Abstract
The rapid increase in the number of novel proteins identified in genome projects necessitates simple and rapid methods for assigning function. We describe a strategy for determining whether novel proteins possess typical sequence-specific DNA-binding activity. Many proteins bind recognition sequences of 5 bp or less. Given that there are 45 possible 5 bp sites, one might expect the length of sequence required to cover all possibilities would be 45 × 5 or 5120 nt. But by allowing overlaps, utilising both strands and using a computer algorithm to generate the minimum sequence, we find the length required is only 516 base pairs. We generated this sequence as six overlapping double-stranded oligonucleotides, termed pentaprobe, and used it in gel retardation experiments to assess DNA binding by both known and putative DNA-binding proteins from several protein families. We have confirmed binding by the zinc finger proteins BKLF, Eos and Pegasus, the Ets domain protein PU.1 and the treble clef N- and C-terminal fingers of GATA-1. We also showed that the N-terminal zinc finger domain of FOG-1 does not behave as a typical DNA-binding domain. Our results suggest that pentaprobe, and related sequences such as hexaprobe, represent useful tools for probing protein function.
INTRODUCTION
Over the last 10 years considerable efforts have been made to understand the molecular mechanisms by which the expression of specific genes is turned on and off. One general principle appears to be that sequence-specific DNA-binding proteins recognise control sequences within their target genes and recruit accessory proteins that either promote or repress gene expression. The importance of sequence-specific DNA-binding proteins to gene regulation means that methods for their identification are of considerable interest.
Currently, it is only straightforward to determine whether a novel protein is a genuine DNA-binding protein if the sequence it recognises is known. In this case, simple experiments such as electrophoretic mobility shift assays and DNase I footprinting can be employed to examine binding to the known recognition sequence. On the other hand, if the cognate DNA sequence is unknown (as is often the case with new proteins) the task is much more challenging. Techniques such as PCR site selection (1) are effective but they can be time consuming. This is particularly true when one is unknowingly dealing with a non-DNA-binding protein. Because PCR site selection is a high sensitivity method, considerable effort is often required to determine whether the sequences obtained are true target sites or artefacts. The magnitude of the undertaking means that many proteins or protein domains are not readily tested for DNA-binding activity.
We have devised a sequence, termed pentaprobe, which provides a one-step method for testing DNA-binding activity of proteins. We have used this sequence in gel retardation experiments to confirm DNA binding by the N-finger of GATA-1 and to show that the N-terminal domain of FOG-1 is not a typical DNA-binding domain. We have also tested other types of DNA-binding domain and confirmed their activity.
The design of pentaprobe was prompted by advances in the understanding of the structural mechanisms by which gene regulatory proteins bind DNA and the realisation that most genes are regulated by combinations of DNA-binding proteins, each recognising surprisingly short motifs. Given that the human genome consists of around 30 000 different genes, embedded in ∼3 × 109 bp of DNA, one might expect that human DNA-binding proteins would be highly selective in their binding so that each could specifically recognise its own target genes but not others. For instance, a DNA-binding protein that recognised a sequence with a length of 16 bp would seem suitable since any single 16 bp motif would be expected to occur in random DNA with a frequency of 1 in 416 bp (∼1 in 4 × 109 bp). Thus, a given 16 bp sequence should, theoretically, occur by chance no more than once in the human genome. Therefore a protein that recognised a 16 bp sequence would be capable of picking out its target against the vast array of irrelevant sequences.
Unexpectedly, however, most known human DNA-binding proteins do not exhibit this degree of specificity. It has been found that many classes of DNA-binding proteins, including homeodomain, Ets domain and various zinc finger type proteins, can bind to very short sequences of 5 bp or less. In other cases, dimeric proteins such as Fos:Jun recognise 6 bp sites, and some zinc finger proteins recognise longer sites, but nevertheless significant binding to 5 bp subsets of the optimal site can usually be observed.
The realisation that many DNA-binding proteins exhibit relatively non-discriminate DNA-binding activity has presented us with the opportunity to construct pentaprobe. We reasoned that if most DNA-binding proteins recognise sequences of no more than 5 bp, then it should be possible to construct a DNA sequence that contains all possible 5 bp target sites (which we term a 5-complete DNA sequence) and use this to determine whether any newly identified protein with unknown function possesses conventional DNA-binding activity. Such a sequence should be of value in identifying sequence-specific DNA-binding proteins from the many novel genes and proteins discovered to date. This is important, given that approximately a third of all recognised genes have no known function and that as many as 5% of all proteins may possess sequence-specific DNA-binding activity. Secondly, such a probe might allow the purification and simultaneous examination of most DNA-binding proteins that are active within a cell.
In this paper we calculate the minimum length of double-stranded (ds)DNA that could conceivably contain all possible 5 bp sequence motifs (and describe general formulae for motifs of length n, where n is any positive integer). We describe an algorithm that we have used to identify actual sequences that are 5-complete and 3-complete (pentaprobe and triprobe, respectively) and are of the minimal theoretical length. Finally, we have synthesised and used these reagents to confirm the DNA-binding activity of a range of test proteins, including the zinc finger proteins BKLF, Eos and Pegasus, the Ets domain protein PU.1 and the treble clef N- and C-terminal fingers of GATA-1. We also showed that the N-terminal zinc finger domain of FOG-1 does not behave as a typical DNA-binding domain. Our results suggest that pentaprobe, and related sequences such as triprobe, represent useful tools for probing protein function.
