Abstract
Plant disease epidemics resulting from introductions of exotic fungal plant pathogens are a well known phenomenon. An associated risk—that accelerated pathogen evolution may be occurring as a consequence of genetic exchange between introduced, or introduced and resident, fungal pathogens—is largely unrecognized. This is, in part, because examples of natural, interspecific hybridization in fungi are very rare. Potential evolutionary developments range from the acquisition of new host specificities to emergence of entirely new pathogen taxa. We present evidence from cytological behavior, additive nucleotide bases in repetitive internal transcribed spacer regions of the rRNA-encoding DNA (rDNA), and amplified fragment length polymorphisms of total DNA that a new, aggressive Phytophthora pathogen of alder trees in Europe comprises a range of heteroploid-interspecific hybrids involving a Phytophthora cambivora-like species and an unknown taxon similar to Phytophthora fragariae. The hybrids’ marked developmental instabilities, unusual morphological variability, and evidence for recombination in their internal transcribed spacer profiles indicates that they are of recent origin and that their evolution is continuing. The likelihood of such evolutionary events may be increasing as world trade in plants intensifies. However, routine diagnostic procedures currently in use are insufficiently sensitive to allow their detection.
Phytophthora is a major genus of plant pathogens within the diploid, alga-like Oomycete fungi. The genus contains about 60 species. Many of these, such as the potato blight pathogen Phytophthora infestans or the forest dieback pathogen Phytophthora cinnamomi, are responsible for destructive crop epidemics or for destabilizing terrestrial ecosystems (1). In such cases the pathogen usually is introduced (2, 3). In 1993, a previously unknown Phytophthora was reported from southern Britain that caused the death of riparian alder (Alnus spp.) (4, 5); more than 10,000 trees were killed in 1996 (J. N. Gibbs, personal communication). Conventionally, Phytophthora species are distinguished on phenotypic characteristics (1). The new alder Phytophthora exhibited female gametangia (oogonia) with distinctive surface ornamentation and two-celled amphigynous (collar-like) male gametangia (antheridia) (Fig. 1e). These features are uniquely characteristic of Phytophthora cambivora, a common pathogen of trees (1) that is nonpathogenic to alder (C.M.B., unpublished data). However, the alder Phytophthora also differed strikingly from P. cambivora. It was self-fertile rather than outcrossing, had a submerged instead of an aerial colony type (Fig. 1b), and had markedly lower cardinal temperatures for growth (5). It also exhibited an unusually high level of zygotic abortion (5).
Figure 1.
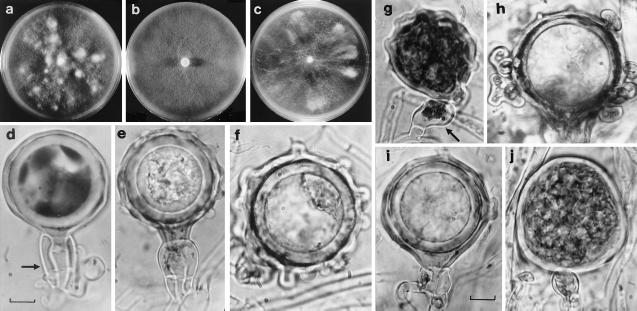
(a–c) Colonies of alder phytophthoras. (a) Swedish variant, showing chimeric white fertile patches. (b) Standard alder Phytophthora with typical appressed colony. (c) Dutch variant, showing irregular fertile sectors. (d–f) Gametangial types. (d) Swedish variant showing smooth oogonium (above) and two-celled amphigynous antheridium (arrowed). (e) Standard alder Phytophthora, showing P. cambivora-like ornamented oogonium and two-celled amphigynous antheridium. (f) Netherlands variant showing extremely ornamented oogonium. (g–j) Diverse gametangial types representative of four different, single hyphal-tip subcultures from U.K. variant P841, including oogonium with paragynous antheridium (g, arrowed) and highly coralloid oogonium (h). (Bar = ≈10 mm.)
Reports of natural hybridization in fungi are very rare (6, 7), but this combination of properties suggested the new Phytophthora might be a hybrid involving P. cambivora as a parent (5). A preliminary cytological study of an alder Phytophthora isolate revealed abnormal nuclear behavior. This observation led us to examine the sequences of the internal transcribed spacer (ITS) regions in its rRNA genes (rDNA). These genes occur in multiple arrays, and mutations in their noncoding regions occur at a rate that approximates the rate of species emergence. Over time, such mutations become fixed through unequal crossing over and gene conversion, a process that is commonly termed concerted evolution (8–12). Within a species, ITS sequences tend to be distinct and monomorphic. They therefore are suitable for species discrimination across a wide range of organisms, including fungi (13). However, in the alder Phytophthora, dimorphic (or additive) ITS arrays were found that were comparable to those recently demonstrated for plant hybrids (11, 12, 14–16).
