Abstract
R6K-encoded π protein can bind to the seven, 22 bp tandem iterons of the γ origin. In this work, we use a variant of π, His-π·F107S, that is hyperactive in replication. In vitro, His-π·F107S-dependent local DNA melting (open complex formation) occurs in the absence of host proteins (IHF/HU or DnaA) and it is positioned in the A + T-rich region adjacent to iterons. Experiments described here examine the effects of ATP, Mg2+ and temperature on the opening reaction. We show that the opening of the γ origin can occur in the presence of ATP as well as AMP-PCP (a non-hydrolyzable ATP analog). This suggests that, for γ origin, ATP hydrolysis may be unnecessary for open complex formation facilitated by His-π·F107S. In the absence of ATP or Mg2+, His-π·F107S yielded data suggestive of distortions in the iteron attributable to DNA bending rather than DNA melting. Our findings also demonstrate that ATP and π stimulate open complex formation over a wide range of temperatures, but not at 0°C. These and other results indicate that ATP and/or Mg2+ are not needed for His-π·F107S binding to iterons and that ATP effects an allosteric change in the protein bound to γ origin.
INTRODUCTION
In many systems, replication proteins (Reps) bind to iterons within their cognate origins (oris) and facilitate the assembly of replisomes on DNA. One of the early steps required for this assembly is formation of the so-called open complex (1).
Open complex formation has been investigated for only a few bacterial Rep/iteron systems. Under most circumstances, the concerted action of both plasmid-encoded Rep protein and host-encoded DnaA is required for the formation of open complexes in the A + T-rich region adjacent to the iterons. One role of DnaA in initiation may be to generate energy for this reaction and, in fact, DnaA possesses ATPase activity whereas the Rep proteins themselves (other than DnaA) have not been examined in this regard. However, ATP hydrolysis is not obligatory for DnaA function in the melting of oriC (2). Rather, available observations support crucial roles for DnaA-ATP and DnaA-ADP complexes in regulating the replication of oriC at the stages following open complex formation (3–6).
Investigations of open complex formation in plasmid systems have been conducted, in vitro, with the aid of hyperactive variants of Rep proteins (copy-up). A copy-up Rep variant was chosen for study because the activity of purified, wild-type Rep is hardly detectable in these experiments (7–9). Data from the Rep variants show a correlation between their elevated replication activity, in vivo, and an enhanced ability to form open complexes, in vitro [mini-F (10), RK2 (11), mini-P1 (12), R6K γ ori (13,14)]. Typically, DnaA and IHF/HU were included in the in vitro experiments.
This laboratory is investigating a minimal replicon that is derived from an antibiotic resistance plasmid, R6K [reviewed in Filutowicz et al. (15) and Filutowicz and Rakowski (16)]. The minimal replicon consists of the γ ori (Fig. 1) and the pir gene that encodes π protein (17). Low levels of π stimulate γ ori replication, whereas, at high levels, the protein inhibits replication (18–20). It has been proposed that π monomers are activators of replication while π dimers are inhibitors (13,21,22). Consistent with this model is the observation that both monomers and dimers of π can bind to iterons in vitro (13,21,22). Experiments using hyperactive and dominant-negative π variants suggest that monomers facilitate open complex formation in γ ori while dimers do not (13). Strand separation in the open complex occurs in the A + T-rich segment of γ ori (Fig. 1) near the start sites for leading strand synthesis and a putative stem–loop structure in the DNA (9,13).
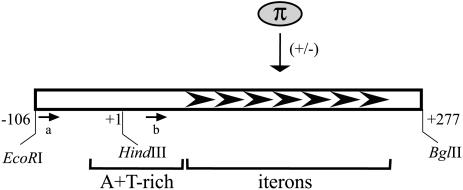
Figure 1.
The segment of the R6K γ ori relevant to this study including: the 106 bp enhancer, the 277 bp core and the 112 bp A + T-rich region (nt –20 to +92). Seven tandem iterons to which the Rep protein (π) binds are shown as black arrowheads. The activation and inhibition of replication by monomers and dimers, respectively, of the protein are indicated by plus and minus signs. Arrows labeled ‘a’ and ‘b’ mark the positions for the primers used in this study. Coordinates shown (–106, 1 and 277) are according to Stalker et al. (23).
The activities of many regulatory proteins are modulated by a ligand. For example, there are systems in which ATP is believed to cause an allosteric change in Rep protein (24,25). Studies of Rep-dependent DNA duplex opening have characteristically included ATP and Mg2+ (10–13). Notably, π lacks any identifiable ATP-binding motif, prompting the question: is ATP, in fact, required for open complex formation at γ ori? The addition of Mg2+ to KMnO4 footprinting reactions examining polymerase–promoter interactions has been shown to increase KMnO4-dependent oxidation of pyrimidines (26,27). It was suggested that the Mg2+ ions might elicit this effect by shielding the negatively charged groups on the DNA surface, thus lowering repulsive interactions with MnO4– anions. Another characteristic of Mg2+ (of particular relevance to this work) is that it may also form metal chelates with ribonucleoside 5′-triphosphates such as ATP (28).
For these reasons, we set out to determine if, and how, ATP and Mg2+ (as well as temperature) affect the reactivity of γ ori DNA to KMnO4 in the presence of π protein. A copy-up π variant, π·F107S, was chosen for this work because (unlike wild-type π) it generates strong signals in our assays and it is known to stimulate open complex formation in the absence of IHF and/or DnaA. This property simplifies data interpretation, particularly in view of the fact that KMnO4 probing experiments have suggested that IHF alone can perturb DNA structure in the A + T-rich segment of γ ori (13). To facilitate protein purification, we previously constructed a His6-tagged form of π·F107S; the properties of His-π·F107S have been shown to be very similar to those of the untagged copy-up protein in vivo and in vitro (9,29).