MATERIALS AND METHODS
Theory and algorithm
n-complete DNA sequences. For the case where n = 1, the sequence motifs that must occur are A, C, G and T and these are obviously contained in the simple 1-complete sequence ACGT (or any other permutation of ACGT). But since DNA is normally double-stranded, the full set of sites is also found in the shorter sequences:
5′-CA-3′and5′-AC-3′
||||
3′-GT-5′3′-TG-5′
When n = 2 the problem of finding minimal sequences becomes more complicated. A 2-complete sequence must contain the full set of 16 possible dinucleotides, namely AA, AC, AG, AT, CA, CC, CG, CT, GA, GC, GG, GT, TA, TC, TG and TT, and obviously the 32 residue sequence AAACAGATCACCCGCTGAGCGGGTTATCTGTT contains all sequences. But this sequence is not the minimal sequence; it contains, for example, the AA dinucleotide twice in the first three residues. Furthermore, when this sequence is made double-stranded for experimental use:
5′-AAACAGATCACCCGCTGAGCGGGTTATCTGTT-3′
||||||||||||||||||||||||||||||||
3′-TTTGTCTAGTGGGCGACTCGCCCAATAGACAA-5′
significant additional repetition occurs. For instance, the dinucleotide AA now occurs twice more, this time on the bottom strand (underlined).
The theoretical minimum length for an n-complete DNA sequence. Consideration of the problem leads to the hypothesis that it may be possible to construct shorter 2-complete sequences (or n-complete, for n equal to any positive integer).
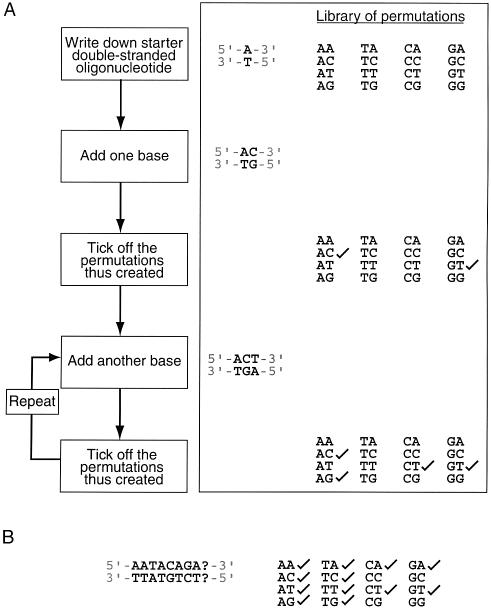
How short could such sequences conceivably be? Consider the two base case (Fig. 1A). Using a double-stranded ‘starter’ oligonucleotide that is 1 bp in length (i.e. n – 1 bp), such as A:T, the addition of a cytosine to the sequence creates the first two base permutation, AC. The existence of the complementary strand means that a second permutation is automatically created, in this case GT (note that it does not also generate TG, since DNA has directionality). Thus, the addition of one base to a starter sequence n – 1 bases in length can generate up to two new permutations of length n. The addition of a third base (giving, for example ACT:TGA) can generate two further permutations, namely CT and AG. If extra permutations can be created every time one additional base pair is added to the original oligonucleotide until the full set of all possible 16 permutations are incorporated into the sequence, then this strategy will generate the shortest conceivable oligonucleotide that contains all two base permutations. The same strategy can be used for all n base cases.
Figure 1.
The creation of an n-complete oligonucleotide. (A) Flow chart showing the process by which an n-complete oligonucleotide might be constructed. Also shown is an example for the 2 bp case. (B) Situation where the addition of any base (A, C, T or G) to the growing oligonucleotide (in position 9) does not give rise to any permutation that has not already been ticked off.
Mathematically, using the above strategy, the shortest conceivable linear oligonucleotide that contains all n base permutations is represented by the equation
N = (n – 1) + (4n/2) for odd n 1
where N is the length (in bp) of the shortest conceivable oligonucleotide containing all n base sequences (where n is odd). This formula is reached by adding the length of the starter oligonucleotide (n – 1 bases) to half of the number of possible permutations (since two extra permutations are created by adding each base pair to the starter oligonucleotide).
One additional issue must be taken into account, however. In cases where n is even, palindromic permutations exist. Thus, if the permutation AT is created during the process of building up the oligonucleotide, no new permutation will be added to the complementary strand (since the complement of AT is AT). This means that the shortest conceivable oligonucleotide containing all n base sequences (where n is even) will be longer than that predicted by equation 1. In fact, because there are 4n/2 palindromic n base sequences (e.g. the four permutations AT, TA, GC and CG in the two base problem), 4n/2 of the 4n one base additions (to the starter oligonucleotide) will generate only one new permutation, rather than two. The 4n/2 palindromic permutations must therefore be created by adding one extra base per permutation. Thus, for even numbered n, the equation describing the length of this species is:
N = (n – 1) + (4n – 4n/2)/2 + 4n/2 for even n 2
= (n – 1) + (4n + 4n/2)/2 for even n 3
Table 1 shows the values of N for a number of values of n. Please note that the above equations are for linear oligonucleotides; cyclic versions of these minimum sequences would contain (n – 1) fewer bases (since there is no need for a starter sequence).
