Abstract
We propose a computing method for the estimation of real-time PCR amplification efficiency. It is based on a statistic delimitation of the beginning of exponentially behaving observations in real-time PCR kinetics. PCR ground fluorescence phase, non-exponential and plateau phase were excluded from the calculation process by separate mathematical algorithms. We validated the method on experimental data on multiple targets obtained on the LightCycler platform. The developed method yields results of higher accuracy than the currently used method of serial dilutions for amplification efficiency estimation. The single reaction set-up estimation is sensitive to differences in starting concentrations of the target sequence in samples. Furthermore, it resists the subjective influence of researchers, and the estimation can therefore be fully instrumentalized.
INTRODUCTION
More than 10 years of PCR-based technologies have found their place in most of the laboratories involved in biomedical science. The application of PCR in gene expression studies is an example of a fast innovating field. So far, real-time PCR in combination with array techniques is the major approach adopted in quantitative gene expression studies. The fact that several nucleic acid molecules can be amplified up to microgram amounts opens the possibility to study gene regulation even in a single cell (1). The recent introduction of various fluorescence-based monitoring detection techniques into PCR (2–7) allowed the documentation of the amplification process in the so-called real-time PCR (8–10). The amplification of nucleic acids within the range of exponential growth of the reaction trajectory can be described by a pure exponential growth (equation 1):
P = I ∗ En 1
where P is the amount of the PCR product of the reaction, I is the input nucleic acid amount, E is the efficiency of the reaction ranging from 1 to 2 and n is the number of PCR cycles.
There is a constant tendency to place the quantification into an early phase of detectable amplification. In such an early portion of PCR trajectory the amplification has the exponential character described in equation 1. The reaction trajectory at later reaction stages significantly diverges from the exponential type, and becomes a more stochastic process. In such an early portion of the amplification kinetics, a threshold fluorescence is set. As soon as the reaction reaches this threshold fluorescence, the information necessary for the quantitative judgment about the input concentration of the target sequence has been gathered (11). The fractional cycle number of threshold value (Ct) or crossing point (CP) is then compared with the CP of control samples. There are two ways of threshold level setting. It can be done either arbitrarily by using a randomly selected threshold or by applying computing algorithms. The maximum of the second or, generally, nth derivative of smoothed amplification kinetics gives a good and justified threshold level within the assay (11).
Since the result of a single quantitative PCR just reflects the relative amount of target sequence in the form of fluorescence units, it must be objectified by some control. Therefore, adequate quantitative information cannot be obtained from a real-time PCR assay unless at least two samples are analyzed. To make sure that RT reactions and amplification reactions proceed in a similar way in both samples, the amplification of another target sequence present in the sample is often introduced into the assay either simultaneously or in separated runs. The expression of the standard, called the housekeeping gene or reference gene, is assumed to be uninfluenced by experimental treatment and a similar detectable amplification product should therefore be obtained (12,13). Yet, there is a lot of evidence for regulation of these genes under defined treatments (14,15).
Recently, problems have been discussed, that different sequences were often amplified with different efficiencies, causing under/overestimation of input template copy numbers in orders of magnitude. The solution is to document the amplification efficiencies (E) of both reactions and to apply a compensating computing algorithm (16–18). The currently used and partly automated method of determination of amplification efficiency is the method of serial dilutions, each analyzed in triplicate (11). Using this method, serial dilutions of the starting template are prepared; in these, the input nucleic acid concentration is varied over several orders of magnitude. Usually, dilution series are prepared by serially diluting the input nucleic acid five to 10 times with sterile water. Subsequently, the CP or Ct values are plotted against the log of the known starting concentration value and from the slope of the regression line the amplification efficiency is estimated (11,16). There are some variations of this method, but the serial dilution is always necessary. The method finally gives only one value of E for all dilution concentrations of the respective sequence. This is, however, a simplified approach, since the E varies considerably as the input concentration changes.