Mobility shift assays, presented here, suggest that iteron DNA is bent in complexes with bound His-π·F107S, and we will discuss the surprising similarity in DNA bending angles produced by protein monomers and dimers. ATP is needed for π·F107S-dependent duplex opening but not for the protein’s ability to bind and bend iteron DNA. Adding these new data to what is already know about Rep/iteron systems, we set forth the hypothesis that ATP probably stimulates the remodeling of His-π·F107S–γ ori complexes from a closed to an open conformation, eliciting its effect via interaction with π monomers.
MATERIALS AND METHODS
Plasmids
The monomeric form of plasmid pMF36 (18) used for KMnO4 probing was obtained by double equilibrium centrifugation in a CsCl–ethidium bromide gradient. Construction of plasmid pRK20, containing an iteron sequence, is described in this work.
Probing DNA reactivity to KMnO4
A typical reaction (25 µl) contained: 25 mM Tris–HCl pH 8.0 (or 40 mM HEPES–KOH pH 8.0), 5 mM MgSO4, 3 mM EDTA, 7.5% glycerol, 40 mM creatine phosphate, 0.1 mg/ml creatine kinase, 0.005% Triton X-100 and 0.3 µg of supercoiled pMF36 template. Amounts of ATP or AMP-PCP (0–32 mM) are indicated in the figure legends. The reactions were incubated for 15 min at 37°C unless otherwise indicated in the figure. KMnO4 was added to a final concentration of 10 mM. After incubation, the reactions were terminated by the addition of 2 µl of β-mercaptoethanol (14 M) and 1 µl of EDTA (0.5 M). Plasmid DNA was recovered using an Ultra Clean 15 DNA purification kit (MolBio) or microspin columns (Qiagen) following the supplier’s recommendations.
Primer extension
The plasmid DNA recovered after KMnO4 treatment was annealed with the following 32P-labeled primers: 5′-CTTT GAGAGGCTCTAAGGG3-3′ (nt _74 to _56; primer ‘a’, Fig. 1) and 5′-TAGAGGCTATTTAAGTTGC-3′ (nt +50 to +68; primer ‘b’, Fig. 1).
PCR incubations were done as follows: 1 min at 95°C, 1 min at 55°C and 1 min at 72°C for a total of 15 cycles. The reactions were stopped by adding stop solution (Sequenase kit, USB), heat denatured and loaded onto an 8% polyacrylamide sequencing gel. Quantification was done using a Phosphor Imager Storm (Molecular Dynamics). The positions of the areas modified by KMnO4 were determined by comparison with a sequencing ladder run in parallel on the same gel. The Taq polymerase used in this work (TaKaRa) for the primer extension reactions contains an exonucleolytic activity. For this reason, primer extension products terminate one or two nucleotides short of the actual sites of KMnO4 modification. This conclusion is based on our comparison of the extension products generated by an exonuclease-free Taq polymerase from Fisher Scientific.
Electrophoretic mobility shift/bending assay
DNA fragments of 140 bp were generated from pRK20, gel purified and labeled with 32P. The labeled fragments were then incubated with His-π·F107S protein; 130 ng of poly(dI–dC):poly(dI–dC) and 500 ng of pBEND5 plasmid were added to minimize non-specific binding. Nucleoprotein complexes were then resolved by electrophoresis on 8% polyacrylamide gels. The position of each electrophoretically retarded band was determined using a PhosphorImager Storm and its Imagequant software. Briefly, signal quantification was done along a line traced in each gel lane and the peak of greatest signal was taken as the band position. The position of each nucleoprotein complex was compared with the migration of the free DNA in its respective lane (relative eletrophoretic mobility). The data were used to determine the µM/µE ratios, and the equation µM/µE = cosθ/2 was employed to calculate the bending angles as described (30).
RESULTS
The reactivity of bases within the A + T-rich segment of γ ori is ATP dependent
Prior to the experiments described here, two groups reported that open complex formation on a superhelical, γ ori DNA template is dependent on π protein (13,14). In the presence of π, enhanced reactivity to KMnO4 [that preferentially modifies unpaired T and C (31)] has been observed for both DNA strands within the A + T-rich segment of the ori. The results that follow are a progression of this earlier work as we explore the contributions of various components to open complex formation at γ ori.
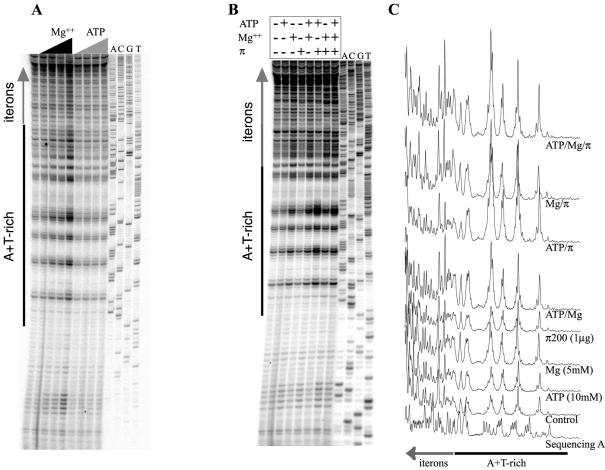
Herein, assays were performed by monitoring the reactivity of DNA to KMnO4 in the presence of His-π·F107S. Primer extension products from KMnO4-modified DNA templates are shown in Figure 2. Incubations were conducted with and without His-π·F107S as well as in the presence and absence of increasing levels of ATP (Fig. 2A and B). We found that His-π·F107S added alone affected the reactivity to KMnO4 within iterons but not within the A + T-rich segment of the template DNA. ATP, added alone, did not change the template’s reactivity to KMnO4. In contrast, His-π·F107S added in combination with ATP did enhance the reactivity of DNA to KMnO4 within the A + T-rich segment of γ ori. The His-π·F107S-dependent enhanced reactivity within iterons together with the ATP-dependent shift in the reactivity towards the A + T-rich segment suggest that the His-π·F107S–γ ori DNA complex probably undergoes a conformational change (remodeling) in an ATP-dependent fashion.
