Abstract
Live attenuated Salmonella strains expressing antigens of pathogens are promising oral vaccine candidates. There is growing evidence that the topology of expression of the foreign antigens can have a dramatic impact on the immunogenicity. We examined the potential of the AIDA-I (Escherichia coli adhesin involved in diffuse adherence) autotransporter domain to display antigenic fragments of the urease A subunit of Helicobacter pylori for the induction of a protective immune response. In the murine H. pylori model, protection is mainly mediated by CD4+ T cells, and we therefore used the AIDA-I expression system to successfully express both nearly full-length UreA and defined T-helper-cell epitopes on the surface of an attenuated Salmonella enterica serovar Typhimurium vaccine strain. Surface exposure of the large UreA fragment or of one UreA T-cell epitope mediated a significant reduction in the level of H. pylori in immunized mice after challenge infection, whereas conventional cytoplasmic expression of UreA in Salmonella had no effect. These results support the concept that surface display increases the immunogenicity of recombinant antigens expressed on oral live vaccine carriers and further demonstrate the feasibility of immunizing against H. pylori with Salmonella vaccine strains expressing CD4+ T-cell epitopes.
The approach of using live Salmonella vaccine strains to deliver recombinant antigens has been generally accepted, and to date several clinical studies have been performed in this field. The results of these studies, although promising, imply that new attenuated strains and improved antigen expression are needed to enhance the immunogenicity of Salmonella vaccine strains (14).
The localization of expressed antigens in bacterial live oral vaccines seems to be very important (20), and therefore many efforts have been made to manipulate surface-exposed proteins to display antigenic determinants (for a review see reference 13). Recently, we observed that an attenuated Salmonella vaccine strain expressing a CD4+ T-cell epitope on its surface via the autotransporter domain of AIDA-I (an adhesin involved in diffuse adherence from Escherichia coli [3]) was able to induce a specific CD4+ T-cell response (30). These findings encouraged us to investigate whether the AIDA-I expression system is able to induce protective immune responses in an animal model of infectious disease in which protection is mainly mediated by CD4+ T cells. We therefore chose the murine Helicobacter pylori infection model, because immunity against this pathogen has been reported to depend mainly on CD4+ T-helper cells (11) and we confirmed this for mice vaccinated with recombinant Salmonella which was effective in IgH−/− mice but not in major histocompatibility complex II gene-deficient mice (Aebischer, unpublished observations).
H. pylori is a gram-negative spiral bacterium that colonizes the human stomach and can cause a variety of diseases, including chronic gastritis, peptic ulcers, gastric adenocarcinoma, and gastric lymphoma (23, 41, 48). Vaccination would be a cost-effective means to control this public health problem faced by one-half of the world's population. Expression of urease subunits A and B from H. pylori in recombinant attenuated Salmonella vaccine strains induced high levels of protection against an H. pylori challenge infection in vaccinated mice (8, 16, 34), and three clinical phase I studies have already been based on this approach (2, 5, 10). Recombinant UreB has been reported to confer protective immunity against Helicobacter felis in different mouse strains (12, 38), whereas variable results have been reported for the protective effects of UreA (12, 38).
In a recent study, spleen-derived oligoclonal CD4+ T-cell lines were isolated from BALB/c mice vaccinated with attenuated Salmonella expressing urease subunits A and B from H. pylori (35). The T cells recognized urease A and could be restimulated with peptides containing predicted H-2d-restricted CD4+ T-cell epitopes (amino acids 28 to 51, 74 to 90, or 209 to 225) (35). Furthermore, adoptive transfer of these T cells into naïve mice partially protected against a H. pylori challenge.
In this study, we expressed translational fusions of a nearly full-length urease A variant or one of the three recognized urease A peptides to the C-terminal autotransporter domain of AIDA-I in attenuated Salmonella and tested these constructs for protective efficacy in the murine Helicobacter infection model.
MATERIALS AND METHODS
Bacterial strains.
All of the bacterial strains employed in this study are listed in Table 1. For all purposes (except preparation of frozen stocks), E. coli and Salmonella strains were grown on Luria-Bertani (LB) agar plates or in liquid medium supplemented with ampicillin (100 μg/ml) and, in the case of recombinant Salmonella, with streptomycin (90 μg/ml). Thymine (50 μg/ml) was added when required. H. pylori P76 was grown on brain heart infusion (BHI) (Difco, Becton Dickinson, Sparks, Md.) serum agar plates (16) supplemented with streptomycin (200 μg/ml) at 37°C under microaerophilic conditions or in BHI culture medium supplemented with 10% fetal calf serum (Gibco, Eggenstein, Germany) and 200 μg of streptomycin per ml with shaking at 37°C.