These observations, together with reports that outbreaks of a new alder disease associated with a Phytophthora were occurring across continental Europe, led us to investigate the hybrid species hypothesis in detail. All available European isolates of the Phytophthora were examined. We report here that the new alder Phytophthora comprises a range of heteroploid hybrids. They exhibit a remarkable range of oogonial and antheridial types, high levels of developmental instability, and unusual combinations of additive and monomorphic ITS polymorphisms. Overall, their properties suggest they represent a hybrid complex that has originated in a recent interspecific hybridization event. The role of interspecific hybridization as a mechanism for rapid evolution of fungal plant pathogens is discussed.
MATERIALS AND METHODS
Fungal Isolates, Culture Methods, and Cytology.
Isolates that were studied are shown in Table 1. Growth medium used and growth temperature tests at 20–35°C were as described elsewhere (5). Nuclei and chromosomes in developing gametangia were examined by the Phytophthora acetoorcein method (17). A minimum of three metaphase plates was counted per isolate, except for Phytophthora fragariae (one only), because its gametangia are rarely produced on agar.
Table 1.
Isolates of the alder Phytophthora, P. cambivora, and P. fragariae and their comparative cytological and cultural properties
| Species | Isolates | Chromosome no.,* inferred karyotype | Meiosis | Colony development | Oogonial type,† | Antheridial type‡ | Optimum/maximum growth temperatures, °C |
|---|---|---|---|---|---|---|---|
| Alder Phytophthora | |||||||
| Standard form | P669¶, P670¶‖, P671‖, P766‖, P772¶ (U.K.); P817¶, P818§¶ (Germany); P834¶‖, P846¶ (France); P844,¶‖, P883, P884 (Austria) | 18–22, ≈4n + 2 | Incomplete | Unstable | Ornamented | 2A | ≈25/30 |
| German variant | P889¶‖, P890¶‖ | 16–18, ≈2n + 7 | Incomplete | Unstable | Ornamented | 2A | ≈27/32 |
| Dutch variant | P770¶‖ | 13–15, ≈2n + 4 | Complete | Very unstable | Extremely ornamented | 1A + P | ≈27/32 |
| Swedish variant | P875¶‖, P876¶‖, P887¶, P888¶ | 11–13, ≈2n + 2 | Complete | Very unstable | Smooth | 2A | ≈27/30 |
| U.K. variant | P841¶‖ | § | |||||
| P. cambivora** | P199¶‖, P208‖, P819‖, P820‖ (U.K.); P821‖ (Italy) | 10–12, 2n | Complete | Stable | Ornamented | 2A | ≈27/34 |
| P. fragariae** | |||||||
| Var. fragariae | P964¶, P965¶ (U.K.) | 10–12, 2n | Complete | Stable | Smooth | 1A + P | ≈25/30 |
| Var. rubi | P822‖ (France); P823‖ (U.K.) |
A range limit normally is given for Phytophthora chromosome numbers.
2A, large, two-celled amphigynous antheridia; 1A + P, predominantly small, single-celled amphigynous plus some paragynous antheridia.
U.K. variant P841 was too unstable to tabulate; see text.
Isolate examined by ITS sequence or by ITS digestion.
Isolate examined cytologically.
In a separate study, a further 20 isolates of P. cambivora and >40 isolates of the two varieties of P. fragariae, respectively, were subjected to ITS digestion; in each case they exhibited the same digest patterns as the isolates of these species listed here.
PCR Amplification and Sequencing of ITS Regions.
Total DNA was extracted from freeze-dried mycelium (Puregene DNA extraction kit; Gentra Systems) and PCR (18) using primers ITS4 (19) and ITS6 (20), which were used to amplify an ≈940-bp product containing the ITS1, 5.8S gene, and ITS2 regions. The PCR products were purified by using Wizard spin columns (Promega), and both strands were sequenced by using a Dye-Terminator Cycle Sequencing Kit and an Applied Biosystems 373 automated sequencer (Perkin–Elmer). Sequences were edited by eye and aligned by using clustalw (21).
Design of Restriction Digest Assays, Cloning, and Analysis of ITS Arrays.