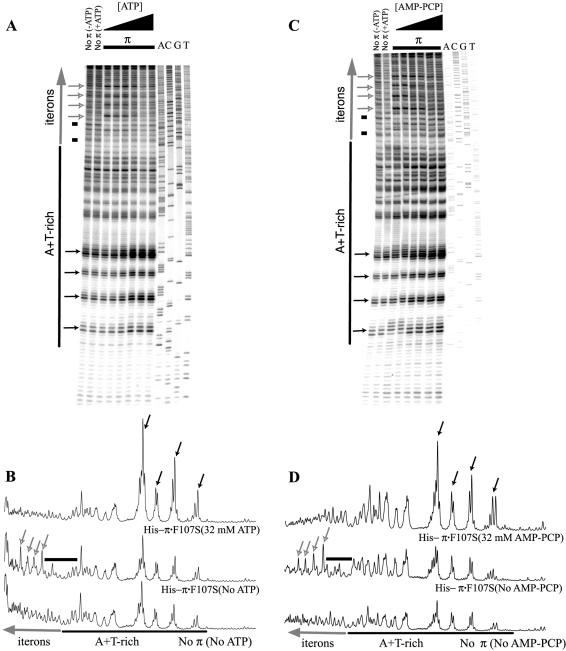
Figure 2.
Reactivity of γ ori DNA to KMnO4 in the presence of His-tagged, copy-up π protein and increasing concentrations of ATP or AMP-PCP. pMF36 DNA template was incubated with 1.00 µg of the copy-up variant protein and with 0, 2, 4, 8, 16 or 32 mM ATP (A), or AMP-PCP (C). For the control reaction, the reactivity of the DNA probe to KMnO4 in the absence of protein and/or ATP/AMP-PCP was examined. Reactive bases were identified using the DNA sequencing ladder displayed in the last four lanes. The positions of the A + T-rich region [black bar and Chen et al. (9)] and iterons (large, vertical, gray arrow) are indicated. Reaction products were processed as described (13). Autoradiographs were quantified (B and D) as described (13). Quantifications of lanes containing no protein (without ATP or AMP-PCP), protein only (without ATP or AMP-PCP) and protein with ATP or AMP-PCP (32 nM) are presented. Bands of interest (singular or in clusters) are indicated as follows: small black arrows highlight bands of increasing intensity; small gray/white arrows highlight bands of decreasing intensity; black bars indicate decreased band intensities at low nucleotide levels in the presence of Rep protein.
AMP-PCP, a non-hydrolyzable analog of ATP, also stimulates remodeling of His-π·F107S–γ ori DNA complexes
We next carried out the KMnO4 probing experiments using AMP-PCP, a non-hydrolyzable ATP analog (32), to determine whether ATP hydrolysis is required for open complex formation. With one exception, no major differences between the effects of ATP and AMP-PCP were observed (Fig. 2A and C, respectively). The exception was confined to the A + T-rich segment that is proximal to the first iteron. From this, we conclude that hydrolysis of ATP is unlikely to be necessary for KMnO4-dependent modification of bases in the A + T-rich region of γ ori.
His-π·F107S, Mg2+ and ATP elicit a variety of effects on the KMnO4 reactivity of γ ori, both singly and in combinations
Since KMnO4 probing typically reveals DNA distortions other than DNA melting (30), we set out to test whether the His-π·F107S-dependent KMnO4 modifications within the iterons occur in one or both DNA strands. KMnO4 reactivity at nearby sites in both strands would be suggestive of DNA melting. As shown in Figure 3, the reactivity of bases to KMnO4 (in the absence of ATP) was observed only on the bottom strand. Thus, the ATP-independent binding of His-π·F107S facilitates changes in the iteron DNA that are probably distinct from DNA melting. We will present a permutation analysis of DNA fragments containing a single iteron that suggests a possible correlation between the reactivity of iteron DNA to KMnO4 and DNA bending.
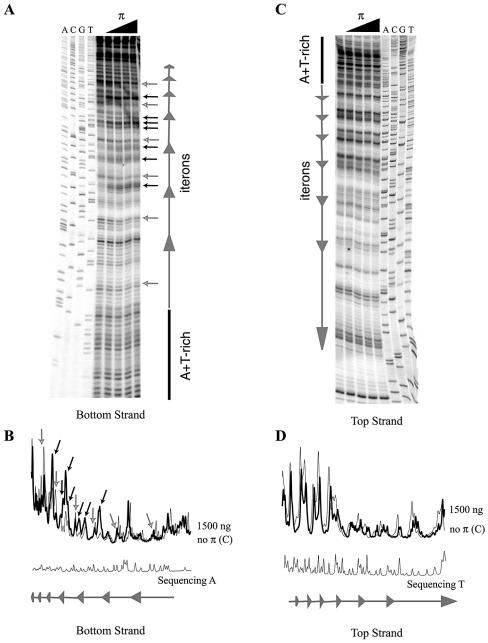
Figure 3.