Table 1. Shortest conceivable oligonucleotides containing all n-base permutations.
| n | N (length of minimal sequence with all possible motifs of length n) |
|---|---|
| 1 | 2 |
| 2 | 11 |
| 3 | 34 |
| 4 | 139 |
| 5 | 516 |
| 6 | 2085 |
| 7 | 8198 |
| 8 | 32 903 |
The discussion above defines only the length of the shortest conceivable linear oligonucleotide that contains all n base permutations; it does not demonstrate that such sequences actually exist, i.e. application of the algorithm described in Figure 1A can reach dead ends, where, irrespective of which base is added to the growing chain, no new permutations are added. This situation is shown in Figure 1B.
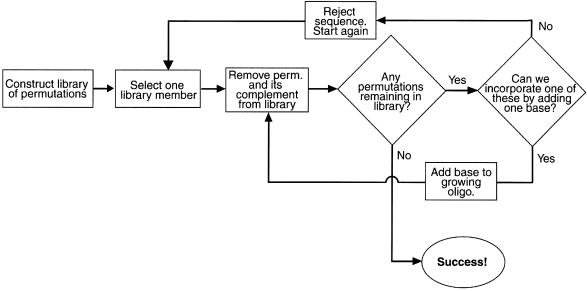
An algorithm for finding minimal n-complete sequences. In order to search for minimal n-complete sequences, we devised a simple computer algorithm (Fig. 2). A library of all n base permutants is constructed for a chosen value of n. One member of the library is selected (e.g. AC for the two base library) and both it and its complement are removed from the library. The library is then examined for permutations that could be included in the growing sequence by the addition of a single base. If such permutants exist (CA, CC, CG and CT in this case), then a single base (e.g. A) is added to the initial sequence (making ACA) to include the new permutant. This process is continued iteratively until either (i) the addition of any of the four bases to the growing sequence can only yield permutations that have already been eliminated from the library in previous rounds or (ii) all library members have been incorporated into the sequence. In the former case, the whole sequence is discarded and the process starts again with a new randomly chosen starting permutation. In the latter case, an n-complete sequence of the minimum theoretical length has been created.
Figure 2.
Computer algorithm used for the identification of n-complete oligonucleotides. A flow chart is shown that outlines the building up of n-complete sequences using a simple process of elimination.
Computer programming
The algorithm described above was implemented as a PERL script (PERL version 5.6.1), running on an Intel-based PC (dual Pentium III chip, 1024 Mb RAM, 133 MHz front-side bus) running the Debian 3.0 (Woody) Linux distribution (with an SMP-enabled version 2.4.14 Linux kernel). This script can be obtained from the authors upon request.
Plasmid construction
The region encoding zinc fingers 1–3 of BKLF (BKLF-F1–3 residues 254–344) was amplified from the murine BKLF cDNA (2) using the primers CGGGATCCACCATGGCAAGGAAGCGCAGGATAC (A19) and CGGAATTCAGACTAGCATGTGGCGTT (A866). The region encoding the Ets domain of PU.1 (residues 169–264) was amplified from a murine cDNA library using primers CGGGATCCAAGAAGAAGATCCGCCTG and GGAATTCTCAGTGGGGCGGGTGG. The region encoding zinc fingers 2–4 of FOG-1 (residues 243–407) was amplified from the murine FOG-1 cDNA using the primers CGGGATCCATGGCATCCATCCTTGCTACC (A90) and GCGAATTCAAAGTTGGCTGCTGGGTGTCC (A91). The resulting fragments were digested with BamHI and EcoRI and inserted into pGEX-2T to generate in-frame fusions with glutathione S-transferase (GST). DNA sequencing was carried out in order to confirm DNA sequences. The clones encoding the N-terminal zinc finger clusters of Eos and Pegasus (3) and the N- and C-terminal zinc fingers of GATA-1 (4) as GST fusion proteins have been previously described.
Protein preparation
All plasmids were transformed into Escherichia coli BL21 (DE3) cells for protein expression. Luria broth was inoculated with the transformed E.coli cells at 37°C. When the A600 reached ∼0.6, protein expression was induced with the addition of IPTG (0.4 mM). After 4 h, the cells were pelleted by centrifugation and stored at –20°C prior to lysis. The cells were resuspended in lysis buffer [50 mM Tris, 50 mM NaCl, 1% Triton X-100, 1.4 mM phenylmethylsulphonyl fluoride (PMSF), 1.4 mM β-mercaptoethanol, pH 8.0] and lysed by gentle sonication. The soluble fraction, separated from the insoluble fraction by centrifugation (15 000 r.p.m., 4°C, 20 min), was loaded onto glutathione–Sepharose beads. Unbound proteins were washed away from the beads with wash buffer (50 mM Tris, 100 mM NaCl, 10% v/v glycerol, 1.4 mM PMSF, 1.4 mM β-mercaptoethanol, pH 8.0) and the GST fusion protein was eluted with a solution containing 5 mM reduced glutathione.
Probe preparation
Oligonucleotides were synthesised by Sigma Genosys. In order to create dsDNA probes, solutions containing complementary oligonucleotides were annealed to create probes for the gel shift experiments. The double-stranded probes were end-labelled according to standard procedures using polynucleotide kinase and purified on native polyacrylamide gels by standard methods (5).
