Abstract
A lectin isolated from the roots of the legume, Dolichos biflorus, binds to Nod factors produced by rhizobial strains that nodulate this plant and has a deduced amino acid sequence with no significant homology to any lectin reported to date. This lectin also is an enzyme that catalyzes the hydrolysis of phosphoanhydride bonds of nucleoside di- and triphosphates; the enzyme activity is increased in the presence of carbohydrate ligands. This lectin–nucleotide phosphohydrolase (LNP) has a substrate specificity characteristic of the apyrase category of phosphohydrolases, and its sequence contains four motifs characteristic of this category of enzymes. LNP is present on the surface of the root hairs, and treatment of roots with antiserum to LNP inhibits their ability to undergo root hair deformation and to form nodules on exposure to rhizobia. These properties suggest that this protein may play a role in the rhizobium–legume symbiosis and/or in a related carbohydrate recognition event endogenous to the plant.
Oligosaccharide signaling events play important roles in the regulation of plant development, defense, and other interactions of plants with the environment (1–4). The establishment of the nitrogen-fixing symbiotic relationship between rhizobia and leguminous plants depends on such a signaling process. The rhizobia produce lipochitooligosaccharidic signals, called Nod factors, that elicit the differentiation of a new organ, the nodule, in the root cortex. The rhizobia bind to and invade the root hairs, where they proceed to the emerging nodule within infection threads produced by the plant (5). Both the initiation of nodule formation and rhizobial entry are host-strain-specific; this specificity is determined by the type of Nod factor produced by a particular rhizobial strain and by the ability of a leguminous species to recognize that signal.
All Nod factors consist of a short, typically tetrapentameric, chitin oligosaccharidic backbone that is N-acylated at the nonreducing end, usually with a common fatty acid, such as cis-vaccenic acid. The Nod factors differ from one another in length of this backbone and the type of substituents that decorate it; these modifications determine the strain specificity of the rhizobium and depend on the set of nodulation genes possessed by that rhizobial strain (4). A related set of signals are the chitin oligosaccharides, themselves, that have been found to elicit defense responses in a wide variety of plants (for review, see ref. 3).
Although the structures of many of these chitooligosaccharides have been characterized, little is known about the plant proteins that bind these signals, whether as receptors for signal transduction or as binding proteins with other functions. High-affinity binding sites for chitin fragments have been found on membranous fractions prepared from tomato (7) and rice (8) suspension-cultured cells, and a 70-kDa protein that binds to chitin fragments was isolated from the rice membranes (9). Particulate fractions from roots of the legume, Medicago truncatula, were found to bind Nod factors produced by the rhizobial symbiont of this plant, and similar binding activity was found in a particulate fraction of tomato roots and the microsomal fraction of Medicago varia cell suspension cultures (10, 11).
In this paper we show that a 46-kDa lectin (LNP), previously isolated from root extracts of the legume Dolichos biflorus and named DB46 (12), is a Nod factor-binding protein as well as a nucleoside di- and triphosphate hydrolase. The presence of this protein on the surface of root hairs, the inhibitory effects of anti-LNP-serum on root hair deformation and nodulation, and the finding that the enzymatic activity of this lectin is elevated on binding to its carbohydrate ligands suggest that it may play a role in the rhizobium–legume symbiosis and/or in a related carbohydrate recognition event endogenous to the plant.
MATERIALS AND METHODS
Preparation of LNP and Anti-LNP-Serum.
LNP was extracted from the roots of 7-day-old D. biflorus plants and isolated by using affinity chromatography on hog blood group A + H -Sepharose (12), followed by ion exchange chromatography. It was iodinated by using the iodine monochloride procedure as described (13), which gave a specific activity of ≈500 × 106 cpm per mg of protein.
Recombinant LNP, representing residues 1–414 of the mature protein, was expressed in Escherichia coli by using the pET expression vector. The protein was extracted from isolated inclusion bodies and further purified by ion exchange chromatography. This protein was used as an immunogen for the production of anti-LNP-serum. Use of this antiserum in immunoblots of 7-day-old D. biflorus root extracts produced a single 46-kDa band.
Carbohydrate-Binding Assays.
Solid-phase binding assays were conducted by using iodinated LNP and purified shrimp chitin powder (Sigma), which was N-acetylated before use with 15 mM acetic anhydride in 0.5 M NaHCO3 for 1 hour at room temperature. The assays were conducted in a final volume of 100 μl of 10 mM Mops buffer, pH 7.2, containing 0.02% Tween 20 and 0.01% NaN3. After incubation at room temperature for 2 hours, binding was measured as described (13).