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype or phenotype | Reference or source |
|---|---|---|
| Escherichia coli JK321 | azi-6 fhuA23 lacY1 leu-6 mtl-1 proC14 purE42 rpsL109 thi-1 trpE38 tsx-67 Δ(ompT-fepC) zih::Tn10 dsbA::Kan | 26 |
| Crea1283 | S. enterica serovar Typhimurium SL3261 ΔaroA | Creatogen AGa |
| Crea1294 | S. enterica serovar Typhimurium SL3261 ΔaroA ΔthyA | Creatogen AG |
| SL3261(pYZ97) | S. enterica serovar Typhimurium SL3261 expressing urease subunits A and B constitutively | 16 |
| H. pylori P76 | Streptomycin-resistant derivative of the mouse-adapted H. pylori strain P49 | 16 |
Modified as described by Hoiseth and Stocker (22).
Genetic manipulations.
E. coli JK321 (26) was used for all cloning procedures. Oligonucleotide sequences used for PCR and plasmid construction are shown in Table 2. For the in vivo experiments transcriptional fusions of AIDA-I and UreA were first constructed in plasmid pLAT238. Plasmid pLAT238 encodes the epitope tag PEYFK derived from the Nef protein from the human immunodeficiency virus fused to a modified cholera toxin B subunit (CTB) gene, followed by the sequence encoding the autotransporter domain of AIDA-I, and it contains a single BglII restriction site between the Nef tag and the signal peptide sequence of CTB (30). Expression of the fusion in pLAT238 is transcriptionally controlled by the constitutive PTK promoter (25). Translocation into the periplasm of this fusion protein is mediated by the leader peptide of CTB. The DNA fragments encoding UreA27-53, UreA74-95, and UreA209-230 were amplified with primers LAT68 and LAT61, with primers LAT181 and LAT182, and with primers LAT183 and LAT184, respectively, by using pYZ97 as the template, treated with BamHI (LAT68-LAT61 product) or BglII and BamHI (LAT181-LAT182 and LAT183-LAT184 products), and inserted into the BglII site of pLAT238. The fusion constructs were then subcloned to obtain plasmids psdUreA27-53, psdUreA27-53, psdUreA209-230, in which expression is transcriptionally controlled by the PpagC promoter and which carry the thymidilate synthase gene thyA for plasmid stabilization purposes (39). A fragment encoding UreA27-53-PEYFK-CTB was amplified with primers LAT68 and LAT198 and treated with BamHI and KpnI; UreA74-95-PEYFK-CTB- and UreA209-230-PEYFK-CTB-encoding fragments were amplified with primers LAT212 and LAT198, treated with BglII and KpnI, and subcloned.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′ > 3′) | Features |
|---|---|---|
| JM54 | GATCTCCTGAATATTTCAAAGGTCCACCTTCTCCAC | Linker encoding PEYFK, sensea |
| JM55 | GATCGTGGAGAAGGTGGACCTTTGAAATATTCAGGA | Linker encoding PEYFK, antisensea |
| LAT61 | GATCGGATCCCTTTTTACCAGCTCTCGb | UreA27-53, antisense, BamHI site |
| LAT68 | GATCGGATCCGGCATTAAGCTTAACTATGb | UreA27-53, sense, BamHI site |
| LAT70 | GATCAGATCTACATATGAAACTGACTCCCAAAGc | UreA, sense, BglII site |
| LAT74 | GATTCCGGTAATACGACTCACTATAGGGd | pRSETb, T7 promoter, sense, HpaII site |
| LAT75 | GTAAGTCGACAAGCTTCGAATTCCATGGTe | pRSETb, multiple cloning site downstream, antisense, SalI site |
| LAT181 | GATCAGATCTGTGGCAAGCATGATCCATGc | UreA74-95, sense, BglII site |
| LAT182 | GATCGGATCCTACGAGTTTAGTCCCATCAb | UreA74-95, antisense, BamHI site |
| LAT183 | GATCAGATCTGAAAGCAAAAAAATTGCTTTACc | UreA209-230, sense, BglII site |
| LAT184 | GATCGGATCCTTTAGCGCCATGAAAACCb | UreA209-230, antisense, BamHI site |
| LAT198 | TCCCAGTATAATTTGACACG | AIDA, antisense |
| LAT212 | AAGGCCTGCTAGCACTAGTAACCCAAAGTCTAGGTGT | pEGE6, sense |
| LAT220 | GATCGGTACCTTAAGATCTCTCCTTAATTGTTTTTACATc | UreA, antisense, BglII site |
| MSC04 | CCTGCTGGTACCTAATCTTTTTCATTTCTTACTCCf | UreA, antisense, Acc651 site |
See reference 36.