Reactivity of γ ori iteron DNA to KMnO4 in the presence of His-tagged, copy-up π protein (and in the absence of ATP). Only the iteron segment was analyzed by primer extension utilizing primers that anneal close to this region (see Materials and Methods.) Bottom strand autoradiogram (A) and quantification (B) panels are on the left. Top strand data (C and D) are on the right. Only quantification from lanes containing no protein or 1.5 µg of the copy-up variant as well as the sequence ladder are shown. Bands of interest (singular or in clusters) are indicated as follows: small black arrows highlight bands of increasing intensity; small gray/white arrows highlight bands of decreasing intensity.
All reactions thus far described were carried out in the presence of Mg2+. We considered the possibility that what we call ‘ATP-dependent remodeling’ could be an artifact caused by ATP’s titration of the metal; the resulting Mg2+ depletion could lead to a change in DNA reactivity to KMnO4 (see Introduction). To test this, we performed KMnO4 probing reactions in the presence and absence of Mg2+ (and with increasing concentrations of ATP). As shown in Figure 4 and demonstrated earlier (Fig. 2A), the presence of ATP alone does not change the susceptibility of the DNA template to KMnO4. However, the presence of increasing concentrations of Mg2+ alone progressively increased the DNA template’s reactivity to the probing agent. Thus our results are in agreement with Mg2+ effects observed by others (26,27).
Figure 4.
KMnO4 reactivity of the bottom (A) and top (C) strands of γ ori in the absence of Mg2+. pMF36 DNA template was incubated with His-tagged, copy-up π protein (250, 500 and 1000 ng) and/or ATP (4, 8 or 16 mM), as indicated by black, gray and dark gray triangles, respectively. All reactions were performed without Mg2+. For the control reaction, the reactivity of the probe to KMnO4 in the absence of protein and ATP was examined. Quantification results for selected lanes are given (B and D). Bands of interest (singular or in clusters) are highlighted by small black arrows.
Next, to better define the individual contributions of His-π·F107S, ATP and Mg2+, we set up KMnO4 reactions containing these components individually and in combinations. As shown in our control sample (Fig. 5), the presence of His-π·F107S stimulated KMnO4 reactivity within the iterons. Again, the presence of Mg2+ with or without ATP increased the overall DNA reactivity to KMnO4. The combination of His-π·F107S and Mg2+ increased the iterons’ susceptibility to KMnO4 modifications when compared with reactions with His-π·F107S alone. In contrast, reactions carried out with His-π·F107S and ATP showed the strongest KMnO4 reactivity within the A + T-rich region. The presence of all three components (His-π·F107S, ATP and Mg2+) enhanced reactivity to KMnO4 within the A + T-rich segment but not within iterons. In summary, regardless of the presence or absence of Mg2+, His-π·F107S (alone) stimulates KMnO4 modifications within the iterons, while His-π·F107S plus ATP stimulates modifications within the A + T-rich segment.
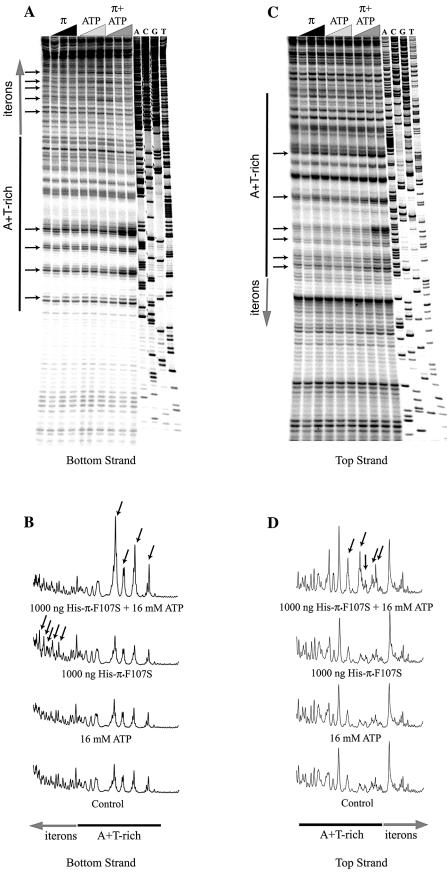
Figure 5.
Contributions of ATP and Mg2+ to the KMnO4 signals within the γ ori. (A) The control lane (unlabeled) is a KMnO4 reaction without ATP or Mg2+. Increasing concentrations of Mg2+ (0, 2.5, 5 and 10 mM) or ATP (0, 4, 8 and 16 mM) were used, as indicated by black and dark gray triangles, respectively. (B) Combinations of His-tagged, copy-up π protein (1 µg), Mg2+ (5 mM) and ATP (10 mM) were used to test the contributions of each of these components. Added components are indicated by ‘+’ and omitted components by ‘–’. Quantification results for (B) are shown in (C).
Bending angles induced by binding of monomers and dimers of His-π·F107S appear to be similar
How does His-π·F107S change the reactivity of iteron DNA to KMnO4? A reasonable hypothesis is that it does so by bending the DNA. Indeed, it has been shown that π, in the form of a 38 kDa fusion protein, bends and unwinds iteron-containing DNA (33). Analyzing and interpreting these data is a challenge given our current understanding of the complex interactions of π monomers and dimers (replication activators and inhibitors, respectively) with a DNA probe containing seven iterons. (13,34,35). To simplify analysis of π-mediated DNA bending, we carried out assays with a series of the DNA probes containing positioning permutations of a single iteron.