Bradyrhizobium japonicum USDA110 Nod factor was isolated as a single peak by reverse-phase HPLC as described, and its concentration was determined by its extinction coefficient (14). The Nod factors from Rhizobium meliloti [≈9:1 NodRm-IV(Ac, S)/NodRm-V(Ac, S)] and Rhizobium sp. NGR234 were graciously provided by J. Denarie (Centre National de la Recherche Scientifique–Institut National de la Recherche Agronomique, Toulouse, France); the purity of these factors was established by using fast atom bombardment mass spectrometry and proton NMR. The concentrations of stock solutions of these Nod factors were based on weight of the samples. Monosaccharides and the chitin disaccharide were purchased from Sigma; the other chitin oligosaccharides were obtained from Seikagaku Kogyo (Tokyo).
Cloning of LNP cDNA.
Total RNA was isolated by using the method of Taylor and Powell (15) from the roots of 1-day-old D. biflorus plants and reverse-transcribed by using Moloney–murine leukemia virus reverse transcriptase and random hexanucleotide primers (16). This cDNA was used as a template in a PCR reaction using Taq polymerase and degenerate sense and antisense primers corresponding to amino acids 6–12 and 244–249 in Fig. 2. The PCR was performed in an automated thermal cycler for 35 cycles of 94°C for 2 min, 37°C for 2 min, and 72°C for 2 min. The predominant 727-bp fragment was isolated on a 1.2% agarose gel, cloned into the pCRII vector (Invitrogen), and sequenced. Gene-specific primers were used in 5′ and 3′ rapid amplification of cDNA ends (RACE) reactions (17), and the products were cloned into the pCRII vector and sequenced. The full-length (1,527-bp) cDNA was assembled by ligating the two RACE products together using an internal SacI site. The sequences of the overlapping regions of the 5′ and 3′ RACE products and the original PCR fragment were identical.
Figure 2.
Deduced amino acid sequence of LNP. Position 1 designates the NH2 terminus of the native protein. Those regions of the deduced sequence verified by amino acid sequences of portions of the protein are underlined. The hydrophobic putative signal sequence is in italics and a ∗ is above the position of each consensus N-glycosylation site. The four apyrase conserved sequence motifs (25) are shown in boxes.
Phosphatase Assays.
Phosphatase assays were conducted in microtiter plates at 25°C in a final volume of 100 μl of 60 mM Mops buffer (pH 6.8) containing 1 mM MgCl2, 20 ng of LNP, and different concentrations of Mg-nucleoside di- and triphosphates. At various time points, 30-μl aliquots were removed and assayed for inorganic phosphate by using a photometric microtiter assay (18), modified by using four parts ammonium molybdate reagent to one part 10% ascorbate for the reagent mixture. Conditions were chosen so that <10% of the total substrate was converted to product. One unit of enzyme activity represents the formation of 1 μmol of inorganic phosphate per minute.
Immunofluorescence Microscopy.
Roots from 7-day-old D. biflorus plants were fixed for 45 minutes at 4°C in 0.01 M phosphate buffer (pH 7.2) containing 0.15 M NaCl and 0.3% paraformaldehyde. After washing, the roots were treated for 20 minutes with a 1:250 dilution of preimmunization serum or antiserum prepared against recombinant LNP. After washing, the roots were treated for 20 minutes with fluorescein-labeled goat anti-rabbit IgG (Sigma), washed, and examined with a Leica TCS NT confocal microscope using a 488-nm laser excitation line and a 560-nm barrier filter. Confocal images were reconstructed with imagespace software.
Nodulation.
D. biflorus seeds were sterilized by shaking for 15 minutes in 70% ethanol and then for 15 minutes in 3% hydrogen peroxide. After extensive washing with sterile H2O, the seeds were germinated and grown in sterile growth pouches. At 3 days, the roots were inoculated with 100 μl of Bradyrhizobium sp. 24A10 (1 × 107 cells per ml). Roots were examined for root hair deformation 62 hours after inoculation. The number of nodules per root was determined after 3 weeks. Antiserum and preimmunization serum used to treat the roots were sterilized by filtration through a 0.45-μm filter. To avoid subjective bias, the roots were examined without knowledge as to which treatment they had undergone.
RESULTS
LNP Is a Nod Factor-Binding Protein.