The BamHI site is underlined.
The BglII site is underlined.
The HpaII site is underlined.
The SalI site is underlined.
The Acc651 site is underlined.
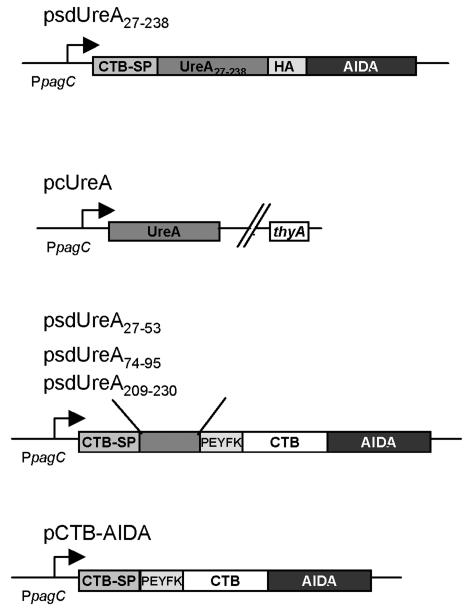
The DNA fragment coding for UreA27-238 was amplified from pYZ97 by using oligonucleotides LAT68 and LAT220, digested with BglII and BamHI, and cloned into a derivative of pJM7 (36) coding for the autotransporter domain of AIDA-I along with additional BglII and MunI restriction sites and an influenza hemagglutinin (HA) epitope tag downstream of the CTB signal peptide under transcriptional control of the PphoP promoter. For the in vivo experiments the fused gene was subcloned to obtain plasmid psdUreA27-238 in order to achieve expression via the PpagC promoter (21). For cytoplasmic expression of UreA plasmid pcUreA was constructed. Briefly, the multiple cloning site from pRSETb (Invitrogen, Carlsbad, Calif.), including the T7 promoter, was amplified by using primers LAT74 and LAT75, treated with HpaII and SalI, and inserted into the ClaI/SalI-digested vector pJM7. The ureA gene amplified from pYZ97 with primers LAT70 and MSC4 was inserted into the single BglII and Acc65I sites of the new multiple cloning site. The ureA fragment was transferred from this vector by digestion with XbaI and SalI to obtain plasmid pcUreA, which mediated UreA expression by the PpagC promoter. The identities of the constructs were verified by dideoxy chain termination sequencing (4base lab GmbH, Reutlingen, Germany). The final plasmids are shown in Fig. 1.
FIG. 1.
Schematic illustrations of plasmids encoding UreA fusion genes used for in vivo vaccination studies. CTB-SP, CTB signal peptide; AIDA, β-barrel of the carboxy-terminal translocation unit of the AIDA-I autotransporter; HA, B-cell epitope of influenza hemagglutinin; PEYFK, NEF epitope of human immunodeficiency virus. In the plasmid designations c indicates that the final localization of the protein products is cytoplasmic and sd indicates that the final localization of the protein products is surface displayed.
Animals.
Specific-pathogen-free female BALB/c mice that were 6 to 8 weeks old were obtained from the Bundesamt für Gesundheitlichen Verbraucherschutz (Berlin, Germany) and were kept under conditions that were in full compliance with German guidelines for animal care. All experiments were approved by the local animal welfare committee.
Preparation of frozen stocks.
Starting from a single colony, each Salmonella vaccine strain was grown on LB agar plates overnight at 37°C. The organisms were harvested on the following day in fresh LB medium, the suspension was used to inoculate culture medium (LB medium containing 90 μg of streptomycin per ml with or without 100 μg of ampicillin per ml) to obtain an optical density at 600 nm (OD600) of 0.1, and the culture was incubated overnight at 28°C and 200 rpm. The culture was harvested and resuspended in a 70% LB medium-30% glycerol mixture at an OD600 of 7 and stored at −80°C. The number of CFU per milliliter in each batch was determined by plating serial dilutions on selective LB agar plates.