Sequencing of the pooled PCR products revealed a number of sites in which 2 bases were present. These were examined with the computer package map (GCG; ref. 22) to identify discriminatory restriction enzymes at such sites. The enzymes AluI, AciI, Eco0109I, MspI, and TaqI distinguished 8 of 12 polymorphic bases (Fig. 2), and, after electrophoresis on 2.5% NuSieve agarose (FMC), the different restriction digest patterns were clearly resolved. This assay was used in two ways: first, to examine total ITS diversity by digestion of PCR products amplified directly from total DNA, and second, to distinguish the bases present in single, cloned copies of the ITS array. In seven isolates (Table 1) the PCR products were cloned by ligation into a pGEM-T vector (Promega) and transformation into Ultracompetent Epicurian coli XL2-Blue Cells (Stratagene) according to the supplier’s instructions. Thirty clones from each isolate were selected at random, and the pattern of ITS bases was established by applying the above restriction assay.
Figure 2.
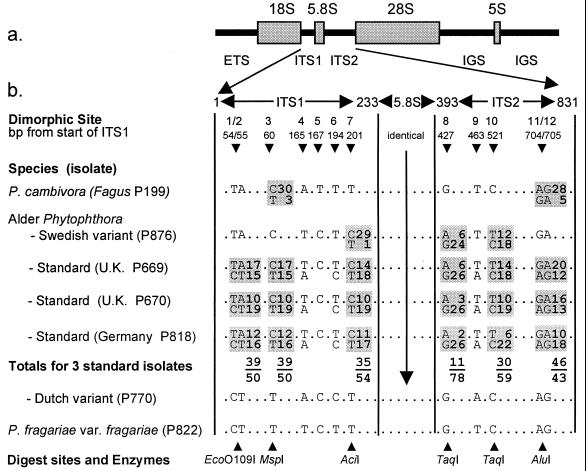
Location of polymorphisms in rDNA of alder Phytophthora isolates. (a) Arrangement within a single repeat of an rRNA gene (rDNA) of the rRNA subunits and spacer regions (ETS, external transcribed spacer; ITS1 and ITS2, internal transcribed spacers; IGS, intergenic spacer; 18S, 5.8S, 28S, and 5S subunits). (b) Arrangement of polymorphic sites within the 831-bp region covering the ITS1 spacer, the 5.8S subunit, and ITS2 spacer: the 5′ end of ITS1 spacer sequence has been designated as 1, and all other sites are numbered accordingly. At dimorphic sites (shaded regions) both bases are shown and boxed. Numbers alongside the boxes refer to the numbers of cloned PCR products containing these bases at these sites; totals for all three standard alder isolates are also shown. Along the bottom are the restriction enzymes used to examine each of the dimorphic sites.
Amplified Fragment Length Polymorphism (AFLP) Analysis of Total DNA.
A modified AFLP protocol (23) was used: DNA (250 ng) was digested with MseI and EcoRI, adapters were ligated, and a preamplification step with E00 and M00 primers used to create a pool of amplified product from which selective PCR with 33P-labeled 2-base overhang primers (E primer labeled in each case) was carried out. Four primer combinations (E11/M14, E11/M16, E19/M15, E19/M17) were used, and each autoradiograph was digitized and the bands were analyzed on gelcompar software (Applied Maths, Kortrijk, Belgium). All four gels were combined end to end, and the Pearson product–moment correlation coefficient was used to estimate levels of similarity between the densitometric curve data for each isolate. The UPGMA (unweighted pair-group method of averages) algorithm was used to construct a dendrogram from this similarity matrix.
RESULTS AND DISCUSSION
Occurrence of Unusual Phenotypic Variants.
A comparison of alder Phytophthora isolates from Europe and the United Kingdom (U.K.) revealed greater variation than expected in a single Phytophthora taxon (Table 1). Seven isolates (from France, Germany, and Austria) closely resembled four “standard” U.K. isolates. However, another eight isolates (from Sweden, Germany, Netherlands, and U.K.), termed here “variants,” had striking phenotypic differences. These included a higher cardinal temperature for growth, colony instabilities, and unusual gametangial development (Table 1). For example, the Swedish variant produced gametangia only within discrete chimeric fertile patches (Fig. 1a). Its oogonia were smooth-walled instead of ornamented (Fig. 1d), and some had an abnormal beak-like form. The Dutch variant also produced oogonia only in fertile colony sectors (Fig. 1c) but these were extremely ornamented (Fig. 1f), often with remarkable coralloid outgrowths (cf. Fig. 1h), a morphological character not recorded previously in a Phytophthora. Many of its oogonia were also highly distorted, some being little more than a primitive, hypha-like tube. In addition, the Dutch variant produced only small, single-celled antheridia. Some of these were not amphigynous but paragynous (laterally attached; cf. Fig. 1j), as the antheridia of P. fragariae (24). The latter is a specialized pathogen of raspberry and strawberry in the same molecular clade as P. cambivora and P. cinnamomi (18).