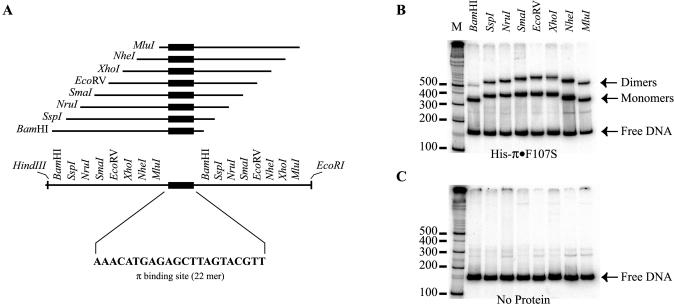
A set of eight restriction fragments was used in π-mediated DNA bending experiments. Fragments were equal in length but differed in the positioning of the iteron sequence relative to the ends of the fragments (Fig. 6A). The electrophoretic mobilities of these fragments were then compared in the presence and absence of His-π·F107S. DNA curvature can have different effects on a fragment’s migration in a gel depending on where the bend occurs (Fig. 6B and C). Our series of permuted fragments showed no migrational differences in the absence of protein. This observation suggests a lack of iteron-induced, intrinsic DNA curvature.
Figure 6.
Iteron DNA binding assay. (A) Construction of a series of DNA fragments with permuted iteron sequence. Blunt end, double-stranded, 22 bp iteron DNA (sequence shown) was generated from synthetic, single-stranded oligos and ligated into HapI-digested pBEND5 (36) giving rise to the plasmid pRK20. Several DNA probes were generated from this plasmid by digestion with restriction enzymes, as indicated, variously placing the iteron sequence near either end or near the middle of the DNA fragment. (B and C) The results of electrophoretic mobility shift assays. The positions of π monomers, dimers and free DNA are indicated by arrows. M is a 32P-labeled 100 bp ladder for DNA fragment migration reference. In (B), DNA fragments were incubated with Rep protein as described in Materials and Methods. (C) is a ‘negative’ control incubated without π protein to reveal any bending that might be intrinsic to the iteron sequence.
As shown in Figure 6, when His-π·F107S was mixed with probe DNA, two electrophoretically retarded bands emerged. These bands contained bound monomers (fast migrating band) and dimers (slow migrating band) of His-π·F107S (13,22). Comparing the results from different DNA fragments, some complexes of the series migrate slower, namely those with the iteron nearest to the center; other fragments migrate faster (those with the iteron nearest to an end). The relative electrophoretic mobilities of DNA–protein complexes for monomers and dimers of His-π·F107S were calculated according to Thompson and Landy (30) from three experiments in which, typically, five fragment permutations were used. The relative electrophoretic mobility (µM/µE) for monomers of His-π·F107S was calculated for EcoRV–BamHI (0.88) and EcoRI–MluI (0.88). This corresponded to an apparent bending angle of 56° (±2.3°). The value of µM/µE for dimers of His-π·F107S was also calculated for EcoRV–BamHI (0.81) and EcoRI–MluI (0.83). This corresponded to an apparent bending angle of 67° (±1.7°). Therefore, the difference between monomer- and dimer-induced DNA bending is surprisingly small. The significance of this observation will be discussed.
His-π·F107S enhances the natural instability of the γ ori A + T-rich region
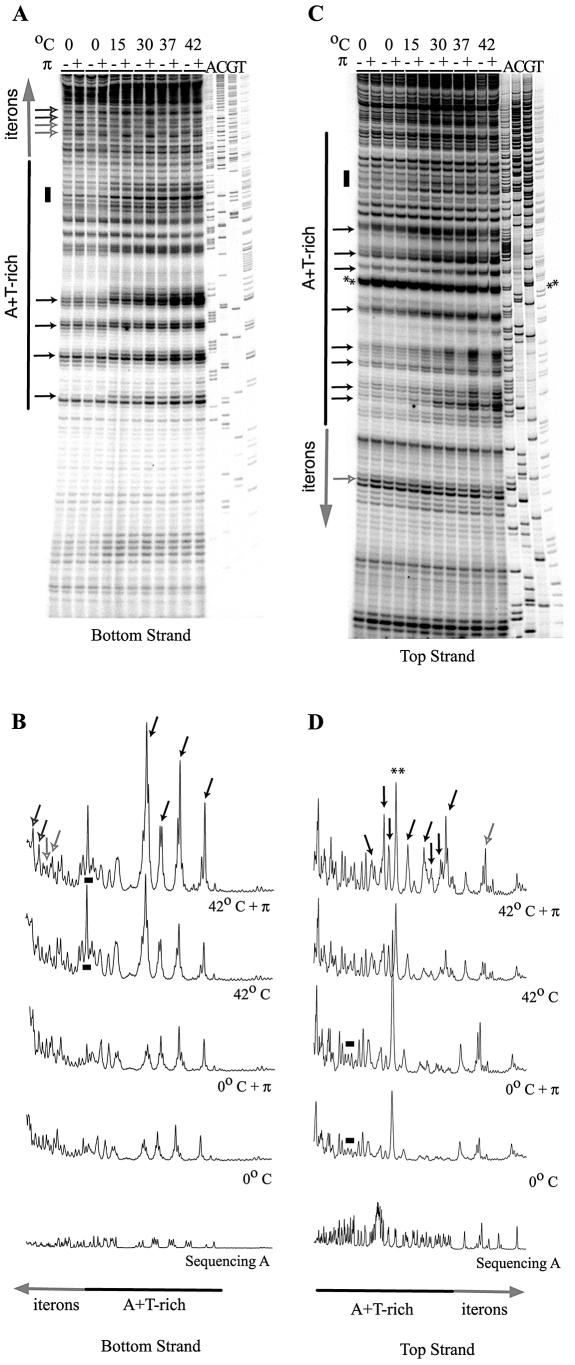
His-π·F107S-dependent DNA melting occurs immediately upstream of the sites where DNA synthesis commences. This poses the question as to how much of the melting is attributable to the ATP-protein–DNA complex and how much is intrinsic to the structure of the A + T-rich DNA itself. This question was addressed by examining the effect of temperature on open complex formation (Fig. 7; KMnO4 probing experiments were performed in the presence of ATP). The results show that raising the temperature of incubation increased the reactivity of the A + T-rich segment to KMnO4 in the absence of His-π·F107S. However, at all temperatures tested except 0°C, the KMnO4 reactivity was enhanced when His-π·F107S (and ATP) was included. Even at 0°C, the iteron DNA, but not the A + T-rich DNA, is clearly reactive to KMnO4 when His-π·F107S is present. Thus, the enhancing effect of ATP is nullified at 0°C but π still elicits its ATP-independent activity.