Immunization experiments.
Prior to oral immunization mice were left overnight without food. Salmonella stocks were thawed, diluted with a 70% LB medium-30% glycerol mixture to obtain a concentration of 1 × 1010 CFU/ml, and then diluted 1:2 with 100 mM NaHCO3 to obtain a concentration of 0.5 × 1010 CFU/ml. The number of CFU per milliliter was confirmed by plating serial dilutions, and 1 × 109 CFU was administered intragastrically by using a round-tip stainless steel needle. Food was returned after immunization.
H. pylori challenge.
Four weeks after oral immunization, mice were challenged orally with 1 × 109 CFU of the streptomycin-resistant strain HP76 (16) by using a round-tip stainless steel needle. Mice were left overnight without solid food and water prior to challenge. H. plyori HP76 was grown on BHI serum agar plates containing 200 μg of streptomycin per ml at 37°C under microaerophilic conditions. After 3 days the organisms were harvested in 3 ml of BHI medium, culture medium consisting of BHI medium supplemented with 10% fetal calf serum and 200 μg of streptomycin per ml was inoculated to obtain an OD590 of 0.1, and then the culture was grown overnight at 37°C under microaerophilic conditions with shaking. Bacteria were harvested by centrifugation, and cells were resuspended in BHI broth to a final OD590 of 5.0. One hundred microliters of 100 mM NaHCO3 was administered intragastrically to neutralize the stomach contents, and this was followed by administration of 100 μl of BHI medium containing 1 × 109 CFU of H. pylori.
Assessment of H. pylori colonization.
Three weeks after challenge mice were anesthetized and killed. The stomachs were removed aseptically, one-half of each stomach was placed in 1 ml of BHI broth and homogenized until the gastric tissue was completely disrupted, and 10-fold serial dilutions were plated on BHI serum agar plates (16) supplemented with 200 μg of streptomycin per ml. Bacterial counts were determined after 5 days of growth under microaerophilic conditions at 37°C.
Determination of in vivo colonization of murine Peyer's patches by Salmonella vaccine strains.
Seven days after immunization mice were sacrificed, and Peyer's patches were removed. Single-cell suspensions were lysed with 0.1% Triton X-100, and serial dilutions were plated on LB medium plates containing 90 μg of streptomycin per ml and 100 μg of ampicillin per ml.
Protein techniques.
Expression of recombinant proteins was analyzed by separation of whole-cell lysates or outer membrane fractions of Salmonella enterica serovar Typhimurium by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE); when appropriate, this was followed by immunoblotting either with a monoclonal antibody against the HA tag conjugated to horseradish peroxidase (Roche Diagnostics, Penzberg, Germany) diluted 1:500 in phosphate-buffered saline (PBS) supplemented with 5% skim milk powder or with a rabbit anti-H. plyori serum (1:2,000) followed by a goat anti-rabbit antibody coupled to horseradish peroxidase (Sigma) (1:2,000). Bound antibodies were detected by enhanced chemoluminescence by using an ECL kit (Amersham) according to the manufacturer's recommendations. Scanning densitometric analyses of immunoblots were performed by using the freely available Scion Image software (http://www.scionCorp.com).
Surface exposure of AIDA-I fusion proteins.
Bacteria were grown overnight on LB agar plates at 37°C and harvested in PBS the following day. The OD575 of the bacterial suspension was adjusted to 10.0, and surface-exposed protein domains were proteolytically cleaved off by incubation of the suspension at 37°C for 10 min with trypsin (50 μg/ml). Cells were washed twice in PBS with gentle centrifugation in order to remove residual trypsin and were subjected subsequently to SDS-PAGE analysis.
Preparation of outer membranes.
Bacterial outer membranes were prepared as described elsewhere (31), with slight modifications. Bacteria grown overnight were harvested from agar plates and resuspended in PBS as described above. The suspension was sonicated with 30 1-s pulses at the maximum intensity by using a Branson Sonifier. Intact cells and large bacterial fragments were separated by centrifugation at 5,000 × g for 5 min. The cleared lysate was supplemented with l-lauryl sarcosinate (Sigma, Deisenhofen, Germany) at a final concentration of 1% to solubilize the inner membrane. Subsequently, the outer membrane was separated from the cytoplasm, periplasm, and inner membrane by centrifugation at 20,000 × g for 30 min at room temperature.