Cytological Evidence for Hybridization.
Phytophthora is diploid, and its chromosomes, though very small, can be observed by light microscopy during gametangial meiosis (17, 25, 26). Owing to their small size, a range normally is given for chromosome numbers at metaphase (17). Chromosome behavior was examined in eight standard alder Phytophthora isolates, one U.K., one Dutch, two German, and two Swedish variants, plus five sexually paired P. cambivora and two P. fragariae var. rubi control isolates.
The previously reported diploid chromosome number for P. cambivora of n = 10–12 (26) was confirmed. A normal meiosis was followed directly by fertilization. P. fragariae similarly was shown to be n = 10–12 (Table 1). In contrast, the chromosome number of nine standard alder Phytophthora isolates was n = 18–22, i.e., approximately tetraploid. There was a normal prophase and chiasma formation, and multivalents were observed at metaphase I. However, frequent chromosome-pairing failures were seen and no second-division stages were observed. The postdivision nuclei were mostly very large, often irregular in size, and frequently failed to dissociate. No evidence of fertilization was seen. These irregularities indicated that the standard alder Phytophthora was a polyploid species hybrid unable to proceed beyond first meiotic division because of difficulties in chromosome pairing. The U.K. and German variants exhibited similar irregularities, but with n = 17–19 and n = 16–18, respectively. The Dutch variant had a lower chromosome number of n = 13–15 and a normal meiosis, although the divisions often resulted in irregularly sized nuclei. The Swedish variant was only n = 11–13, i.e., close to diploid, with normal meiosis and fertilization stages. The variants, therefore, ranged from abnormal to normal in their nuclear behavior.
Evidence from ITS Sequences.
The ITS regions of six alder Phytophthora isolates (three standard and three variant forms) were PCR-amplified and sequenced. These sequences were aligned with those published previously for 21 other Phytophthora species (18, 27, 28) and unpublished data for a further 15 species. In a phylogenetic analysis, the standard and variant alder Phytophthoras grouped in a compact clade that comprised only P. cambivora, P. fragariae, and the more distantly related P. cinnamomi (results not shown). The sequences of most alder isolates differed from P. fragariae and P. cambivora by only 12 bases (Fig. 2). The standard isolates (e.g., P669, P670, and P818) had a highly unusual pattern not reported previously in any other Phytophthora species: two overlapping peaks occurred in the electropherogram at 11 of 12 distinguishing bases, indicating a combination of different ITS types in the pooled PCR product, i.e., additivity.
Such additivity recently has been demonstrated in the rDNA of allopolyploid species hybrids in plants (11, 12, 14–16). After a hybridization event, bidirectional concerted evolution generally leads to homogenization and the emergence of only one of the parental ITS types (14). However, long-term maintenance of both types also may occur (15), possibly because of centromeric location of the rRNA genes (29). In the absence of either crossing over or chromosome loss, parental ITS arrays on homeologous chromosomes would be expected to remain discrete and in their parental form, i.e., in a 1:1 ratio. To examine the extent of homogenization in the alder Phytophthora, the base combinations at 6 of 12 polymorphic sites, representing 8 bp covering both ITS regions (Fig. 2), were determined by using the previously described restriction digest assay. Approximately 30 clones were analyzed for each of 3 standard and 2 variant alder isolates, P. cambivora and P. fragariae. All the P. fragariae clones were of one ITS type, whereas the majority (28/31) of the P. cambivora clones were of another (Fig. 2). In 89 clones from the standard isolates, two different base options were present in approximately 1:1 ratios at positions 1–2, 3, 7, and 11–12 (Fig. 2). At positions 8 and 10, however, the base frequencies were unbalanced: at position 8 the A/G ratio was 11:78, and at position 10 the T/C ratio was 30:59.
In the Swedish variant, the bases were monomorphic at all but three sites and the overall sequence was more similar to that of P. cambivora than to the other variants (see below). Of the three dimorphic sites, site 7 was rarely so (1 in 29 clones) but dimorphism was more frequent at sites 8 (6:24 ratio) and 10 (12:18). This matched the imbalance in the ratios observed at sites 8 and 10 in the standard isolates, indicating either that the standard isolates have contributed to the genome of the Swedish variant or vice versa. In contrast, the Dutch, German, and U.K. variants were monomorphic throughout, with ITS sequences very similar to that of P. fragariae (only three differences). Examination of a further eight standard and variant isolates by digestion of the pooled PCR product corroborated the above patterns.