Therefore, what is required is a method of amplification efficiency determination that uses the reaction kinetics of a single sample. Since the amplification fluorescence raw data are available by data export from LightCycler (19) or ABI Prism Sequence Detection System (20) software, the efficiency estimation can be based on these data. Liu and Saint (21) suggested a method of amplification efficiency estimation based on absolute fluorescence increase in single reaction kinetics data. In this method, the portion of the data array believed to be exponentially behaving is taken, log- transformed and plotted. The authors consider the slope of the regression line the amplification efficiency. The idea behind this method is correct, but the crucial disadvantage consists of the researcher’s subjective judgment; what data are exponentially behaving and what data are not. Furthermore, the necessary subjective delimitation procedure can not be instrumentalized. Delimitation of the exponential portion of the data is done precisely at the end of it, as the reaction kinetics here strongly depart from the exponential. A similar published method (22) is also based on the absolute fluorescence increase, but it takes place around the point of inflection of the quantification trajectory where a strong decaying trend of the amplification efficiency already occurs. This method is therefore underestimating the ‘real efficiency’.
Here, we report a new method for a reliable estimation of real-time PCR efficiency, which is based on the fluorescence history of just a single reaction set-up and it resists any subjective manipulation. This method was applied on raw fluorescence data from the LightCycler real-time PCR platform.
MATERIALS AND METHODS
SRY plasmid DNA construction
The bovine SRY (male sex determining) gene coding sequence was cloned into pCR®4-TOPO® vector using the TOPO TA Cloning Kit for Sequencing (Invitrogen, Karlsruhe, Germany). This circular DNA construct was linearized with restriction digest and its purity was inspected on a 2% agarose gel. Exact quantification of the DNA content was done at OD260 nm on a spectrophotometer (BioPhotometer®; Eppendorf, Hamburg, Germany) with UVettes (Eppendorf) in various dilutions and repeats (n = 12), to circumvent any source of error. For standard curve acquisition, six serial dilutions of linearized plasmid DNA ranging were prepared, representing 2.65 × 102–2.65 × 107 single-stranded (ss) SRY DNA molecules, serving as DNA templates for real-time PCR.
Real-time PCR on LightCycler
A primer pair flanking sequence within bovine SRY gene was constructed and synthesized (MWG Biotech, Ebersberg, Germany) as follows: forward primer, 5′-GAA CGC CTT CAT TGT GTG GTC-3′; reverse primer, 5′-TGG CTA GTA GTC TCT GTG CCT CCT-3′. The conditions for PCR were optimized in a gradient cycler (TGradient; Biometra, Göttingen, Germany) and subsequently in LightCycler (Roche Diagnostics, Mannheim, Germany) analyzing the melting curves of the products acquired (23). This was done with respect to primer annealing temperatures, primer concentrations, template concentrations and number of cycles applied. Real-time PCR using SYBR Green I technology (Roche Diagnostics) (10,19) with the above-mentioned primers was carried out amplifying cloned sequence in triplicate for each respective concentration. Master-mix for each PCR run was prepared as follows: 6.4 µl of water, 1.2 µl of MgCl2 (4 mM), 0.2 µl of each primer (4 µM), 1.0 µl of Fast Start DNA Master SYBR Green I and 2.65 × 102–2. 65 × 107 copies of ss SRY linearized plasmid DNA. The following amplification program was applied: after 10 min of denaturation at 95°C, 40 cycles of four-segment amplification were accomplished with: (i) 15 s at 95°C for denaturation, (ii) 10 s at 60°C for annealing, (iii) 20 s at 72°C for elongation and (iv) determination of fluorescence at an elevated temperature of 83°C (22). Subsequently, a melting curve program was applied with continuous fluorescence measurement.