Statistical analysis.
Statistical analysis was performed by using the GraphPad Prism program (version 3.0; GraphPad Software, San Diego, Calif.). The level of significance used was P < 0.05.
RESULTS
Construction of Salmonella vaccine strains displaying UreA fragments on the cell surface.
The autotransporter domain of AIDA-I adhesin has been used in several studies to target epitopes, CTB, and β-lactamase to the surface of E. coli cells (29, 31, 36). In this study, we used AIDA-I to target H. pylori UreA fragments to the surface of an attenuated Salmonella vaccine strain. The UreA epitopes UreA27-53, UreA74-95, and UreA209-230 were translationally fused to the N terminus of a fusion protein of the AIDA-I autotransporter domain and CTB, whereas a large UreA27-238 fragment was translationally fused to the N terminus of the AIDA-I autotransporter domain without CTB but contained an HA tag sequence separating UreA27-238 and AIDA-I. Plasmids psdUreA27-53, psdUreA74-95, psdUreA209-230, and psdUreA27-238 are shown in Fig. 1. Intermediate constructs contained the transcriptonal fusions under control of the constitutive PTK promoter (25), which were electroporated into an aroA Salmonella carrier strain for biochemical localization of the products (see below). For in vivo studies, the thyA gene coding for thymidilate synthase was also included in the plasmid backbone of the final constructs (Fig. 1) for plasmid stabilization purposes (39). The Salmonella vaccine strain Crea1294, which contained a chromosomal deletion of the thyA gene and was derived from the aroA-deficient strain Crea1283 (unpublished data), was transformed with the corresponding plasmids. Complete UreA was expressed from the PpagC promoter (pcUreA) as a cytoplasmic antigen localization control (Fig. 1).
Localization of AIDA-I fusion proteins in the outer membrane.
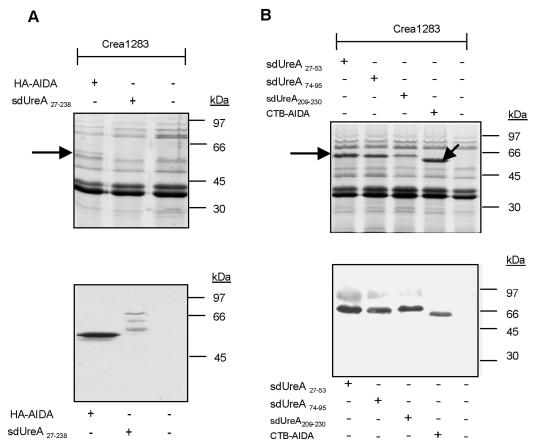
Autotransporter proteins localize to the outer membrane of gram-negative bacteria. To evaluate expression and outer membrane targeting of the various UreA fusion proteins in the attenuated S. enterica serovar Typhimurium aroA strains, outer membrane fractions from the Salmonella strains were analyzed by SDS-PAGE and Western blotting by using a monoclonal antibody to HA or a polyclonal antibody to cholera toxin (Fig. 2). All AIDA-I fusion proteins were detected in the outer membrane fraction. Decreased expression of the UreA27-238-AIDA-I fusion protein compared to the expression of AIDA-I without UreA27-238 was detected, and we also detected some degradation which might have been caused by partial proteolysis in the periplasm or at the outer membrane. This partial proteolysis most likely affected the UreA part of the fusion protein, since the degradation products were recognized by the HA antibody and remained membrane associated.
FIG. 2.
AIDA-I fusion proteins are present in the outer membrane. Outer membrane preparations of Salmonella vaccine strains expressing UreA fusion proteins with either HA-tagged (A) or CTB-tagged (B) AIDA-I were subjected to SDS-PAGE and analyzed by Coomassie brilliant blue staining (upper panels) (the arrows indicate fusion proteins) and by Western blot analysis by using an antibody against HA (A) or cholera toxin (B) (lower panels).