Previous studies established that LNP (previously named DB46) is a lectin that binds to hog gastric mucin blood group A + H substance (12). This protein also binds to chitin, a polymer of N-acetyl-d-glucosamine residues; this binding is saturable with a capacity of 28 nmol of LNP per gram of chitin and a Kd of 48 nM. By using chitin as a solid phase, a competitive binding assay was used to examine the carbohydrate binding specificity of this protein (Fig. 1). Inhibition of binding was obtained with high concentrations of N-acetyl-d-glucosamine but not with similar concentrations of N-acetyl-d-galactosamine, the C-4 epimer of this sugar, nor with other common monosaccharides. The chitin disaccharide gave approximately 10-fold better inhibition than the monosaccharide, whereas the chitin penta- and hexasaccharides were slightly better inhibitors than the disaccharide. No inhibition was obtained with de-N-acetylated chitin oligosaccharides when tested at low concentrations; however, when tested in the millimolar range of concentrations, several of these oligosaccharides precipitated the lectin, even under highly buffered conditions. Whether this precipitation is specific or nonspecific has not been determined.
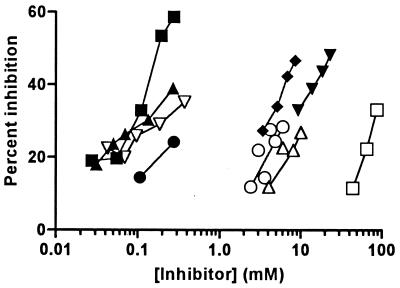
Figure 1.
Inhibition of binding of 125I-LNP to chitin. Various concentrations of mono- and oligosaccharides were combined with 109 ng of 125I-LNP and 250 μg of chitin in a total volume of 100 μl. B. japonicum USDA110 Nod factor (▪); R. sp. NGR234(NGRA) Nod factor (▴); R. sp. NGR234(NGRB) Nod factor (▿); R. meliloti Nod factor (●); N-acetylglucosamine (□); chitin disaccharide (▾); chitin tetrasaccharide (▵); chitin pentasaccharide (♦); chitin hexasaccharide (○). Most points represent single determinations. A sufficient amount of R. meliloti Nod factor was available only for the above assay; all other ligands showed similar results in assays with other chitin preparations.
Of a variety of oligosaccharides tested, the best inhibition was obtained with the Nod factor isolated from B. japonicum USDA110 (Fig. 1). This Nod factor consists of a chitin pentasaccharide backbone, modified by a 2-O-methyl α-l-fucose at C-6 of the sugar at the reducing end and the substitution of the acetyl group on the sugar at the nonreducing end with cis-vaccenic acid (14), an 18:1 fatty acid. No significant inhibition of LNP binding to chitin was obtained with cis-vaccenic acid when tested at concentrations up to 1.2 mM or with l-fucose at concentrations up to 50 mM.
Two Nod factors from Rhizobium sp. NGR234 also inhibited the binding of LNP to chitin (Fig. 1). These Nod factors differ from the USDA110 Nod factor in that they have a sulfate on C-3 (NGRA) or an acetate on C-4 (NGRB) of the 2-O-methyl-l-fucose; they are also methylated on the amide nitrogen, carbamoylated at C-6, and partially carbamoylated at C-3 or C-4 of the sugar at the nonreducing end (19). In contrast to the above Nod factors, the Nod factor from R. meliloti gave weaker inhibition when tested at equivalent concentrations (Fig. 1), although it should be noted that sufficient material was available for only two points. The predominant form of this Nod factor has a chitin tetrasaccharide backbone with a sulfate on C-6 of the sugar at the reducing end, an acetate at C-6 on the sugar at the nonreducing end, and a 16:2 fatty acid (20).
Both B. japonicum USDA110 and Rhizobium sp. NGR234 elicit the formation of nodules on D. biflorus roots; however, both of these rhizobial strains are only weak nodulators of this plant, and the nodules formed with the USDA110 strain do not fix nitrogen. No nodulation of the roots of this plant is obtained with R. meliloti. Nod factors from rhizobial strains that are strong nodulators of D. biflorus have not yet been purified or characterized.
Cloning and Characterization of LNP cDNA.
Peptides of LNP were obtained by reverse-phase HPLC of trypsin digests of the reduced, alkylated protein by using procedures described (21), and sequenced. The sequences of these peptides and of the LNP NH2 terminus (12) were used to design degenerate antisense and sense oligonucleotides for priming a PCR by using reverse-transcribed D. biflorus root RNA as a template. Sequence analysis of the cloned 727-bp product obtained in this reaction showed a single ORF encoding a polypeptide sequence that included the sequences of portions of the peptides adjacent to the regions used in the design of the primers as well as the sequence of a third tryptic peptide. This cDNA fragment was then extended by using RACE technology (17) to obtain the entire 1,597-bp cDNA sequence. To guard against the possibility of errors introduced during the PCR amplification, two clones were independently generated from each end, and both strands were sequenced in entirety. The 5′ and 3′ RACE products showed identical sequence overlaps with the original 727-bp fragment by 430 bp and 663 bp, respectively. Hybridization of these clones with total RNA prepared from young D. biflorus roots yielded a single band of ≈1,700 nt, a size large enough to encode the LNP protein.