RESULTS
After optimization of the real-time PCR assay with SRY, the gene sequence could be routinely run generating specific amplicons showing no primer dimers, a single sharp peak, identical melting points and an expected length of 164 bp in gel electrophoresis. The sensitivity of the LightCycler RT–PCR was evaluated using different starting amounts of cDNA in a standard curve. SYBR Green I fluorescence determination at the elevated temperature resulted in a reliable and sensitive cDNA quantification assay with high linearity (r = 0. 99) over six orders of magnitude from 2.65 × 102 to 2.65 × 107 recombinant standard DNA start molecules.
Determination of fluorescence ground phase in PCR
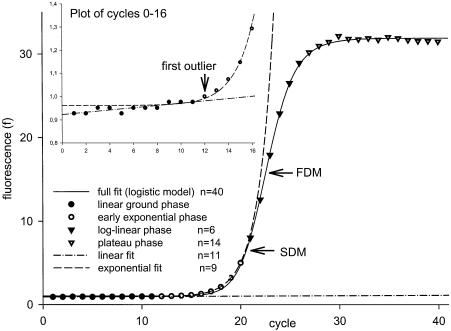
The earliest observation of detectable growth phase above the ground phase with sufficient n is well suited for estimation of E (Fig. 1, inlay).
Figure 1.
Plot of fluorescence observations from LightCycler (Roche Diagnostics). Forty observations give a sigmoid trajectory that can be described by a full data fit (FPLM). The ground phase can be linearly regressed (inlay). The following data of n > 7 are considered to behave exponentially and can be fitted using an exponential model. Various model fits are described in the legend within the figure. FDM and SDM denote the position of the FDM and SDM within the full data fit.
To objectively detect the beginning of the exponential phase and to skip down the prior ground phase, a statistical method is applied. The ground phase is considered to behave linearly (equation 2) and linear regression with intercept ilin and slope β:
ylin = ilin + β × x 2
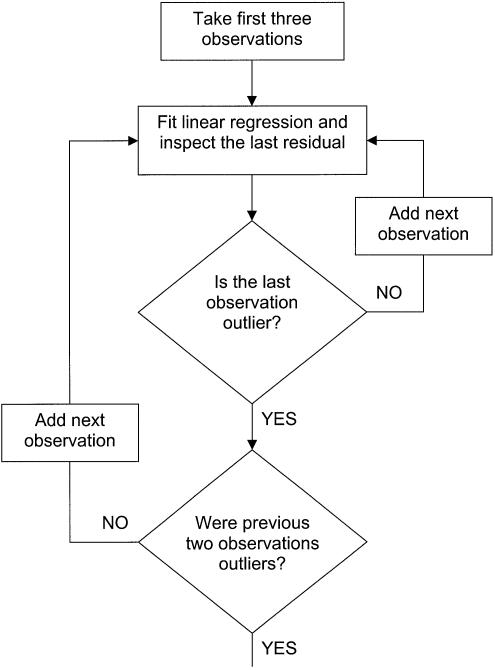
Therefore, it can fit observations as long as there is no sudden significant increment of fluorescence due to reaction product generation. At the moment when the increment of fluorescence becomes a consistent trend, the beginning of the exponential phase takes place. To inspect whether each successively inspected observation still belongs to the linear ground phase or not, standardized residuals of the linear regression are computed. The last one of the regressed observations is always inspected as to whether it does or does not deviate from the linear trend. This procedure starts with the first three observations and proceeds in the way shown in the flowchart in Figure 2.
Figure 2.
Flowchart of the statistical estimation of the beginning of the exponential phase based on inspection of externally studentized residuals.
Computation of the studentized residual statistics is a way to obtain a test on the distribution of particular residuals. To test statistically the probability that a given residual value is an outlier we must ensure that the residual value is comparable with some defined pre-existing probability distribution (here a tn–1–p distribution; see later).
Since observation of xi from the data set of varying n is always inspected, it must be taken into account that observations further from the x̄ (mean value) have stronger influence [hii (leverage)] on the slope of the regression line:
Therefore, hii (equation 3) is the measure of a particular influence of the respective observation xi on the slope of the regression line.