DNA-binding assays
Electrophoretic mobility shift assay (EMSA) reactions were set up in a total volume of 30 µl, comprising ∼0.2 pmol of 32P-labelled probe (final concentration ∼6 nM), ∼500 ng of recombinant protein (final concentration ∼500 µM), 10 mM HEPES, pH 7.8, 50 mM KCl, 5 mM MgCl2, 1 mM EDTA and 5% glycerol and 25 µg/ml poly(dI·dC). Note that lowering the concentration of poly(dI·dC) or omitting it altogether may be preferable with some preparations of protein. After incubation on ice for 10 min, the samples were loaded onto a 6% native polyacrylamide gel made up in 0.5× TBE. The gel was then subjected to electrophoresis at 15 V/cm and 4°C for 3 h, dried, analysed and quantified when necessary using a PhosphorImager (Molecular Dynamics).
RESULTS
Finding minimal n-complete sequences
Using the algorithm described above, minimal 1- and 2-complete sequences were obtained instantaneously (in fact, minimal 1- and 2-complete sequences may be written down from inspection). Minimal 3-complete, 5-complete and 7-complete sequences were obtained in less than 0.01, 0.3 and 104 min of CPU time, respectively. In fact, for the two, three, five and seven base cases, multiple solutions were easily obtained. In contrast, no solution was found for the four base case after more than 150 h of CPU time. Sequences that were a few bases longer in length could, however, be easily generated. For example, a 144 bp 4-complete sequence (c.f. the theoretical minimum of 139 bp) is obtained after a few minutes of CPU time. For practical purposes, such sequences would be quite adequate.
Pentaprobe
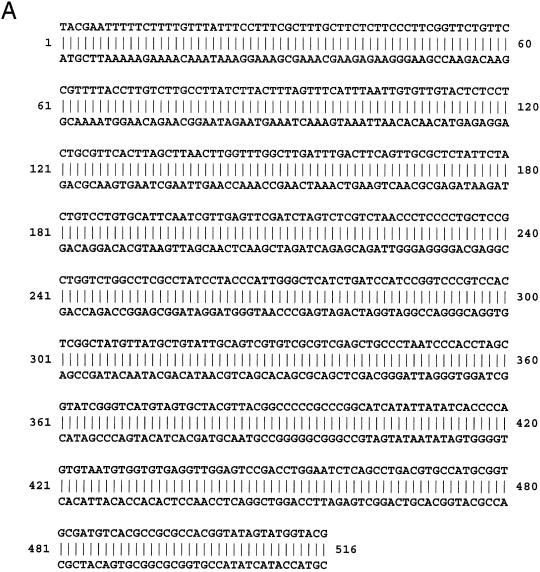
Figure 3A shows one sequence solution for n = 5. Instead of making a single sequence of 516 bp of dsDNA, six overlapping sets of oligonucleotides were synthesised. Six overlapping probes are more useful than a single long DNA fragment for several reasons: they can readily be synthesised using standard solid-phase chemical synthesis; they can readily be purified away from incomplete products of synthesis; better separations are obtained during gel retardation experiments, especially when low molecular weight DNA-binding proteins are analysed. Although the overlaps add slightly to the total length of DNA required, they are necessary to ensure that no sequence motifs are lost at junctions. The actual six overlapping double-stranded oligonucleotides used as pentaprobe in the current work are listed in Figure 3B and are termed pentaprobe.1 to pentaprobe.6.
Figure 3.
Sequences of a pentaprobe and a triprobe. (A) One of the sequence solutions of a single double-stranded oligonucleotide (pentaprobe) that contains all 5 base elements exactly once. (B) The sequences of six overlapping double-stranded oligonucleotides (pentaprobe.1–pentaprobe.6) that together comprise a 5-complete solution. All possible 5 base sequences can be found exactly once in these oligonucleotides, with the exception of those sequences found in the overlap regions, which are represented twice. (C) The sequence (in bold) of a single double-stranded oligonucleotide (triprobe) that contains all 3 base elements exactly once. Flanking sequences to include BamHI and EcoRI restriction sites are shown in italic.
Testing pentaprobe against known DNA-binding proteins
In order to assess the effectiveness of pentaprobe for the detection of DNA-binding activities, we tested it against several different known and putative DNA-binding domains. We chose domains with several different folds. Since many DNA-binding proteins are classical zinc finger proteins (perhaps half of all eukaryotic DNA-binding proteins, which equates to more than 1000 in the human genome) we chose three typical zinc finger proteins. The first, Basic Krüppel-like Factor/Krüppel-like Factor 3 (BKLF/KLF3), is a member of the well-characterised Sp1/KLF family (6), contains three classical C2H2 zinc fingers and is known to bind to sites of the general form NCNCACCCN (where N is any nucleotide) (2). The second, Pegasus, is a divergent member of the Ikaros family (3). This protein also contains a DNA-binding domain consisting of three zinc fingers, but has little overall homology with BKLF and recognises sequences with the consensus GNNTGTNG (3). The third, Eos, is a typical member of the Ikaros family (3). Eos has four zinc fingers in its DNA-binding domain and recognises sequences of the form TGGGAA (3). We also examined the Ets domain of PU.1, which binds sequences of the form GAGGAA (7), and the two treble clef zinc-binding domains (the N- and C-terminal fingers) of the erythroid transcription factor GATA-1, which have been shown to bind sites such as A/TGATAA/G and A/C/TGATC, respectively (4,8).