Figure 7.
The effect of temperature on the KMnO4 reactivity of the γ ori. Reactions were incubated at 0, 15, 30, 37 or 42°C as indicated. (A and B) The bottom strand autoradiogram and selected quantification results, respectively. (C and D) Data from the top strand. Added His-tagged, copy-up π protein is indicated by ‘+’ and omitted protein by ‘–’. Asterisks show the two most KMnO4-reactive bases (T49 and T50) within the A + T-rich region; the reactivity is Rep independent. The same reactivity was evident in the top strand autoradiograph from the experiment documented in Figure 4. Bands of interest (singular or in clusters) are indicated as follows: small black arrows highlight Rep-sensitive bands of increasing intensity; small gray/white arrows highlight Rep-sensitive bands that are temperature independent; small black/gray arrows highlight Rep-sensitive bands with peak intensities at intermediate temperatures; black bars highlight temperature-sensitive, Rep-independent bands.
An interesting side note to this series of experiments is the observation of strong KMnO4 reactivity at bases T49 and T50 within the A + T-rich region (Fig. 7, marked with asterisks) that occurred in a π-independent fashion. This reactivity suggests that these bases are unstacked in supercoiled plasmid DNA. Previously, we speculated that a putative stem–loop structure could form within the A + T-rich segment; T49 and T50 fall within the predicted loop structure which lies near the start sites for leading strand synthesis (9).
DISCUSSION
ATP stimulates the remodeling of π–γ ori DNA complexes
The results described here support a possible role for ATP in open complex formation at γ ori. An ATP dependence of open complex formation might be expected if, for example, the nucleotide was needed for the binding of π to iterons. However, at least two observations argue against this idea. First, ATP is not required, in vitro, for π binding to linear DNA (37) or superhelical DNA (34) containing iterons. Secondly, the ability of His-π·F107S to bind a superhelical template can be inferred from the primer extension assays that are reported here. In fact, in samples containing His-π·F107S, specific bases within the iterons (as opposed to the A + T-rich segment) exhibit enhanced reactivity to KMnO4 only when ATP is absent or supplied at low levels. Diminished reactivity in the presence of ATP is also evident in the A + T-rich segment proximal to the first iteron. Nonetheless, as the ATP levels increase and the enhancements in, and proximal to, the iterons gradually diminish, the enhancements well within the A + T-rich segment become prominent.
DNA strand separation versus DNA distortions
π-dependent bending of iteron DNA has been reported by Mukherjee et al. (33). Extending this observation, we have found that monomers and dimers of π bend a single iteron-containing DNA fragment by ∼56° and 67°, respectively. This observation was unexpected and suggests that DNA bending alone cannot explain why π monomers are replication activators and π dimers are not. Also, it is unclear whether the strand-specific KMnO4 modifications within the iteron-bearing region are the result of monomers or dimers of His-π·F107S binding and bending the DNA. Two dominant-negative π variants that bind and appear to bend (preliminary data, not shown) iteron DNA do not stimulate open complex formation in the presence of ATP (13). Because these two proteins bind iterons solely as dimers [not as monomers (13)], we hypothesize that dimers probably cannot undergo an allosteric change in the presence of ATP (leading to open complex formation) while monomers seemingly can.
We have proposed that protein–protein interactions between neighboring π monomers and π dimers bound to tandem iterons are not identical (22). This assumption is strongly supported by the solved crystallographic structures of two other members of the Rep protein family, plasmid F-encoded RepE (38) and plasmid pPS10-encoded RepA (39). The structure of monomeric RepE54 with its cognate iteron revealed a pseudo-symmetric protein comprised of two winged-helix domains (WH). The WH is a fold consisting of a helix–turn–helix DNA-binding motif with one or two β-hairpin wings. Monomers of RepE proteins undergo a remodeling upon dimerization as revealed by the crystallographic structure of the dimerization domain of RepA (39,40). If the Rep protein of R6K behaves similarly, it would not be unreasonable to expect that the architecture of γ ori complexes containing bound π monomers differs from the architecture when π dimers are bound (22). It appears that the binding of iterons by π monomers, but not dimers, is required and yet insufficient for stimulating open complex formation at the nearby A + T-rich segment. ATP, but not its hydrolysis, also seems to be required for the double-stranded DNA to open.
The relationship of our results to other systems
With regard to the stimulatory role of ATP in π-dependent, open complex formation (at γ ori), both similarities and differences are evident when comparisons are made with other systems. For instance, in the Introduction, we noted that ATP hydrolysis is not essential for oriC melting (41), a characteristic that γ ori seems to share. However, in a departure from what we see with γ ori, duplex DNA at oriV (plasmid RK2) could be opened in the absence of ATP although these experiments used a hyperactive form of Rep (His-TrfA254D/267L) in conjunction with HU and/or DnaA (11). Even under these conditions, however, the open complex appeared to be formed in a larger fraction of DNA molecules when ATP was included in the reactions.