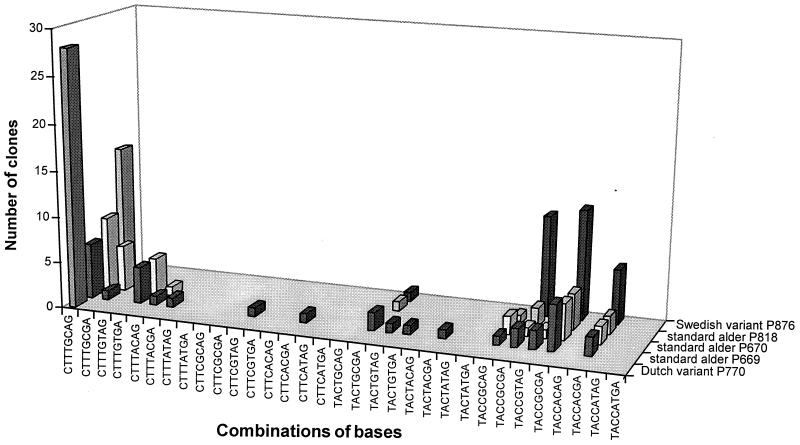
Although in the standard isolates the ratio of polymorphic bases at the majority of sites was ≈1:1 (Fig. 2), the polymorphisms occurred in many different combinations among the clones, with no particular combinations being dominant (Fig. 3). Thus, among the 89 clones examined, 16 different combinations (of a possible 64) occurred. This indicates that crossing over is continuing between the parental rDNA arrays, i.e., the process of homogenization. The absence of CT/T or TA/T combinations at sites 1/2 and 3 (Fig. 3) probably reflects their close linkage (only 5 bp apart; Fig. 2). It also suggests crossing over rather than gene conversion as the explanation for the new base combinations.
Figure 3.
Distribution of ITS sequence combinations across five alder Phytophthora isolates (three standard, one Dutch, and one Swedish variant) as determined by restriction digestion at six points across the cloned ITS PCR products. More ITS combinations are evident in the standard hybrid types than in the variants.
In P. cambivora, the additive bases were also in imbalance at site 3 (30/3) and sites 11–12 (28/5). Restriction digests indicated that additive bases always occurred at the same positions and, judging from band intensities, in similar proportions in all five isolates of P. cambivora examined. These sites may represent the remnants of an earlier reticulation event in the evolution of this species (compare also the significant AFLP variation in P. cambivora; Fig. 4).
Figure 4.
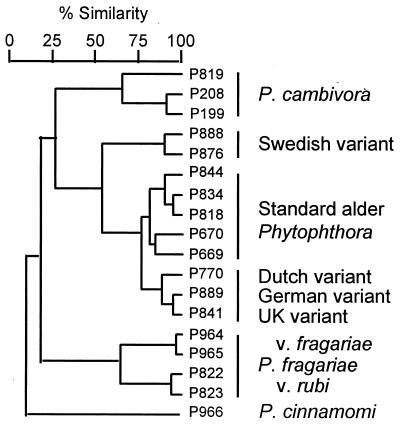
Dendrogram prepared from AFLP fingerprints of genomic DNA of the standard alder Phytophthora (five isolates); the Dutch (one), German (one), U.K. (one), and Swedish variants (two); P. cambivora (three); P. fragariae [var. fragariae (two) and var. rubi (two)]; and P. cinnamomi (one).
Evidence from Total DNA.
AFLP fingerprinting of total DNA provides a useful measure of fine-scale genetic variation in fungi (30). Fingerprints from four primer combinations were used to generate a dendrogram of relationships among isolates representing the alder phytophthoras, P. cambivora and P. fragariae, with P. cinnamomi as an outgroup. The result confirmed the high degree of relatedness within the alder phytophthoras (Fig. 4). The Dutch, German, and U.K. variants formed a discrete cluster and were more closely related to the standard isolates than the Swedish variants. These groupings therefore were in agreement with those based on ITS evidence. In each of the four individual primer combinations, P. cambivora shared more bands with the alder phytophthoras than did P. fragariae, and in one primer combination P. cambivora and the Swedish variant clustered together (results not shown). Overall, therefore, these results were consistent with a common origin for all the alder isolates and with the involvement of P. cambivora in their genesis.