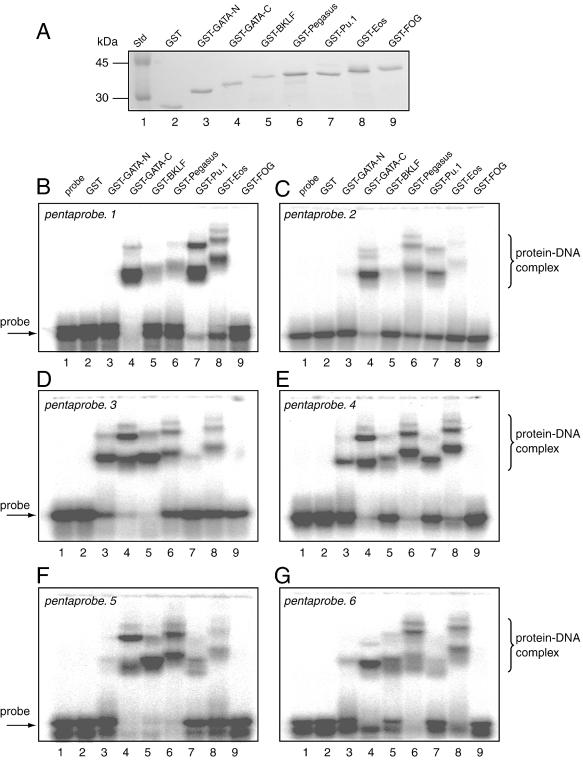
Each domain was expressed as a GST fusion protein, and GST alone and probe alone were also included as negative controls. The GST fusion proteins were expressed in E.coli and purified using glutathione affinity chromatography. Coomassie stained SDS–PAGE was used to demonstrate that all proteins were recovered in comparable amounts (Fig. 4A). These proteins were then used in six separate EMSAs, one with each of pentaprobe.1 to pentaprobe.6 (Fig. 3B). The results are shown in Figure 4B–G. In Figure 4B it can be seen that in the negative control lanes (containing either probe alone or GST alone; lanes 1 and 2) there is very little evidence of retarded species, while in contrast GST–GATA-C, GST–BKLF, GST–Pegasus, GST–PU.1 and GST–Eos (lanes 4–8, respectively) all show the presence of retarded species. In some cases (lanes 4, 7 and 8), a number of retarded bands with different mobilities can be seen, suggesting that the fusion protein has bound to several sites within the probe. Similar results were obtained for pentaprobe.2 to pentaprobe.6 (Fig. 4C–G). In each case a subset of the known DNA-binding proteins produces retarded bands.
Figure 4.
Analysis of the DNA-binding activity of chosen GST fusion proteins. (A) Coomassie stained SDS–PAGE showing that each of the protein solutions used in subsequent EMSA experiments was similar in concentration. (B–G) EMSAs carried out using pentaprobe.1–pentaprobe.6 and the GST fusion proteins shown in (A). Lane 1, probe only; lane 2, GST; lane 3, GST fusion with GATA-N; lane 4, GST fusion with GATA-C; lane 5, GST fusion with BKLF; lane 6, GST fusion with Pegasus; lane 7, GST fusion with PU.1; lane 8, GST fusion with Eos; lane 9, GST fusion with FOG.
Testing pentaprobe against a putative DNA-binding protein
The results presented above demonstrate that pentaprobe can be used to detect the DNA-binding activities of a range of DNA-binding proteins. We therefore sought to determine whether a domain for which no function is known, but which has been proposed to be a DNA-binding domain, was indeed capable of recognising DNA. The N-terminal zinc finger domain of FOG-1 (9) was chosen for analysis. This domain contains four classical zinc fingers and fingers 2–4 are arranged in tandem in the characteristic three finger array found in many zinc finger DNA-binding proteins (including BKLF and Pegasus). Thus it was initially thought that this domain of FOG-1 might also be a typical sequence-specific DNA-binding domain (9); data supporting or refuting this hypothesis have never been presented.
Figure 4B shows no evidence of retarded bands in the lane containing GST–FOG-1–4. Similar results were obtained for pentaprobe.2 to pentaprobe.6 (Fig. 4C–G, lane 9). In each case a subset of the known DNA-binding proteins produces retarded bands, but in no case is retardation observed with GST–FOG-1–4. GST–FOG-1–4 was also tested against all probes in the absence of the competitive inhibitor poly(dI·dC) (which is routinely included in the reaction mixture). While other DNA-binding proteins bound well under these conditions (data not shown), no binding was detected by GST–FOG-1–4. No significant binding by the negative control probe alone or GST alone was detected. Finally, we tested GST–FOG-1–4 against each of the probes in its single-stranded form, but again no binding was detected (data not shown). As a positive control we tested the binding of PU.1 to single-stranded (ss)DNA. This protein has previously been shown to bind single-stranded nucleic acids (10), and binding was also evident in our assays (data not shown). We conclude that unlike the related classical C2H2 zinc finger proteins BKLF, Pegasus and Eos, FOG-1–4 does not function as a typical dsDNA-binding domain. Further, it does not appear to function as a ssDNA-binding domain.