The 1,597-bp sequence of the entire cDNA contains an ORF encoding a polypeptide of 462 aa (Fig. 2). The glutamic acid at position 1 initiates a 414-aa segment, which has an NH2 terminus identical to the NH2 terminus previously determined for LNP (12) and contains all of the sequence information obtained on tryptic peptides of this protein isolated above. The calculated molecular mass of 45,595 Da and the amino acid composition of this 414-aa segment are in close agreement with the values previously obtained on the isolated protein (12), providing further confirmation that this cDNA indeed represents LNP. Hydropathic plot analysis (data not shown) of the complete deduced amino acid sequence showed only one predicted transmembrane segment at residues −38 to −20. This 19-aa segment, which begins with a methionine, is characteristic of a hydrophobic signal sequence necessary for translocation across the endoplasmic reticulum membrane (22). The presence of such a signal is consistent with our unpublished findings that LNP is a glycoprotein that apparently traverses the secretory pathway. The 10 amino acids preceding this segment may also be part of this signal, although it is more likely that they represent a cytoplasmic tail of the nascent protein or may perhaps not even be translated. The signal sequence is separated from the NH2 terminus of the isolated protein by a 19-aa segment (residues −19 to −1). Although this segment may be a propeptide that is cleaved from the nascent protein after translation, we cannot rule out the alternative possibility that this cleavage is an artifact that occurs during extraction of the protein. Should this latter case be correct, it is possible that LNP may exist as a membrane protein in vivo, anchored to the membrane by way of its signal sequence.
Two consensus N-glycosylation sites are present in the sequence of the mature protein at residues 111 and 276 (Fig. 2). We have established that LNP is indeed glycosylated at at least one of these sites (data not shown). It should be noted, however, that we do not yet know whether other posttranslational modifications of this protein may occur, such as the COOH-terminal proteolysis that modifies two other lectins from this plant (21). A search of protein and nucleotide databases using the NCB1 tblastn and blastn programs (23) showed no significant similarities of LNP to the amino acid or cDNA sequences of any other plant or animal lectin yet described. It did, however, show 65.6% and 47.6% amino acid identity and 70.7% and 58.7% nucleotide identity with the sequences of a pea nucleotide triphosphatase (24) and an apyrase isolated from potato tubers (25), respectively. Considerably less, but significant, similarity also was found with the sequences of several other animal and yeast phosphohydrolases. Of particular interest in this comparison is the presence in all of these sequences of four motifs (designated by the boxes in Fig. 2) identified as conserved regions among a variety of plant and animal apyrases (25).
LNP Is an Apyrase.
The sequence similarities found among LNP and the above enzymes prompted us to test LNP for phosphohydrolase activity. LNP catalyzes the hydrolysis of phosphate from both ATP and ADP but showed no activity with AMP, pyrophosphate, or glucose 6-phosphate. The Km of LNP for Mg-ADP is 615 μM. The lectin has a broad specificity for nucleotide triphosphates, including GTP, CTP, and UTP (data not shown). This substrate specificity is characteristic of the apyrase category of phosphohydrolases (EC 3.6.1.5).
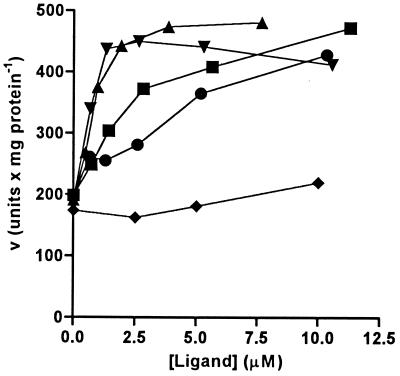
Preincubation of LNP with ligands that are recognized by its carbohydrate-binding site results in an increase in the Vmax of this enzyme. Low micromolar concentrations of the above Nod factors stimulate this increase in activity, with slightly higher concentrations necessary for the R. meliloti Nod factor than for the Nod factors produced by rhizobia that nodulate the plant (Fig. 3). Such an increase in enzyme activity also was obtained with low millimolar concentrations of the chitin oligosaccharides and N-acetylglucosamine, but not with N-acetylgalactosamine (data not shown). These results suggest that there is interaction between the carbohydrate-binding and phosphatase sites of LNP. Whether this interaction represents a direct stimulation of the enzyme activity or perhaps a stabilization of the enzyme under the assay conditions remains to be determined.