Furthermore, the so called ‘externally studentized’ residual (24) is computed as follows:
where εi is the raw residual value, etc., the difference between the observed fluorescence (yi) value and fitted fluorescence (ŷi) value, si(n–1) is the deviance of residuals in the regression model fitted over data with the deleted inspected observation (n – 1). This is computed as follows:
MSEi(n–1) (equation 5) is the mean square residual of the regression model with the deleted inspected data point. n – 2 in the denominator denotes the residual degrees of freedom of the regression model.
Each ri(n–1) is distributed as tn–1–p under the model. Therefore, we can test the hypothesis whether a single observation deviates from the model by comparing ri(n–1) with the tn–1–p distribution (equation 6) where F(·) is the cumulative distribution function of the tn–1–p distribution:
P-value = 2 × [1 – F(1 – |ri(n – 1)|)] 6
Note, that even if the model holds for every observation (i.e. there are no outliers), one expects ∼5% of the observations to have P-values <0.05. Therefore, we cannot automatically call every observation with a P < 0.05 an outlier, especially when n is large. If the observation is really an outlier and the fluorescence data points are entering the exponential phase, the following observations will also be detected as residuals. Based on experience, two more data points should be inspected after the first outlier is indicated to make sure that a consistent trend takes place (Fig. 1).
Determination of exponential observations
The start of the exponential behavior of the kinetic PCR is estimated by the described ‘externally studentized’ residual algorithm. We considered the end of the exponentially behaving observation to be just under the second derivative maximum (SDM) value as generated by LightCycler software 3.3 (Roche Diagnostics). Alternatively, from a four parametric logistic model (FPLM) with the parameters y0, a, x0 and b (equation 7), fitting all fluorescence observations without any background correction gives:
where f is the value of function computed (fluorescence at cycles x), y0 is the ground fluorescence, a is the difference between the maximal fluorescence acquired in the run and the ground fluorescence, x is the actual cycle number, x0 is the first derivative maximum (FDM) of the function or the inflexion point of the curve and b describes the slope of the curve at x0. The FPLM maximal value of its second derivative (SDM) is computed as follows. First, second and third derivatives of the model are calculated (data not shown). To result in an SDM, the third derivative must be null, which can be achieved by computing equation 8. Two maxima are obtained; only the first ‘positive maximum’ is relevant for the approximation of the CP:
Other ways of computing the SDM were tested: these were based on just a distinct part of amplification trajectory around the expected SDM (25) or on a four parametric sigmoidal model (FPSM). These methods yield similar results to the SDM of FPLM values obtained (26) herein.
Estimation of amplification efficiency (E)
Once the beginning and the end of the exponential phase are defined, the exponential model is fitted over these data (equation 9):
f = γ0 + αEn 9
The fluorescence value is represented by f, γ0 is the upward shift due to ground fluorescence, α is the fluorescence due to the nucleic acid input, n is the cycle number and E is the efficiency of amplification in the early exponential phase of real-time PCR.
Verification of the method
Real-time PCR amplification efficiency was calculated from the given slopes in LightCycler Software 3.3 (Roche Diagnostics) (11). In the DNA calibration curve model, the efficiency per cycle was E1fp = 1.95, using the ‘fit-point method’ (Table 1). The threshold fluorescence Y of the amplified real-time PCR product was calculated according to equation 10:
Table 1. Comparison of five different methods for the calculation of real-time PCR efficiencies.