Eliminating weak signals caused by contaminants
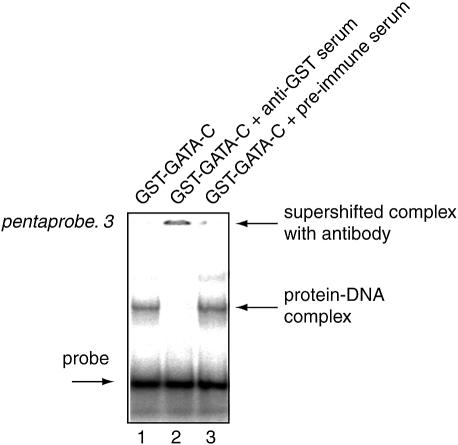
We noted that when testing non-DNA-binding proteins, such as GST or GST–FOG-1–4, very weak bands are sometimes observed (substantially weaker than those observed for most DNA-binding domains). The presence of these weak bands may suggest that the test protein is binding pentaprobe. It is important to be able to distinguish whether these faint signals indicate weak but genuine DNA-binding activity by the test protein or represent artefacts caused by contaminating DNA-binding proteins originating from the E.coli expression system. We find that this problem occurs most often when the GST fusion protein is expressed at low levels and has to be purified from very large preparations of E.coli. We have used anti-GST serum to distinguish between genuine and spurious signals. As shown in Figure 5, the anti-GST serum interferes with the retarded complexes generated by the GST–GATA-C preparation (lane 2). This experiment shows that simply adding anti-GST serum to the reaction mixture can minimise doubts as to the identity of the observed bands.
Figure 5.
Detection of artefacts from contaminating bacterial DNA-binding proteins using anti-GST serum. EMSA showing the DNA-binding activity of GST–GATA-C (lane 1). Lane 2 additionally contains anti-GST serum, while lane 3 contains preimmune serum. The supershifting of the retarded bands seen for lane 2 indicates that the GST fusion protein, rather than an irrelevant contaminant, is responsible for the retardation.
Assessing triprobe as a rapid first screen
The proteins tested above are all conventional mammalian transcription factors with recognition sites that are generally thought to encompass more than 5 bp. For example, classical zinc finger proteins are thought to contact 3 bp per finger, so a four finger domain like that of Eos might have been expected to bind a 12 bp sequence. Further, previous work suggests that Pegasus binds to the 8 bp consensus site GNNGTGNG (3).
It was striking therefore that each of the DNA-binding domains tested gave rise to retarded bands with each of pentaprobe.1–pentaprobe.6, when one might have expected that proteins with theoretically 12 and 8 bp recognition sites might have failed to bind at all. In fact, we estimate that Eos, for example, binds pentaprobe at around 15 different sites, as does Pegasus. It seems likely that we are observing binding to non-optimal sites. This shows that while many proteins (including zinc finger proteins) may have optimal recognition sites that are longer than 5 bp, it is likely that DNA binding will still be detected using pentaprobe, i.e. some degeneracy often exists in the DNA recognition site for each protein. In addition, the high concentrations of recombinant protein used in the assay probably allow us to observe binding to non-optimal, lower affinity sites.
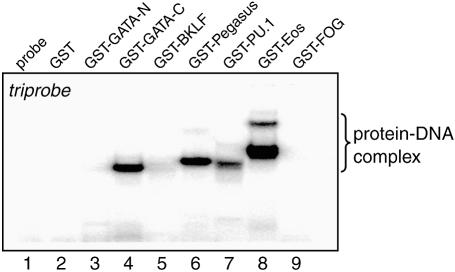
Given that we detected such extensive binding to pentaprobe, we next tested whether binding could also be detected to triprobe, a 34 bp sequence that contains all possible 3 bp recognition elements (Fig. 3C). Given the view that DNA-binding proteins are reasonably selective about which sequences they bind, we did not expect all proteins to bind. As shown in Figure 6, there is no evidence of retarded species with either probe alone, GST alone, GST–FOG-1–4 or GST–GATA-N. However, binding is observed with GST–PU.1, GST–GATA-C, GST–Pegasus and GST–Eos. Thus many, but not all, proteins will bind the triprobe sequence, making it a simple first screen that requires the labelling of only a single oligonucleotide probe.
Figure 6.
Triprobe acts as an effective reagent for detecting DNA-binding activity. EMSAs carried out using triprobe and the GST fusion proteins shown in Figure 4A. Lane 1, probe only; lane 2, GST; lane 3, GST fusion with GATA-N; lane 4, GST fusion with GATA-C; lane 5, GST fusion with BKLF; lane 6, GST fusion with Pegasus; lane 7, GST fusion with PU.1; lane 8, GST fusion with Eos; lane 9, GST fusion with FOG.
DISCUSSION
We have presented a formula for the calculation of the minimum length of dsDNA that is required to contain all n bp elements exactly once. We have also described an algorithm that can be used to identify such sequences, at least in the cases where either n = 2 or n is an odd integer.