More AFLP markers were observed in the near tetraploid standard isolates (mean, 68 markers) than in either the Dutch or German variants (60 each), the Swedish variant (55), or in the diploid P. cambivora and P. fragariae (58 and 55). This is consistent with the expectation that in an allopolyploid species hybrid, the number of markers per primer combination would increase with the number of homeologous chromosomes. In addition, the banding pattern of the standard isolates was almost entirely a composite of the bands in the Dutch/German and Swedish variants, providing further proof that these variants are either parents or breakdown products of the standard form.
Extreme Phenotypic Instability of U.K. Variant P841.
A recently acquired U.K. isolate, P841, is the only variant among more than 40 isolates of the alder Phytophthora sampled in the U.K. Since it was first isolated, P841 has been unstable in culture. Fifty single hyphal-tip subcultures were taken in an attempt to “stabilize” it. These cultures exhibited numerous unique combinations of colony patterns, growth rates, and growth–temperature relationships. They also ranged from being highly fertile to sterile; from producing very small, highly coralloid oogonia to very large, smooth-walled oogonia; and forming either two-celled amphigynous antheridia, single-celled amphigynous antheridia, or a mixture of single-celled amphigynous and paragynous antheridia (Fig. 1 g–j). This again represented extremes of variation not usually seen in Phytophthora species. The gametangial types alone were a bizarre range of P. cambivora- and P. fragariae-like characters. The ITS profiles of six such phenotypically diverse subcultures, together with the “original” P841 culture, were examined and found to share the same monomorphic profile as Dutch variant P770 (Fig. 2).
Origins and Status of the Alder Phytophthoras.
Our analysis shows that the new “alder Phytophthora” is not a single taxonomic entity but an array of phenotypically highly diverse heteroploid genotypes. The cytological evidence, ITS sequence data, and AFLP banding patterns all indicate that the common standard form is an allopolyploid species hybrid. It is also evident that the standard and variant forms are closely related, though the precise evolutionary basis of their relationship remains unclear. In the past, interspecific hybridization has been suggested as an explanation for polyploidy, phylogenetic complexity, and sterility in Phytophthora (31–33), and synthetic hybrids have been created in the laboratory (34, 35). However, to our knowledge there has been no previous demonstration of a naturally occurring hybrid complex in the genus.
An outstanding issue is the identity of the hybrids’ progenitors. The possibility that P. cambivora and P. fragariae are the immediate progenitors of the standard hybrid fits the observed phenotypic variation well, but is not fully consistent with the pattern of ITS dimorphism and is not well supported by the AFLP data. Although the latter suggest that the Swedish and Dutch variants could be the progenitors, the phenotypic diversity, developmental instability, and intermediate chromosome numbers of the variants as a whole indicate that they are most probably products of recombination and chromosome loss in the standard hybrid that have reverted toward one or the other parental genotype and, in some cases, regained a normal meiosis. The possibility that they could have arisen via somatic, rather than sexual, segregation is suggested by the extreme variability of the hyphal-tip subcultures taken from U.K. variant P841. On present evidence, therefore, we favor the view that the progenitors of the standard hybrid are P. cambivora and an unknown taxon close to P. fragariae. Nonetheless, the additional possibility that more than one hybridization event was involved cannot yet be excluded.
The novel combinations of ITS polymorphisms in the standard isolates contrast with the ITS uniformity in other phytophthoras (18, 27, 28) and other fungal groups. It indicates their rDNA is in the process of fixation, providing clear evidence for ongoing, concerted evolution. The monomorphic ITS sequence of the Dutch, German, and U.K. variants and the limited dimorphism in the Swedish variant may be a result of partial or complete homogenization of the rDNA or loss of the relevant parental chromosomes before recombination. These features, plus the unusual phenotypic instability already described, indicate that the standard hybrid is in a nascent state and that major genetic controls contributed by its parent chromosome sets remain in conflict. Such instability is likely to result in rapid genomic reorganization. The latter is a process by which allopolyploid hybrids can evolve in sympatry (37, 38). It also will result in the appearance of novel AFLP markers, blurring the phylogenetic relationship between hybrids and their parent species.
Wider Implications for the Evolution and Detection of Plant Pathogens.
The hybrid status of the alder phytophthoras raises many issues and challenges. One is the role of interspecific hybridization in the population biology of fungal plant pathogens. In fungi of all types, examples of natural interspecific hybridization are very rare, with as few as six clear examples before 1990 (6, 7). In consequence, the potential for gene flow between fungal plant pathogens and the associated risk that these processes could lead to the emergence of new plant diseases have been largely overlooked (7).