| E1fp | E1SDM | E2FDM | E2SDM | Enew | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Conc. | n | CPfp | CPSDM | Eall | Y | CV% (Y) | Eall | Y | CV% (Y) | E | CV% (E) | Y | CV% (Y) | E | CV% (E) | Y | CV% (Y) | E | CV% (E) | Y | CV% (Y) |
| 2.65E+07 | 3 | 11.02 | 14.10 | 8.58E+10 | 138.40 | 2.67E+11 | 5.40 | 1.37 | 0.23 | 2.59E+09 | 5.49 | 1.47 | 0.19 | 1.04E+09 | 1.47 | 1.84 | 0.40 | 1.43E+11 | 7.46 | ||
| 2.65E+06 | 3 | 15.93 | 17.20 | 1.10E+11 | 28.62 | 2.03E+11 | 0.38 | 1.37 | 0.16 | 6.74E+08 | 1.99 | 1.47 | 0.17 | 1.35E+08 | 0.42 | 1.85 | 0.67 | 1.04E+11 | 11.96 | ||
| 2.65E+05 | 3 | 18.47 | 20.53 | 5.82E+10 | 16.70 | 1.79E+11 | 5.12 | 1.37 | 0.22 | 2.02E+08 | 7.92 | 1.48 | 0.25 | 1.72E+07 | 1.59 | 1.85 | 0.28 | 7.88E+10 | 5.64 | ||
| 2.65E+04 | 3 | 21.45 | 24.88 | 4.24E+10 | 15.15 | 3.09E+11 | 13.33 | 1.37 | 0.37 | 7.25E+07 | 7.52 | 1.47 | 0.14 | 2.20E+06 | 1.33 | 1.86 | 1.59 | 1.36E+11 | 30.54 | ||
| 2.65E+03 | 3 | 26.08 | 28.18 | 1.25E+11 | 69.40 | 2.67E+11 | 14.56 | 1.36 | 0.48 | 1.83E+07 | 7.45 | 1.46 | 0.81 | 2.55E+05 | 1.21 | 1.84 | 1.34 | 7.71E+10 | 24.79 | ||
| 2.65E+02 | 3 | 30.31 | 32.66 | 1.74E+11 | 65.65 | 5.09E+11 | 24.13 | 1.36 | 0.38 | 6.28E+06 | 7.91 | 1.46 | 0.58 | 3.04E+04 | 1.09 | 1.83 | 0.15 | 9.25E+10 | 24.72 | ||
| Summary for n = 18 | 1.95 | 9.91E+10 | 79.65 | 1.92 | 2.89E+11 | 41.48 | 1.37 | 0.46 | 5.93E+08 | 159.77 | 1.47 | 0.71 | 1.99E+08 | 195.92 | 1.84 | 0.62 | 1.05E+11 | 30.80 | |||
Conc., input concentration of nucleic acid in sample; CPfp, CP based on the fit-point method; CPSDM, CP based on the SDM computing method by LightCycler software 3.3 (Roche Diagnostics); E1fp, amplification efficiency computed from calibration curve (11) where CPs are obtained as fit-points; E1SDM, amplification efficiency computed from calibration curve where CPs are computed as the SDM; E2FDM, amplification efficiency computed from absolute fluorescence increment in point of inflexion (FDM) of amplification trajectory (22); E2SDM, amplification efficiency computed from absolute fluorescence increment in the SDM of amplification trajectory model; Enew, amplification efficiency computed according to the method suggested here; E, the mean value(s) of efficiency for n = 3; Y, fluorescence product computed from equation 10 for respective E for n = 3; CV, coefficient of variation for n = 3; Summary, either the overall mean or overall CV for n = 18.
Y = I ∗ ECP 10
This resulted in a distinct product threshold fluorescence Y at a mean concentration (n = 18) of 9.91 × 1010 ss SRY molecules/set-up for E1fp, with a coefficient of variance (CV) of Y of 79.65%. Additionally, the SDM in the LightCycler Software 3.3 (Roche Diagnostics) (11) was performed, and resulted in 2.89 × 1011 ss SRY molecules/set-up for E1SDM and in lower real-time PCR efficiency (E1SDM = 1.92) and variation (CV = 41.48%).