Figure 3.
Effect of carbohydrate ligands on phosphatase activity of LNP isolated from D. biflorus roots. LNP (402 ng/ml) was preincubated for 1 hour in 10 mM MOPS buffer, pH 7.2, containing various concentrations of B. japonicum USDA110 Nod factor (▪), Rhizobium sp. NGR234(NGRA) Nod factor (▴), Rhizobium sp. NGR234(NGRB) Nod factor (▾), R. meliloti Nod factor (●), or cis-vaccenic acid (♦) and then assayed for phosphatase activity by using a final concentration of 3 mM Mg-ADP. Points are averages of duplicate determinations.
Localization of LNP and Effects of Anti-LNP-Serum on Root Hair Deformation and Nodulation.
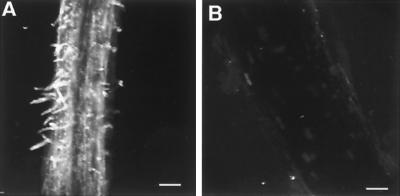
Immunofluorescence confocal microscopy of whole mounts of 7-day-old D. biflorus roots that had been fixed before staining showed that LNP is present on the epidermal cell surface of young roots and is particularly predominant on the surface of the root hairs (Fig. 4). The roots of 3-day-old D. biflorus plants were immersed for 1 hour in sterile medium or 1:100 dilutions of preimmunization serum or antiserum prepared against recombinant LNP. After washing, the plants were transferred to growth pouches. Half of the plants in each set were each inoculated with Bradyrhizobium sp. 24A10, a symbiont of D. biflorus. After 62 hours, the roots of some of these plants were analyzed for root hair deformation. A significant inhibition of root hair deformation was seen in those plants that had been treated with the antiserum (Table 1 and Fig. 5).
Figure 4.
Localization of LNP on D. biflorus roots. Immunofluorescence confocal microscopy of fixed whole mounts of the nodulation zone of 7-day-old D. biflorus roots treated with antiserum prepared against recombinant LNP (A) or preimmunization serum (B). Each image is a three-dimensional composite of 26–29 optical sections. (Bars = 100 μm.)
Table 1.
Effect of anti-LNP-serum on root hair deformation and nodulation of D. biflorus roots
| Treatment | Root hairs deformed, %
|
Nodules on roots inoculated with rhizobia
|
||
|---|---|---|---|---|
| Inoculated | Uninoculated | Treated region of root | Region of root emerged after treatment | |
| Untreated | 69.5 ± 4.6 | 6.8 ± 1.3 | 4.0 ± 0.2 | 1.7 ± 0.2 |
| Preimmunization serum | 76.6 ± 7.4 | 3.2 ± 0.6 | 3.8 ± 0.5 | 2.4 ± 0.5 |
| Anti-LNP-serum | 17.2 ± 3.6 | 4.4 ± 0.7 | 0.3 ± 0.1 | 1.3 ± 0.3 |
The roots of three sets of 3-day-old D. biflorus plants were immersed for 1 hour in 1:100 dilutions of preimmunization serum, anti-LNP-serum or serum-free medium, washed, and transferred to growth pouches. Half of each set of plants was inoculated with Bradyrhizobium sp. 24A10. After 62 hours, the roots of six plants from each of the above sets were examined for root hair deformation. After 3 weeks, the number of nodules in the treated region as well as in the region of root that emerged after treatment were recorded. No nodules were observed on the roots that had not been inoculated with rhizobia. The nodulation data represent the results of two separate experiments, using 10 and 16 plants per set, respectively. Values given are averages ± SE.
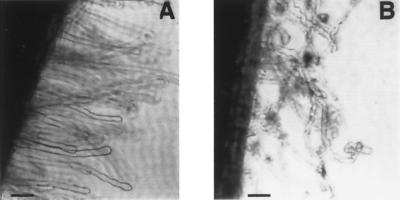
Figure 5.
Inhibition of root hair deformation by treatment of roots with anti-LNP-serum. A comparison of root hairs 62 hours after inoculation of 3-day-old D. biflorus roots with the symbiont, Bradyrhizobium sp. 24A10. Before exposure to rhizobia, the roots of the plants were incubated for 1 hour in 1:100 dilutions of anti-LNP-serum (A) or preimmunization serum (B). Whole mounts of the roots were examined by phase-contrast microscopy. (Bar = 50 μm.)