Our algorithm has not been successful in finding minimal sequences where n is an even natural number greater than two. The existence of palindromic permutations appears to place substantial restrictions on the number of n-complete sequences. Note, however, that the 1 week search that we carried out for a 4-complete sequence still only sampled ∼2.5 × 107 sequences out of the total of 4.9 × 1083 permutations that are possible for a 139 bp oligonucleotide. The issue of whether minimal 4-complete sequences exist is therefore still undetermined. It is possible that the rules of base pairing and the problem of palindromes preclude the creation of minimal n-complete sequences for even numbers n greater than two. Nevertheless, our algorithm was able to generate many sequences in which only 6 of the 136 unique four base permutations could not be incorporated into the growing chain by the addition of a single base. These permutations could easily be added to the end of the chain by simple contantenation (and following the strategy that these remaining permutations are introduced firstly by the addition of a single base if possible, or else, progressively by two, three and then four base additions) and the resulting oligonucleotide, while slightly longer than the shortest theoretical sequence (for example, 144 versus 139 bp in the 4-complete case), would still be of value experimentally. Interestingly, a probe that contains all 136 four base permutations has been independently derived and has proved useful in determining the sites at which certain antibiotics bind DNA (11).
In order to test whether pentaprobe is useful for the detection of DNA-binding activity we tested it against a variety of known and putative DNA-binding domains. We detected binding with domains of BKLF, Eos, Pegasus, PU.1 and GATA-1. We did not detect binding with FOG-1. The former proteins are recognised as genuine DNA-binding proteins that regulate gene expression in vivo but there is no evidence that FOG-1 can bind DNA alone, and attempts to determine a specific DNA-binding site using conventional methods, such as PCR site selections, have been unsuccessful (data not shown). Given that the technique was successful in detecting DNA binding by related classical zinc finger proteins BKLF, Eos and Pegasus, our data suggest that FOG-1 is not a conventional zinc finger DNA-binding protein.
What limits are there to detecting DNA-binding activities with pentaprobe?
Current results from independent studies together with the data shown here suggest that in vitro most DNA-binding proteins, including those of the classical zinc finger, Ets domain and treble clef finger classes tested here, will recognise short DNA motifs of 5 bp or less. Other highly represented proteins, such as homeodomain proteins, are also known to contact short motifs and it is likely that they too will bind pentaprobe. In general, we expect that most if not all monomeric eukaryotic DNA-binding proteins will display detectable binding to pentaprobe, at least under the conditions used here (i.e. in the presence of relatively high concentrations of protein). It is possible that some proteins, such as those that bind as homodimers to elongated recognition sites, may not show binding to pentaprobe. For example, nuclear receptors often bind to sites composed of a pair of (often degenerate) 6 bp repeats, sometimes separated by 3 bp and sometimes inverted (12). The full set of such sequences is not contained in pentaprobe. Nevertheless, it has been reported that nuclear receptors can function through so-called ‘half-sites’ (13) and it is possible that binding would be detected in vitro. Other dimeric proteins, such as leucine zipper proteins, typically bind repeated 3 bp sites but binding is still observed when one residue is mutated. Hence, we would expect that at least some leucine zipper proteins would bind pentaprobe. It is notable that the Ikaros family proteins, such as Eos, can homodimerise, and dimerisation is thought to enhance DNA-binding activity. The portion of Eos used in our experiments lacks the dimerisation domain and so exists only as a monomer but nevertheless displays strong binding to pentaprobe, indicating that binding by monomer subunits can be detected by this method.
The relevance to in vivo DNA binding
Our results are also relevant to current methods of studying DNA-binding proteins and the biological mechanisms through which gene expression is regulated. It is common practice to test the binding of recombinant proteins in vitro and to use in vitro techniques to derive their binding sites. In general, rather loose consensus sequences are found. In other words, the core binding site of essential residues is generally very short (typically, we believe, 5 bp or less). It is not surprising that weak and degenerate consensus sequences are often generated by PCR site selection experiments, given that PCR site selection relies on high concentrations of purified protein and binding to low affinity sites can therefore be observed. Similarly, we used relatively high concentrations of both DNA and protein in order to maximise the possibility of detecting weak binding events. It is possible that using lower concentrations would allow better discrimination and that only the higher affinity sites bound by high affinity binding proteins would be detected. It is notable, however, that some DNA-binding domains, such as that of BKLF (14), bind DNA with affinities of ∼107 M–1, so that high concentrations of reagents are necessary if binding is to be robustly detected.
Our work does not directly address the physiological relevance of the range of observed sequences but when in vivo binding studies have been compared with the in vitro findings there has often been good agreement (15). It is possible that, even in vivo, DNA-binding proteins recognise very short and sometimes sub-optimal sites. In some instances, it is even possible that low affinity, sub-optimal sites are ideal for the purposes of regulation, as occupancy can be regulated by subtle changes in the concentration of the binding protein. The conventional view is that short binding sites are common in gene regulatory elements and combinations of different proteins interacting cooperatively are required for gene regulation in vivo. Our observations of both multiple binding to pentaprobe and the binding of many proteins to triprobe clearly indicate that DNA-binding proteins are relatively indiscriminate in their binding activity in vitro.
We have used the GST fusion system as it is convenient for these types of experiments. Firstly, it provides a very good one-step purification procedure and, secondly, antibodies to GST exist and can be used to verify that the retarded species are generated by the GST fusion protein and not by irrelevant contaminants. We have used purified proteins in our experiments rather than either complete nuclear extracts or proteins overexpressed in mammalian cells. We expect that the vast number of DNA-binding proteins that would be present in the latter systems (even if some purification step were included) would generate a more complicated pattern of binding and may render the results non-interpretable.