Theoretically, the risk is greatest when closely related but previously geographically isolated taxa, which lack strong reproductive barriers, come into contact in the same niche (7). This is the case with P. cambivora and P. fragariae: both species are believed to be exotic to Europe (39, 40) and both can occur together on infected Rubus (41). Such conditions are likely to increase as world trade in plants intensifies and more plants and their associated pathogens are introduced into new biogeographic environments. Further support for this viewpoint comes from evidence that hybrids are appearing between the resident and recently immigrant Dutch elm disease pathogens Ophiostoma ulmi and Ophiostoma novo-ulmi in Europe (42) and evidence that a new Melampsora rust in New Zealand is a hybrid between two introduced species (43). In addition, two recent, molecular-based studies show that hybridization has played a significant role in the origins of modern Fusarium and Epichloe taxa (44, 45).
By increasing levels of global environmental disturbance, therefore, we may be enhancing the potential for rapid evolution of fungal pathogens. Our ability to assess the full extent of these processes, however, will depend on our ability to detect them. Many hybrids or introgressants are unlikely to be detected by the conventional, mainly morphologically based diagnostic methods used in international quarantine. They may not be detectable even by ITS-based methods if homogenization of the rDNA arrays has occurred. Furthermore, a closely related issue, which has yet to be tested in mycology, is how to taxonomically define a hybrid or a hybrid complex in a way that is of practical use in quarantine legislation and diagnosis (36). This issue is well exemplified by the diverse properties of the alder phytophthoras.
Interspecific hybrids are more likely to survive if they have a fitness advantage over the parent species, such as increased aggressiveness or the ability to exploit a new host (7). Although Alnus species play an important role in the stability of riparian ecosystems across the northern hemisphere, they have not been reported previously to be susceptible to any Phytophthora (1, 5). In a separate study, we have demonstrated that neither P. fragariae nor P. cambivora is pathogenic to Alnus. Remarkably, progenies with novel host specificities recently have been recovered among laboratory-generated Phytophthora hybrids (35). The specificity for Alnus shown by the alder phytophthoras therefore may have arisen through the hybridization process. Such possibilities, together with the other issues already raised, indicate that the role of interspecific hybridization in fungal plant pathogens should be more critically assayed and its evolutionary significance more thoroughly investigated.
Acknowledgments
We thank Susan Kirk and Naomi Williams for technical assistance and European colleagues for providing Phytophthora isolates from dying alder.
ABBREVIATIONS
- ITS
internal transcribed spacer
- AFLP
amplified fragment length polymorphism
- rDNA
rRNA-encoding DNA.
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF13966, AF13967, AF13968, AF13969, and AF13970).
References
- 1.Erwin D C, Ribeiro O K. Phytophthora Diseases Worldwide. St. Paul, MN: American Phytopathological Society; 1996. [Google Scholar]
- 2.Zentmyer G A. Trans Br Mycol Soc. 1988;91:367–378. [Google Scholar]
- 3.Goodwin S B, Cohen B A, Fry W E. Proc Natl Acad Sci USA. 1994;91:11591–11595. doi: 10.1073/pnas.91.24.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibbs J N. EPPO Bull. 1995;25:661–664. [Google Scholar]
- 5.Brasier C M, Rose J, Gibbs J N. Plant Pathol. 1995;44:999–1007. [Google Scholar]
- 6.Burnett J H. Trans Br Mycol Soc. 1983;81:1–14. [Google Scholar]
- 7.Brasier C M. Can J Bot. 1995;73:S1213–S1221. [Google Scholar]
- 8.Dover G. Nature (London) 1982;299:111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- 9.Bowen T, Dover G A. Mol Ecol. 1995;4:419–427. doi: 10.1111/j.1365-294x.1995.tb00235.x. [DOI] [PubMed] [Google Scholar]