Other plants, treated as above, were allowed to grow for 3 weeks and then examined for nodule formation. Because new root growth occurred during that time interval, the treated and newly emerged root regions were examined separately. As shown in Table 1, the treatment of roots with antiserum to LNP inhibited the ability of these roots to be nodulated by the rhizobia; this inhibition occurred only in the region of the root that had been exposed to the antiserum. No nodules were observed on any of the roots that had not been inoculated with rhizobia. It is unlikely that the above inhibition is an indirect effect because of the interaction of the antibodies with the rhizobia because the treated roots were washed before the exposure to the rhizobia, and we have found that the antiserum does not react with these rhizobia (data not shown). Although the possibility of steric hindrance of a site adjacent to LNP on the root hair surface cannot be excluded, these inhibition results suggest that LNP may be involved in the events leading to the initiation of root hair deformation and nodulation.
DISCUSSION
Because oligosaccharides are important signals for a variety of developmental events in plants, the proteins that bind them are an essential target of study. In recent years, considerable attention has been focused on lectins, which are defined as proteins of nonimmune origin that bind to carbohydrate ligands without altering the covalent structures of the ligands (26). Although extensive studies have been conducted on these carbohydrate-binding proteins from the seeds of plants (26), relatively little attention has been focused on lectins produced by the plant vegetative tissues in which many of the oligosaccharide-signaling events are known to occur. In contrast to the abundant quantities of lectins found in the seeds of legumes, the vegetative tissues of these plants have been found to contain small amounts of lectins that are encoded by separate genes that are differentially expressed, both spatially and temporally (27). As shown in this paper, the lectin from the roots of the legume, D. biflorus, has no similarity in sequence to any plant, animal, or bacterial lectin yet described, and has properties that suggest it may play a role in the rhizobium–legume symbiosis and/or a related oligosaccharide-signaling event.
LNP was first identified as a lectin based on its ability to bind to hog blood group A + H substance (12). The heterogeneity of this complex carbohydrate (28) has enabled it to be used for the purification of a number of lectins with a wide range of specificities, including lectins, such as the wheat germ agglutinin and the tomato lectin, that bind to chitin (26). LNP differs from all other chitin binding proteins described to date (29) in that it does not have a hevein domain in its sequence. The low capacity of chitin for this lectin suggests that LNP may be recognizing either a large or a modified determinant on this polysaccharide.
The higher relative affinities of LNP for the Nod factors used in this study than for the chitin oligosaccharides indicate that the decorations of the chitin backbone contribute to the affinity of the lectin for these ligands. Although a slight preference in binding to Nod factors produced by rhizobia that nodulate the plant appears to be exhibited by this lectin, the limited amounts of Nod factor available for this study and possible differences in purity among the Nod factor preparations do not enable us to establish whether and to what extent LNP may exhibit host/strain specificity for Nod factors; such studies await the availability of purified Nod factors from normal rhizobial symbionts of the D. biflorus plant.
The ability of LNP to bind Nod factors, its presence on the root hair surface, and the inhibition of root hair deformation and nodulation by treatment of the roots with antiserum to the lectin suggest that LNP plays a role in the rhizobium–legume symbiosis. Previous attempts to implicate lectins in this symbiosis have been focused on the legume seed lectins (30, 31), which have not been reported to bind Nod factors. As a Nod factor-binding protein, LNP could possibly function in Nod factor transport, as a host/strain specific receptor, as a second, less stringent receptor postulated for this process (32), or even perhaps participate in events leading to a modification of the ligand. It also is possible that LNP or homologs of this protein that we have recently identified in the plant may function in the recognition of endogenous Nod-factor like signals that have been proposed to play a role in the regulation of plant growth and organogenesis (33). It is also possible that the expression of LNP may not be limited to the epidermal and root hair cells and that the protein may have a more generic role.
The low concentrations (10−12 M) of Nod factor that have been found to induce physiological responses in legumes (4) predict that Nod-factor receptors have high affinity for their ligands. Indeed, high-affinity binding sites for Nod factors have been found on particulate fractions from roots of the legume, M. truncatula, and the microsomal fraction of Medicago cell-suspension cultures (10, 11). Although the inhibition data in Fig. 1 show the relative affinities of LNP for its ligands, they do not permit the determination of the absolute affinities of this lectin for the Nod factors. The concentrations of Nod factors required for the stimulation of increased apyrase activity of the protein (Fig. 3) suggest that the Kds may be in the high nanomolar-to-low micromolar range. It should be noted, however, that these binding assays were conducted in solution in which LNP exists primarily as a monomer (12) rather than with membranes or cells, where LNP on the cell surface could function as a multivalent protein. As established with antibodies (34), such multivalence could increase the apparent affinity of LNP for multivalent ligands such as Nod factor micelles or Nod factor on the surface of rhizobia by several orders of magnitude. It should also be noted that increased enzymatic activity of CD39, an animal apyrase, has recently been shown to be associated with the degree of oligomerization of this membrane protein (35).
