 5. Nature, 363, 185–188. [DOI] [PubMed] [Google Scholar]
- 48.Selby C.P., Witkin,E.M. and Sancar,A. (1991) Escherichia coli mfd mutant deficient in ‘mutation frequency decline’ lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc. Natl Acad. Sci. USA, 88, 11574–11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selby C.P. and Sancar,A. (1993) Molecular mechanism of transcription–repair coupling. Science, 260, 53–58. [DOI] [PubMed] [Google Scholar]
- 50.Hanawalt P.C. (1994) Transcription-coupled repair and human disease. Science, 266, 1957–1958. [DOI] [PubMed] [Google Scholar]
- 51.Selby C.P. and Sancar,A. (1990) Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J. Biol. Chem., 265, 21330–21336. [PubMed] [Google Scholar]
- 52.Tornaletti S., Reines,D. and Hanawalt,P.C. (1999) Structural characterization of RNA polymerase II complexes arrested by a cyclobutane pyrimidine dimer in the transcribed strand of template DNA. J. Biol. Chem., 274, 24124–24130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frayne E.G., Leys,E.J., Crouse,G.F., Hood,A.G. and Kellems,R.E. (1984) Transcription of the mouse dihydrofolate reductase gene proceeds unabated through seven polyadenylation sites and terminates near a region of repeated DNA. Mol. Cell. Biol., 4, 2921–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hara R. and Sancar,A. (2002) The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol. Cell. Biol., 22, 6779–6787. [DOI] [PMC free article] [PubMed] [Google Scholar]