ATP hydrolysis-independent effects have also been observed in two other systems and proposed to depend on the allosteric changes of Rep proteins. Monomers of the SV40-encoded T antigen, in the presence of ATP, assemble into a double hexamer on the core repeats and facilitate DNA duplex opening (24,31). In the absence of ATP, the opening does not occur and a tetrameric structure is the largest found at the core origin (24). Additionally, ATP-dependent changes in both DNA conformation and helix opening have been reported for the above-mentioned RepE54 (25). In this system, ATP seems to affect the oligomerization of RepE54 monomers on their cognate iterons. An opening of ∼3 bp was detected within the A + T-rich region of mini-F ori DNA, and this opening was shown to be dependent on RepE and ATP (25).
We have been unable to identify a nucleotide-binding motif, such as a P-loop (42,43), in the amino acid sequence of π, whereas such a motif is present in DnaA protein (41). These results suggest that a novel nucleotide-binding motif might be utilized by π protein. Experiments are in progress to identify the ATP-binding site.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Sheryl Rakowski for her contributions of editing and manuscript preparation. This work was supported by National Institute of Health Grant GM 40314 to M.F. Support for R.K. was provided by CAPES/Brasilia/Brazil.
REFERENCES
- 1.Bramhill D. and Kornberg,A. (1988) Duplex opening by DnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell, 52, 743–755. [DOI] [PubMed] [Google Scholar]
- 2.Skovgaard O., Olesen,K. and Wright,A. (1998) The central lysine in the P-loop motif of the Escherichia coli DnaA protein is essential for initiating DNA replication from the chromosomal origin, oriC, and the F factor origin, oriS, but is dispensable for initiation from the P1 plasmid origin, oriR. Plasmid, 40, 91–99. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima T., Nishida,S., Kurokawa,K., Katayama,T., Miki,T. and Sekimizu,K. (1997) Negative control of DNA replication by hydrolysis of ATP bound to DnaA protein, the initiator of chromosomal DNA replication in Escherichia coli. EMBO J., 16, 3724–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurokawa K., Nishida,S., Emoto,A., Sekimizu,K. and Katayama,T. (1999) Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J., 18, 6642–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishida S., Fujimitsu,K., Sekimizu,K., Ohmura,T., Ueda,T. and Katayama,T. (2002) A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidence from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J. Biol. Chem., 277, 14986–14995. [DOI] [PubMed] [Google Scholar]
- 6.Katayama T., Kubota,T., Kurokawa,K., Crooke,E. and Sekimizu,K. (1998) The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell, 94, 61–71. [DOI] [PubMed] [Google Scholar]
- 7.Blasina A., Kittell,B.L., Toukdarian,A.E. and Helinski,D.R. (1996) Copy-up mutants of the plasmid RK2 replication initiation protein are defective in coupling RK2 replication origins. Proc. Natl Acad. Sci. USA, 93, 3559–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levchenko I., Inman,R.B. and Filutowicz,M. (1997) Replication of the R6K γ origin in vitro: dependence on wt π and hyperactive πS87N protein variant. Gene, 193, 97–103. [DOI] [PubMed] [Google Scholar]
- 9.Chen D., Feng,J., Kruger,R., Urh,M., Inman,R.B. and Filutowicz,M. (1998) Replication of R6K γ origin in vitro: discrete start sites for DNA synthesis dependent on π and its copy-up variants. J. Mol. Biol., 282, 775–787. [DOI] [PubMed] [Google Scholar]
- 10.Kawasaki Y., Matsunaga,F., Kano,Y., Yura,T. and Wada,C. (1996) The localized melting of mini-F origin by the combined action of the mini-F initiator protein (RepE) and HU and DnaA of Escherichia coli. Mol. Gen. Genet., 253, 42–49. [DOI] [PubMed] [Google Scholar]
- 11.Konieczny I., Doran,K.S., Helinski,D.R. and Blasina,A. (1997) Role of TrfA and DnaA proteins in origin opening during initiation of DNA replication of the broad host range plasmid RK2. J. Biol. Chem., 272, 20173–20178. [DOI] [PubMed] [Google Scholar]
- 12.Park K., Mukhopadhyay,S. and Chattoraj,D.K. (1998) Requirements for and regulation of origin opening of plasmid P1. J. Biol. Chem., 273, 24906–24911. [DOI] [PubMed] [Google Scholar]
- 13.Krüger R., Konieczny,I. and Filutowicz,M. (2001) Monomer/dimer ratios of replication protein modulate the DNA strand-opening in a replication origin. J. Mol. Biol., 306, 945–955. [DOI] [PubMed] [Google Scholar]
- 14.Lu Y.B., Datta,H.J. and Bastia,D. (1998) Mechanistic studies of initiator–initiator interaction and replication initiation. EMBO J., 17, 5192–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filutowicz M., Dellis,S., Levchenko,I., Urh,M., Wu,F. and York,D. (1994) Regulation of replication of an iteron-containing DNA molecule. Prog. Nucleic Acid Res. Mol. Biol., 48, 239–273. [DOI] [PubMed] [Google Scholar]
- 16.Filutowicz M. and Rakowski,S.A. (1998) Regulatory implications of protein assemblies at the γ origin of plasmid R6K—a review. Gene, 223, 195–204. [DOI] [PubMed] [Google Scholar]
- 17.Kolter R., Inuzuka,M. and Helinski,D.R. (1978) Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell, 15, 1199–1208. [DOI] [PubMed] [Google Scholar]