Whether LNP exists in vivo as an integral membrane protein or a peripheral protein remains to be determined. The sequence of this protein and the absence of any predicted transmembrane segment other than its signal sequence suggests that if indeed LNP is bound to the membrane via its signal, it would have little if any cytoplasmic tail. Should this protein function as a receptor, its ability to activate downstream events would thus most likely involve its association with other membrane proteins.
The presence of both carbohydrate-binding and apyrase activity on the same protein has prompted us to rename this lectin nucleotide phosphohydrolase LNP. The apparent interaction of these sites suggest that, on binding its carbohydrate ligand, LNP may play a role in activating downstream events either directly by signal transduction or indirectly, perhaps by serving as a motor for transport of the Nod factor or rhizobia with Nod factor on their surface. In this context, it is of interest that the human CD39 lymphoid cell activation antigen, one of the apyrases found to have some sequence similarity to LNP, is thought to be involved in the regulation of B cell adhesion (36). Although these other apyrases have not been tested for lectin activity, it is possible that such dual activities of these proteins may have been conserved throughout evolution.
The unique amino acid sequence and apyrase activity of LNP distinguish this lectin from the conventional lectins found in abundance in the seeds of legumes (37). The possibility that other such plant lectin/enzymes exist is suggested by the recent finding of a cDNA from Arabidopsis thaliana that encodes a receptor-like serine/threonine kinase as well as a legume seed lectin-like domain (38). LNP may thus be one of many multifunctional carbohydrate binding proteins that may function in plant oligosaccharide-signaling events. A variety of transgenic experiments are underway to elaborate its role in such processes.
Acknowledgments
We thank Dr. Jean Denarie for his generous gift of some of the Nod factors used in this study. We thank Jill Brigham and Anuja Ramil for their expert technical assistance and Dr. Irwin Segel and Dr. Gary Stacey for helpful discussions. This research was supported by National Institute of General Medical Sciences–National Institutes of Health Grant GM21882 and U.S. Department of Agriculture Grant 97-35305-4630. R.B.D. was supported by Department of Energy Grant DE-FG02-97ER-20260.
ABBREVIATION
- LNP
lectin–nucleotide phosphohydrolase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF139807).
References
- 1.Ryan C A, Farmer E E. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:651–674. [Google Scholar]
- 2.Mohnen D, Hahn M G. Semin Cell Biol. 1993;4:93–102. doi: 10.1006/scel.1993.1012. [DOI] [PubMed] [Google Scholar]
- 3.Cote F, Hahn M G. Plant Mol Biol. 1994;26:1379–1411. doi: 10.1007/BF00016481. [DOI] [PubMed] [Google Scholar]
- 4.Denarie J, Debelle F, Prome J-C. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 5.Mylona P, Pawlowski K, Bisseling T. Plant Cell. 1995;7:869–885. doi: 10.1105/tpc.7.7.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt J, Rohrig H, John M, Wieneke U, Stacey G, Koncz C, Schell J. Plant J. 1993;4:651–658. [Google Scholar]
- 7.Baureithel K, Felix G, Boller T. J Biol Chem. 1994;269:17931–17938. [PubMed] [Google Scholar]
- 8.Shibuya N, Ebisu N, Kamada Y, Kaku H, Cohn J, Ito Y. Plant Cell Physiol. 1996;37:894–898. doi: 10.1093/oxfordjournals.pcp.a029002. [DOI] [PubMed] [Google Scholar]
- 9.Shibuya N, Ito Y, Kaku H. In: Biology of Plant–Microbe Interactions. Stacey G, Mullin B, Gresshoff P M, editors. St. Paul, MN: Soc. Mol. Plant–Microbe Int.; 1996. pp. 83–88. [Google Scholar]