- 10.Schlotterer C, Tautz D. Curr Biol. 1994;4:777–783. doi: 10.1016/s0960-9822(00)00175-5. [DOI] [PubMed] [Google Scholar]
- 11.Sang T, Crawford D J, Stuessy T F. Proc Natl Acad Sci USA. 1995;92:6813–6817. doi: 10.1073/pnas.92.15.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cluster P D, Calderini O, Pupilli F, Crea F, Damiani F, Arcioni S. Theor Appl Genet. 1996;93:801–808. doi: 10.1007/BF00224079. [DOI] [PubMed] [Google Scholar]
- 13.Bruns T D, White T J, Taylor J W. Annu Rev Ecol Syst. 1991;22:525–564. [Google Scholar]
- 14.Wallace A V, Jansen R K. Plant Syst Evol. 1995;198:253–265. [Google Scholar]
- 15.Wendel J F, Schnabel A, Seelanan T. Proc Natl Acad Sci USA. 1995;92:280–284. doi: 10.1073/pnas.92.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quijada A, Liston A, Robinson W, Alvarez-Buylla E. Mol Ecol. 1997;6:995–996. [Google Scholar]
- 17.Sansome E R, Brasier C M. Trans Br Mycol Soc. 1974;63:461–467. [Google Scholar]
- 18.Cooke D E L, Duncan J M. Mycol Res. 1997;101:667–677. [Google Scholar]
- 19.White T J, Bruns T, Lee S, Taylor J. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 315–322. [Google Scholar]
- 20.Bonants P, Hagenaar-de Weerdt M, van Gent-Pelzer M, Lacourt I, Cooke D, Duncan J. Eur J Plant Pathol. 1997;103:345–355. [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilcox W F, Scott P H, Hamm P B, Kennedy D M, Duncan J M, Brasier C M, Hansen E M. Mycol Res. 1993;97:817–881. [Google Scholar]
- 25.Sansome E, Brasier C M. Nature (London) 1973;241:344–345. [Google Scholar]
- 26.Sansome E. In: Evolutionary Biology of the Fungi. Rayner A D M, Brasier C M, Moore D, editors. Cambridge, U.K.: Cambridge Univ. Press; 1987. pp. 97–111. [Google Scholar]
- 27.Lee S B, Taylor J W. J Mol Evol. 1992;9:636–653. doi: 10.1093/oxfordjournals.molbev.a040750. [DOI] [PubMed] [Google Scholar]
- 28.Crawford A R, Bassam B J, Drenth A, Maclean D J, Irwin J A G. Mycol Res. 1996;100:437–443. [Google Scholar]
- 29.Cronn R, Zhao X, Paterson A H, Wendel J F. J Mol Evol. 1996;42:685–705. doi: 10.1007/BF02338802. [DOI] [PubMed] [Google Scholar]
- 30.Majer D, Mithen R, Lewis B G, Vos P, Oliver P P. Mycol Res. 1996;100:1107–1111. [Google Scholar]
- 31.Boccas B, Zentmyer G A. Phytopathology. 1976;66:477–484. [Google Scholar]
- 32.Sansome E R, Brasier C M, Hamm P B. Mycol Res. 1991;95:273–277. [Google Scholar]
- 33.Brasier C M. In: Phytophthora. Lucas J A, Shattock R C, Shaw D S, Cooke L R, editors. Cambridge, U.K.: Cambridge Univ. Press; 1991. pp. 104–128. [Google Scholar]
- 34.Goodwin S B, Fry W E. Exp Mycol. 1994;18:20–32. [Google Scholar]
- 35.Ersek T, English J T, Schoelz T E. Phytopathology. 1995;85:1343–1347. [Google Scholar]
- 36.Brasier C M. In: Species: The Units of Biodiversity. Claridge M F, Dawah H A, Wilson M R, editors. London: Chapman & Hall; 1997. pp. 135–170. [Google Scholar]
- 37.Soltis D E, Soltis P S. Proc Natl Acad Sci USA. 1995;92:8089–8091. doi: 10.1073/pnas.92.18.8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rieseberg L H, Vanfossen C, Desrochers A M. Nature (London) 1995;375:313–316. [Google Scholar]
- 39.Duncan J M, Kennedy D M, Scott P H. In: Phytophthora. Lucas J A, Shattock R C, Shaw D S, Cooke L R, editors. Cambridge, U.K.: Cambridge Univ. Press; 1991. pp. 129–147. [Google Scholar]
- 40.Oudemans P, Coffey M D. Mycol Res. 1983;81:1–14. [Google Scholar]
- 41.Duncan J M. EPPO Bull. 1990;20:107–115. [Google Scholar]
- 42.Brasier C M, Kirk S A, Pipe N J, Buck K W. Mycol Res. 1998;102:45–57. [Google Scholar]
- 43.Spiers A G, Hopcroft D H. Mycol Res. 1994;98:889–903. [Google Scholar]
- 44.Tsai H F, Liu J S, Staben C, Christensen M J, Latch G C M, Siegel M R, Schardl C L. Proc Natl Acad Sci USA. 1994;91:2542–2546. doi: 10.1073/pnas.91.7.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Donnell K, Cigelnik E. Mol Phylogenet Evol. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]