- 18.Filutowicz M., McEachern,M.J. and Helinski,D.R. (1986) Positive and negative roles of an initiator protein at an origin of replication. Proc. Natl Acad. Sci. USA, 83, 9645–9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dellis S. and Filutowicz,M. (1991) Integration host factor of Escherichia coli reverses the inhibition of R6K plasmid replication by π initiator protein. J. Bacteriol., 173, 1279–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu F., Levchenko,I. and Filutowicz,M. (1994) Binding of DnaA protein to a replication enhancer counteracts the inhibition of plasmid R6K γ origin replication mediated by elevated levels of R6K π protein. J. Bacteriol., 176, 6795–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J., Sektas,M., Chen,D. and Filutowicz,M. (1997) Two forms of replication initiator protein: positive and negative controls. Proc. Natl Acad. Sci. USA, 94, 13967–13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urh M., Wu,J., Forest,K., Inman,R.B. and Filutowicz,M. (1998) Assemblies of replication initiator protein on symmetric and asymmetric DNA sequences depend on multiple protein oligomerization surfaces. J. Mol. Biol., 283, 619–631. [DOI] [PubMed] [Google Scholar]
- 23.Stalker D.M., Kolter,R. and Helinski,D.R. (1979) Nucleotide sequence of the region of an origin of replication of the antibiotic resistance plasmid R6K. Proc. Natl Acad. Sci. USA, 76, 1150–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mastrangelo I.A., Hough,P.V., Wall,J.S., Dodson,M., Dean,F.B. and Hurwitz,J. (1989) ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature, 338, 658–662. [DOI] [PubMed] [Google Scholar]
- 25.Yoshimura S.H., Ohniwa,R.L., Sato,M.H., Matsunaga,F., Kobayashi,G., Uga,H., Wada,C. and Takeyasu,K. (2000) DNA phase transition promoted by replication initiator. Biochemistry, 39, 9139–9145. [DOI] [PubMed] [Google Scholar]
- 26.Lozinski T. and Wierzchowski,K.L. (2001) Mg2+ ions do not induce expansion of the melted DNA region in the open complex formed by Escherichia coli RNA polymerase at a cognate synthetic Pa promoter. A quantitative KMnO4 footprinting study. Acta Biochim. Pol., 48, 495–510. [PubMed] [Google Scholar]
- 27.Lozinski T. and Wierzchowski,K.L. (2001) Effect of Mg2+ on kinetics of oxidation of pyrimidines in duplex DNA by potassium permanganate. Acta Biochim. Pol., 48, 511–523. [PubMed] [Google Scholar]
- 28.Wu C.W. and Goldthwait,D.A. (1969) Studies of nucleotide binding to the ribonucleic acid polymerase by a fluoresence technique. Biochemistry, 8, 4450–4458. [DOI] [PubMed] [Google Scholar]
- 29.Krüger R. and Filutowicz,M. (2003) Characterization of His-tagged, R6K-encoded π protein variants. Plasmid, 50, 80–85. [DOI] [PubMed] [Google Scholar]
- 30.Thompson J.F. and Landy,A. (1988) Empirical estimation of protein-induced DNA bending angles: applications to λ site-specific recombination complexes. Nucleic Acids Res., 16, 9687–9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borowiec J.A. and Hurwitz,J. (1988) Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J., 7, 3149–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yount R.G. (1975) ATP analogs. Adv. Enzymol. Relat. Areas Mol. Biol., 43, 1–56. [DOI] [PubMed] [Google Scholar]
- 33.Mukherjee S., Patel,I. and Bastia,D. (1985) Conformational changes in a replication origin induced by an initiator protein. Cell, 43, 189–197. [DOI] [PubMed] [Google Scholar]
- 34.Filutowicz M., Uhlenhopp,E. and Helinski,D.R. (1986) Binding of purified wild-type and mutant π initiation proteins to a replication origin region of plasmid R6K. J. Mol. Biol., 187, 225–239. [DOI] [PubMed] [Google Scholar]
- 35.Urh M., York,D. and Filutowicz,M. (1995) Buffer composition mediates a switch between cooperative and independent binding of an initiator protein to DNA. Gene, 164, 1–7. [DOI] [PubMed] [Google Scholar]
- 36.Kim J., Zwieb,C., Wu,C. and Adhya,S. (1989) Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene, 85, 15–23. [DOI] [PubMed] [Google Scholar]
- 37.Germino J. and Bastia,D. (1983) Interaction of the plasmid R6K-encoded replication initiator protein with its binding sites on DNA. Cell, 34, 125–134. [DOI] [PubMed] [Google Scholar]
- 38.Komori H., Matsunaga,F., Higuchi,Y., Ishiai,M., Wada,C. and Miki,K. (1999) Crystal structure of a prokaryotic replication initiator protein bound to DNA at 2.6 Å resolution. EMBO J., 18, 4597–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giraldo R., Fernandez-Tornero,C., Evans,P.R., Díaz-Orejas,R. and Romero,A. (2003) A conformational switch between transcriptional repression and replication initiation in the RepA dimerization domain. Nature Struct. Biol., 10, 565–571. [DOI] [PubMed] [Google Scholar]
- 40.Forest K.T. and Filutowicz,M.S. (2003) Remodeling of replication initiator proteins. Nature Struct. Biol., 10, 496–498. [DOI] [PubMed] [Google Scholar]
- 41.Sekimizu K., Bramhill,D. and Kornberg,A. (1987) ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell, 50, 259–265. [DOI] [PubMed] [Google Scholar]
- 42.Finch P.W. and Emmerson,P.T. (1984) The nucleotide sequence of the uvrD gene of E.coli. Nucleic Acids Res., 12, 5789–5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]