- 10.Bono J-J, Riond J, Nicolaou K C, Bockovich N J, Estevez V A, Cullimore J V, Ranjeva R. Plant J. 1995;7:253–260. doi: 10.1046/j.1365-313x.1995.7020253.x. [DOI] [PubMed] [Google Scholar]
- 11.Niebel A, Bono J-J, Ranjeva R, Cullimore J. Mol Plant–Microbe Interact. 1997;10:132–134. [Google Scholar]
- 12.Quinn J M, Etzler M E. Arch Biochem Biophys. 1987;258:535–544. doi: 10.1016/0003-9861(87)90375-4. [DOI] [PubMed] [Google Scholar]
- 13.Etzler M E. Glycoconj J. 1994;11:395–399. doi: 10.1007/BF00731274. [DOI] [PubMed] [Google Scholar]
- 14.Sanjuan J, Carlson R W, Spaink H P, Bhat U R, Barbour W M, Glushka J, Stacey G. Proc Natl Acad Sci USA. 1992;89:8789–8793. doi: 10.1073/pnas.89.18.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor B, Powell A. Focus. 1982;4:4–6. [Google Scholar]
- 16.Ausubel F, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current Protocols in Molecular Biology. Vol. 1. New York: Wiley; 1988. pp. 3.7.1–3.7.3. [Google Scholar]
- 17.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drueckes P, Schinzel R, Palm D. Anal Biochem. 1995;230:173–177. doi: 10.1006/abio.1995.1453. [DOI] [PubMed] [Google Scholar]
- 19.Price N P J, Talmont F, Wieruszeski J-M, Prome D, Prome J C. Carbohydr Res. 1996;289:115–136. doi: 10.1016/0008-6215(96)00119-x. [DOI] [PubMed] [Google Scholar]
- 20.Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Prome J C, Denarie J. Nature (London) 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 21.Etzler M E. Biochemistry. 1994;33:9778–9783. doi: 10.1021/bi00198a049. [DOI] [PubMed] [Google Scholar]
- 22.von Heijne G. Nucleic Acids Res. 1986;11:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh H-L, Tong C-G, Thomas C, Roux S J. Plant Mol Biol. 1996;30:135–147. doi: 10.1007/BF00017808. [DOI] [PubMed] [Google Scholar]
- 25.Handa M, Guidotti G. Biochem Biophys Res Comm. 1996;218:916–923. doi: 10.1006/bbrc.1996.0162. [DOI] [PubMed] [Google Scholar]
- 26.Goldstein I J, Poretz R D. In: The Lectins: Properties, Functions, and Applications in Biology and Medicine. Liener I E, Sharon N, Goldstein I J, editors. San Diego: Academic; 1986. pp. 33–247. [Google Scholar]
- 27.Etzler M E. Trends Glycosci Glycotechnol. 1998;10:247–255. [Google Scholar]
- 28.Van Halbeek H, Dorland L, Vliegenthart J F G, Kochetkov N K, Arbatsky N P, Derevitskaya V A. Eur J Biochem. 1983;127:21–29. doi: 10.1111/j.1432-1033.1982.tb06832.x. [DOI] [PubMed] [Google Scholar]
- 29.Beintema J J. FEBS Lett. 1994;350:159–163. doi: 10.1016/0014-5793(94)00753-5. [DOI] [PubMed] [Google Scholar]
- 30.Diaz C L, Melchers L S, Hooykass P J J, Lugtenberg B J J, Kijne J W. Nature (London) 1989;338:579–581. [Google Scholar]
- 31.van Rhijn P, Goldberg R B, Hirsch A M. Plant Cell. 1998;10:1233–1249. doi: 10.1105/tpc.10.8.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ardourel M, Demont N, Debelle F, Maillet F, de Billy F, Prome J-C, Denairie J, Truchet G. Plant Cell. 1994;6:1357–1374. doi: 10.1105/tpc.6.10.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spaink H P, Wijfjes A H M, Van Vliet T B, Kijne J W, Lugtenberg B J J. Aust J Plant Physiol. 1993;20:381–392. [Google Scholar]
- 34.Hornick C L, Karush F. Immunochemistry. 1972;9:325–340. doi: 10.1016/0019-2791(72)90096-1. [DOI] [PubMed] [Google Scholar]
- 35.Wang T-F, Ou Y, Guidotti G. J Biol Chem. 1998;272:24814–24821. doi: 10.1074/jbc.273.38.24814. [DOI] [PubMed] [Google Scholar]
- 36.Kansas G S, Wood G S, Tedder T F. J Immunol. 1991;146:2235–2244. [PubMed] [Google Scholar]
- 37.Sharon N, Lis H. FASEB J. 1990;4:3198–3208. doi: 10.1096/fasebj.4.14.2227211. [DOI] [PubMed] [Google Scholar]
- 38.Herve C, Dabos P, Galaud J-P, Rouge P, Lescure B. J Mol Biol. 1996;258:778–788. doi: 10.1006/jmbi.1996.0286. [DOI] [PubMed] [Google Scholar]