Other uses for pentaprobe
In this work we have concentrated on the idea that pentaprobe can be used for detecting dsDNA-binding activity, but it may also have other uses. Firstly, it could be used to detect ssDNA-binding activity and we have used it to confirm the previously reported ssDNA-binding activity of PU.1 (10). The addition of a T7 promoter sequence would allow the generation of RNA and this could be used to test for RNA-binding activities. Additionally, the sites recognised could readily be mapped and binding sites could be defined. In the case of dsDNA-binding proteins, standard techniques such as DNase I footprinting and methylation interference could be carried out to define the exact residues contacted by the protein. If the test protein binds more than once (as many of the proteins tested here did) then it would be possible to determine an initial consensus sequence.
Pentaprobe could also be used as a tool for the purification of DNA-binding proteins. In many recent applications of proteomics it is desirable to look at large sets of proteins simultaneously, but the complexity of the cell is such that it is not possible to look at all proteins. Using nuclear fractionation followed by chromatography through a column containing pentaprobe it should be possible to purify and then examine a large subset of all DNA-binding proteins. In this way, one might be able to monitor changes in DNA-binding protein profiles observed during cellular differentiation or disease progression.
In summary, while there are many ways of testing whether a newly identified protein has DNA-binding activity, the fact that assessment by pentaprobe is simple (requiring nothing beyond standard electrophoresis equipment) and rapid (the process involves a single gel retardation experiment) means that this should prove to be a practical way of examining novel proteins for DNA-binding functions. Moreover, it is relatively inexpensive, in that once the set of oligonucleotides is generated, numerous experiments can be carried out without additional costs other than the minor costs of running gel retardation experiments.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jose Perdomo for providing clones of the N-terminal zinc finger clusters of Eos and Pegasus, Natalie Bartle for providing a clone of fingers 2–4 of FOG-1, Wendy Yung for providing a clone of the Ets domain of PU.1 and Stephen Graham and Dr Mitchell Guss for expert maintenance of our computing facility. This work was supported by a University of Sydney Sesqui Grant to M.C., a grant from the Australian Research Council to J.P.M. and M.C. and an Australian Postgraduate Award to A.H.Y.K.
REFERENCES
- 1.Pollock R. and Treisman,R. (1990) A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res., 18, 6197–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crossley M., Whitelaw,E., Perkins,A., Williams,G., Fujiwara,Y. and Orkin,S.H. (1996) Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol., 16, 1695–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perdomo J., Holmes,M., Chong,B. and Crossley,M. (2000) Eos and pegasus, two members of the Ikaros family of proteins with distinct DNA binding activities. J. Biol. Chem., 275, 38347–38354. [DOI] [PubMed] [Google Scholar]
- 4.Newton A., Mackay,J. and Crossley,M. (2001) The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J. Biol. Chem., 276, 35794–35801. [DOI] [PubMed] [Google Scholar]
- 5.Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 6.Turner J. and Crossley,M. (1999) Mammalian Kruppel-like transcription factors: more than just a pretty finger. Trends Biochem. Sci., 24, 236–240. [DOI] [PubMed] [Google Scholar]
- 7.Kodandapani R., Pio,F., Ni,C.Z., Piccialli,G., Klemsz,M., McKercher,S., Maki,R.A. and Ely,K.R. (1996) A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature, 380, 456–460. [DOI] [PubMed] [Google Scholar]
- 8.Merika M. and Orkin,S.H. (1993) DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol., 13, 3999–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsang A.P., Visvader,J.E., Turner,C.A., Fujiwara,Y., Yu,C., Weiss,M.J., Crossley,M. and Orkin,S.H. (1997) FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell, 90, 109–119. [DOI] [PubMed] [Google Scholar]
- 10.Hallier M., Tavitian,A. and Moreau-Gachelin,F. (1996) The transcription factor Spi-1/PU.1 binds RNA and interferes with the RNA-binding protein p54nrb. J. Biol. Chem., 271, 11177–11181. [DOI] [PubMed] [Google Scholar]
- 11.Lavesa M. and Fox,K.R. (2001) Preferred binding sites for [N-MeCYs(3), N-MeCys(7)]TANDEM determined using a universal footprinting substrate. Anal. Biochem., 293, 246–250. [DOI] [PubMed] [Google Scholar]
- 12.Evans R.M. (1988) The steroid and thyroid hormone receptor superfamily. Science, 240, 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato S., Tora,L., Yamauchi,J., Masushige,S., Bellard,M. and Chambon,P. (1992) A far upstream estrogen response element of the ovalbumin gene contains several half-palindromic 5′-TGACC-3′ motifs acting synergistically. Cell, 68, 731–742. [DOI] [PubMed] [Google Scholar]
- 14.Simpson R.J., Cram,E., Czolij,R., Matthews,J.M., Crossley,M. and Mackay,J.P. (2003) CCHX zinc finger derivatives retain the ability to bind Zn(II) and mediate protein-DNA interactions. J. Biol. Chem., 278, 28011–28018. [DOI] [PubMed] [Google Scholar]
- 15.Horak C.E., Mahajan,M.C., Luscombe,N.M., Gerstein,M., Weissman,S.M. and Snyder,M. (2002) GATA-1 binding sites mapped in the beta-globin locus by using mammalian chIp-chip analysis. Proc. Natl Acad. Sci. USA, 99, 2924–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]