Furthermore, the method of absolute fluorescence increase in the FDM (or point of inflection) of the amplification trajectory E2FDM (22) and in its SDM E2SDM was applied to compute the amplification efficiency E2. Briefly, the slope (or the first derivative) of the model curve at the respective maximum point is divided by the absolute fluorescence value reached at this point. These efficiencies varied between 1.351 < E2FDM < 1.377 and 1.448 < E2SDM < 1.484, with CVs of 159.77 and 195.92%, respectively. Y was also calculated and resulted in significant lower concentrations of 5.93 × 108 ss SRY molecules/set-up for E2FDM and 1.99 × 108 ss SRY molecules/set-up for E2SDM. The difference between the general efficiency calculation methods E1 and E2 is approximately three orders of magnitude.
Finally, in the new single curve estimation method by FPLM, as suggested here, the mean product threshold fluorescence was 1.05 × 1011 ss SRY molecules/set-up with a variation of 30.80%, comparable with E1 methods. The calculated efficiency values varied in the range 1.822 < Enew < 1.884, and lay between the evaluations described previously.
The verification method was straightforward and was based on equation 10. At the same threshold level, the amount of nucleic acids must also be identical in samples with a different known input concentration of nucleic acid. Here, the fractional value of n is known as the CP. If equation 10 is computed for each sample with the respective value of I, n and E, identical P-values should be theoretically obtained. The Y values were computed for each three concentrations of the dilution series used. As the E values obtained from different computing methods were entered, the method with the lowest variance of computed Y was considered the most accurate (Table 1).
DISCUSSION
As shown in Figure 1, fluorescence observations acquired from real-time PCR fluorescence monitoring are generally of a logistic or sigmoid shape (21,26), indicating that the PCR kinetics (27) consist of early ground phase, exponential growth phase, linear growth phase, and plateau phase. In the ground phase, the fluorescence acquisition is not detectable or just barely detectable due to the fluorescence passively emitted by the initial reaction system itself. At a certain cycle, the fluorescence emitted by the reaction product steps over the ground phase and enters the phase of growth. This phase takes several cycles and possesses a non-linear character (11). At the very beginning of this phase, the nature of the product increment can be well approximated as exponential (r > 0.999, P < 0.001). The rate of product generation slows down until the plateau phase is entered. In this phase, no more significant specific product is generated, as a consequence of reaction exhaustion (28).
Herein, we propose a method of real-time PCR amplification efficiency estimation based on single reaction kinetic observations. As shown in the theoretical work of Peccoud and Jacob (29), if the raw fluorescence observations on the PCR trajectory are available, they contain information about the amplification efficiency in itself. The pitfall in such an amplification efficiency estimation from fluorescence observations is that just a few of the reaction observations represent the initial exponential mode of the reaction. To detect where the reaction leaves its undetectable ground phase, a statistical method of residual inspections was applied. This method was robust enough to detect the first observation significantly diverging from the ground phase. In this method, no influence of the number of observations (n) was present, as long as very few observations were not inspected (n = 4). Such reaction kinetics, where the exponential phase is entered after the first three cycles, are, however, far from real usage.
The end of the exponentially behaved observations was placed into the last observation just before the SDM. This is not an arbitrary decision. After the reaction reaches the SDM, it weakens and looses its exponential character. The computing of the fractional value of the SDM for the purpose of efficiency estimation need not be of the precision demanded for threshold placement, because just discrete observations are used for the efficiency computing. In this respect, the fit of the full-observation model such as the FPLM can be used for computing the SDM. Taking LightCycler computed values of SDM (23) yields similar results. Such a delimitation of observations representing the exponentially behaved part of the PCR yields a set of observations that can be fitted by an exponential model (equation 10) with high significance (r2 > 0.999), where the number of raw data fluorescence observations per set-up was n > 7.