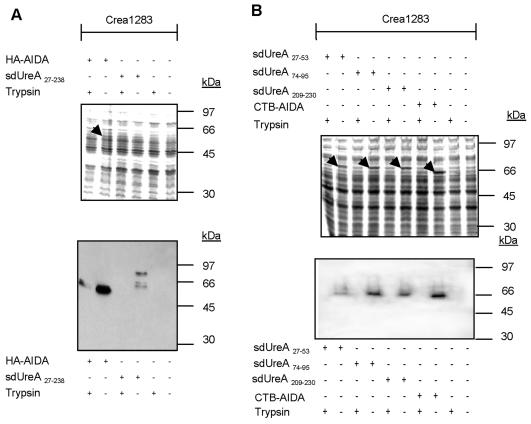
Surface localization of proteins can be investigated by trypsin treatment of intact cells, because surface-exposed protein structures that are sensitive to trypsin are cleaved off during exposure to the protease, whereas cytoplasmic, periplasmic, or protein domains embedded in the outer membrane are not affected (31, 37). This method is therefore suitable for monitoring the surface exposure of passenger domains fused to autotransporters. Physiologically intact cells, expressing the various AIDA-I fusions proteins, were subjected to trypsin digestion or were left untreated. In cells treated with trypsin UreA-AIDA-I fusion proteins were undetectable in immunoblot analyses of whole-cell lysates but were present in untreated control cells (Fig. 3). As only surface-exposed proteins were accessible to trypsin under these conditions, the results corroborated the localization data obtained by cell fractionation (Fig. 2) and further demonstrated that the vast majority of the fusion proteins were surface exposed.
FIG. 3.
AIDA-I fusion proteins are accessible to exogenously added trypsin. Intact bacteria expressing AIDA-I fusion proteins were treated with trypsin as described in Materials and Methods. Total cell lysates of trypsin-treated or untreated Salmonella strains expressing UreA fusion proteins with either HA-tagged (A) or CTB-tagged (B) AIDA-I were prepared, subjected to SDS-PAGE, and stained with Coomassie brilliant blue (upper panel) (the arrows indicate fusion proteins detectable by Coomassie brilliant blue staining) or analyzed by immunoblotting as described in the legend to Fig. 2. Trypsin treatment eliminated fusion protein detection in whole-cell lysates, indicating that the vast majority of the fusion proteins were surface exposed.
Immunization with Salmonella expressing UreA27-238-AIDA-I protects mice against an H. pylori challenge.
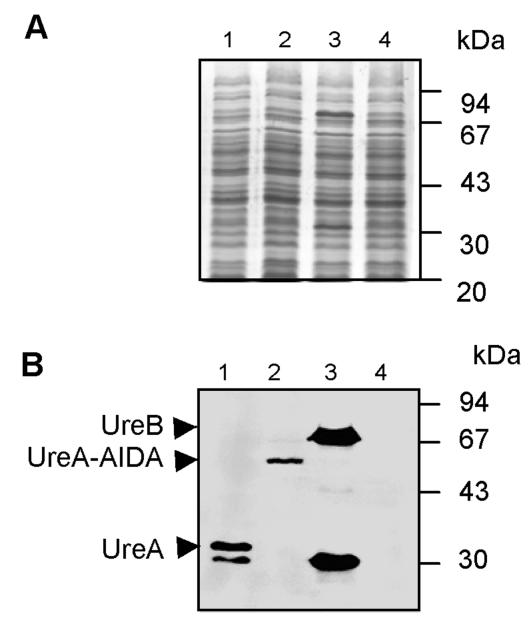
In order to analyze the vaccine potential of aroA-attenuated Salmonella expressing either cytoplasmic UreA [Crea1294(pcUreA)] or surface-exposed UreA27-238-AIDA-I fusion protein [Crea1294(psdUreA27-238)], protein expression was first compared in the corresponding vaccine strains. The UreA levels expressed under in vitro conditions were assessed by immunoblot analysis by using a Helicobacter-specific antiserum raised in rabbits (Fig. 4). The level of UreA expression was higher in the strain expressing UreA in the cytoplasm, Crea1294(pcUreA) (603 arbitrary staining intensity units), than in Crea1294(psdUreA27-238) (150 arbitrary staining intensity units). For reference, UreA expression in our standard vaccine strain, SL3261(pYZ97) expressing both UreA and UreB subunits (16), was also analyzed, and the level of UreA expression in this strain was 829 optical density units.
FIG. 4.