This efficiency calculation method was tested on a further four bovine target sequences of IGF-1, TNFα, prion protein and 18S rRNA amplified in several independent runs on the LightCycler platform (Roche Diagnostics), and resulted in similar findings (data not shown). Furthermore, the method was applied to real-time fluorescence data generated during the amplification of the recombinant sequence of the Pyrenophora teres 18S rRNA gene on an ABI-Prism 7700 instrument (Applied Biosystems, Branchburg, USA), using either SYBR Green I dye or a FIC-labeled minor groove binding 18S rRNA probe. Good results comparable with the dilution series method were also obtained here (data not shown). Altogether, 145 reaction kinetics of various samples have been analyzed in this way, all giving consistent results.
This shows that the presented algorithm is independent of the used platform, the used fluorescence dye (SYBR Green I or FIC), the analyzed target gene and, furthermore, independent of any arbitrary decisions made by the investigator.
In conclusion, verification recalculation of the product amount at a constant threshold level of fluorescence with known efficiency showed that such computed efficiency is more accurate than the method currently used. This is above all clear in the dilution series of the same sequence as the method shows the resolution for various input target concentrations in the sample. Such a computed amplification efficiency can be output from automated platforms, as well as CP values for each sample. This is the major advantage in contrast to other real-time PCR efficiency calculation methods (11,21,22). Efficiency estimations done after the SDM are therefore underestimating the real-time PCR efficiency, whereas previously described methods using a dilution series overestimate it. The newly developed method, with values lying between those of the conventional methods, in our opinion, reflects the ‘real PCR efficiency’. The CV values for the variation of Y might seem to be too large (e.g. CV for E1fp = 79.65%; Table 1). Here, the fact must be taken into account that a great deal of the Y variance is caused by initial vertical shifts in the ground phase. That is, different samples have different fluorescence products already at the very beginning, before any cycling starts. This discrepancy between different samples contributes to the overall CV value for a given method (Table 1). Therefore, not the absolute CV values, but rather its order, is a measure of the applicability of a given method.
Although we want to stress the possibility of determining the amplification efficiency from just a single sample, a statistical approach with more replicates can be adopted. Herein, three replicates were investigated to confirm the stability of the described model.
REFERENCES
- 1.Liss B. (2002) Improved quantitative real-time RT–PCR for expression profiling of individual cells. Nucleic Acids Res., 30, E89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Higuchi R., Fockler,C., Dollinger,G. and Watson,R. (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology, 11, 1026–1030. [DOI] [PubMed] [Google Scholar]
- 3.de Silva D. and Wittwer,C.T. (2000) Monitoring hybridization during polymerase chain reaction. J. Chromatogr. B Biomed. Sci. Appl., 741, 3–13. [DOI] [PubMed] [Google Scholar]
- 4.Holland P.M., Abramson,R.D., Watson,R. and Gelfand,D.H. (1991) Detection of specific polymerase chain reaction product by utilizing the 5′–3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl Acad. Sci. USA, 88, 7276–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitcombe D., Theaker,J., Guy,S.P., Brown,T. and Little,S. (1999) Detection of PCR products using self-probing amplicons and flourescence. Nature, 17, 804–807. [DOI] [PubMed] [Google Scholar]
- 6.Morrison T.B., Weis,J.J. and Wittwer,C.T. (1998) Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques, 24, 954–958. [PubMed] [Google Scholar]
- 7.Marras S.A.E., Kramer,F.R. and Tyagi,S. (1999) Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal., 14, 151–156. [DOI] [PubMed] [Google Scholar]
- 8.Schmittgen T.D. (2001) Real-time quantitative PCR. Methods, 25, 383–385. [DOI] [PubMed] [Google Scholar]
- 9.Klein D. (2002) Quantification using real-time PCR technology: applications and limitations. Trends Mol. Med., 8, 257–260. [DOI] [PubMed] [Google Scholar]
- 10.Meuer S., Wittwer,C. and Nakagawara,K. (2001) Rapid Cycle Real-time PCR: Methods and Applications. Springer, Heidelberg. [Google Scholar]
- 11.Rasmussen R. (2001) Quantification on the LightCycler instrument. In Meuer,S., Wittwer,C. and Nakagawara,K. (eds), Rapid Cycle Real-time PCR: Methods and Applications. Springer, Heidelberg, pp. 21–34. [Google Scholar]
- 12.Thellin O., Zorzi,W., Lakaye,B., De Borman,B., Coumans,B., Hennen,G., Grisar,T., Igout,A. and Heinen,E. (1999) Housekeeping genes as internal standards: use and limits. J. Biotechnol., 75, 291–295. [DOI] [PubMed] [Google Scholar]
- 13.Warrington J.A., Nair,A., Mahadevappa,M. and Tsygantskaya,M. (2000) Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol. Genomics, 2, 143–147. [DOI] [PubMed] [Google Scholar]
- 14.Schmittgen T.D. and Zakrajsek,B.A. (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT–PCR. J. Biochem. Biophys. Methods, 46, 69–81. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T., Higgins,P.J. and Crawford,D.R. (2000) Control selection for RNA quantitation. Biotechniques, 29, 332–337. [DOI] [PubMed] [Google Scholar]
- 16.Pfaffl M.W. (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res., 29, E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfaffl M.W., Horgan,G.W. and Dempfle,L. (2002) Relative Expression Software Tool (REST©) for group wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res., 30, E36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meijerink J., Mandigers,C., van de Locht,L., Tonnissen,E., Goodsaid,F. and Raemaekers,J. (2001) A novel method to compensate for different amplification efficiencies between patient DNA samples in quantitative real-time PCR. J. Mol. Diagn., 3, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wittwer C.T., Ririe,K.M., Andrew,R.V., David,D.A., Gundry,R.A. and Balis,U.J. (1997) The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. Biotechniques, 22, 176–181. [DOI] [PubMed] [Google Scholar]
- 20.Livak K.J. (2001) ABI Prism 7700 Sequence Detection System User Bulletin #2. Relative quantification of gene expression. http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf
- 21.Liu W. and Saint,D.A. (2002) A new quantitative method of real time reverse transcription polymerase chain reaction assay based on simulation of polymerase chain reaction kinetics. Anal. Biochem., 302, 52–59. [DOI] [PubMed] [Google Scholar]
- 22.Pfaffl M.W. (2001) Development and validation of externally standardised quantitative insulin like growth factor-1 (IGF-1) RT–PCR using LightCycler SYBR® Green I technology. In Meuer,S., Wittwer,C. and Nakagawara,K. (eds), Rapid Cycle Real-time PCR: Methods and Applications. Springer, Heidelberg, pp. 21–34. [Google Scholar]
- 23.Ririe K.M., Rasmussen,R. and Wittwer,C.T. (1997) Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem., 245, 154–160. [DOI] [PubMed] [Google Scholar]
- 24.Neter J., Kutner,M.H., Nachtsheim,C.J. and Wasserman,W. (1996) Applied Linear Statistical Models, 4th Edn. Irwin, Chicago. [Google Scholar]
- 25.Wittwer C.T., Gutekunst,M. and Lohmann,S. (1999) Method for quantification of an analyte. United States Patent No. US 6,303,305 B1.
- 26.Tichopad A., Dzidic,A. and Pfaffl,M.W. (2002) Improving quantitative real-time RT–PCR reproducibility by boosting primer-linked amplification efficiency. Biotechnol. Lett., 24, 2053–2057. [Google Scholar]
- 27.Schnell S. and Mendoza,C. (1997) Theoretical description of the polymerase chain reaction. J. Theor. Biol., 188, 313–318. [DOI] [PubMed] [Google Scholar]
- 28.Kainz P. (2000) The PCR plateau phase—towards an understanding of its limitations. Biochim. Biophys. Acta, 1494, 23–27. [DOI] [PubMed] [Google Scholar]
- 29.Peccoud J. and Jacob,C. (1998) Statistical estimation of PCR amplification rates. In Ferré,F. (ed.), Gene Quantification. Birkhauser, Boston, pp. 111–128. [Google Scholar]