Levels of expression of UreA in recombinant Salmonella vaccine strains. The diverse Salmonella vaccine strains were grown overnight on selective LB agar plates at 37°C, and whole-cell lysates of Crea1294(pcUreA) (lane 1) and Crea1294(psdUreA27-238) (lane 2) expressing UreA in the cytosol and on the cell surface, respectively, were separated by SDS-PAGE and analyzed by Coomassie brilliant blue staining (A) or immunoblotting with an anti-H. pylori serum (B). The standard laboratory vaccine strain SL3261(pYZ97) expressing UreA and UreB (lane 3) was used as a positive control, and the empty carrier strain Crea1283 (lane 4) was used as a negative control.
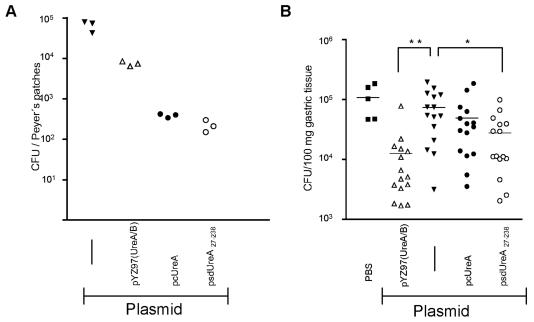
A second important parameter of live vaccine strains is the relative ability to colonize the host. To estimate the bacterial fitness, groups of three BALB/c mice were inoculated orally with single intragastric doses of 109 CFU of either Crea1294(psdUreA27-238), Crea1294(pcUreA), the plasmidless carrier strain, or the positive control strain SL3261(pYZ97) (16). Bacterial loads were compared on day 7, when SL3261 colonization usually reached peak levels. The new UreA-expressing strains Crea1294(pcUreA) and Crea1294(psdUreA27-238) showed reduced colonization of the Peyer's patches on day 7 postimmunization (2.2 × 102 ± 4.3 × 102 and 3.9 × 102 ± 2.4 × 102 CFU, respectively) compared to the colonization by the plasmidless strain (6.6 × 104 ± 1.1 × 104 CFU) and the control strain, SL3261(pYZ97) (7.5 × 103 ± 6.1 × 103 CFU) (Fig. 5A). However, for the present study it was important that the new vaccine strains expressed comparable amounts of UreA and had similar colonization capabilities.
FIG. 5.
Colonization capabilities of Salmonella vaccine strains expressing UreA localized differently and efficacies against H. pylori. Groups of BALB/c mice were immunized orally with 1 × 109 cells of recombinant Salmonella strain Crea1283 (carrier only) (solid lines), SL3261(pYZ97) expressing H. pylori UreA and UreB, Crea1294(pcUreA), or Crea1294(psdUreA27-238). Seven days postinfection three mice were sacrificed, and the Peyer's patches were removed, homogenized, and plated on selective LB agar plates (A). Alternatively, immunized mice were challenged 4 weeks later with H. pylori, and colonization by this pathogen was analyzed 3 weeks later (B). Combined results from two independent experiments are shown [n = 5 for the sham-immunized group, n = 15 for Crea1294(psdUreA27-238), Crea1294(pcUreA), and Crea1283, and n = 14 for SL3261(pYZ97)]; the horizontal line represents the mean H. pylori burden. Statistically significant differences compared to the results for the Crea1283-immunized group were analyzed by using the unpaired t test [one asterisk, P = 0.0089 for Crea1294(psdUreA27-238); two asterisks, P = 0.0005 for SL3261(pYZ97)].
To determine vaccine efficacy, the four different strains [the carrier control, positive control SL3261(pYZ97), cytoplasmic UreA, and surface-exposed UreA strains] were orally administered to mice, and 4 weeks later the mice were challenged with 1 × 109 CFU of H. pylori. Three weeks after the challenge infection, mice were sacrificed, and the H. pylori burden in the stomach was determined. Vaccination with the Salmonella strain expressing UreA at the cell surface resulted in significantly reduced H. pylori colonization compared to the colonization of the carrier-immunized control group (P = 0.0089, as determined by the Student t test) (Fig. 5B). In contrast, the Salmonella strain expressing cytoplasmic UreA failed to induce a significant reduction in the H. pylori burden compared to the burden observed after administration of the carrier strain control, although the bacterial counts were lower on average.
Salmonella sp. expressing a UreA27-53 peptide on the surface partially protects against H. pylori.
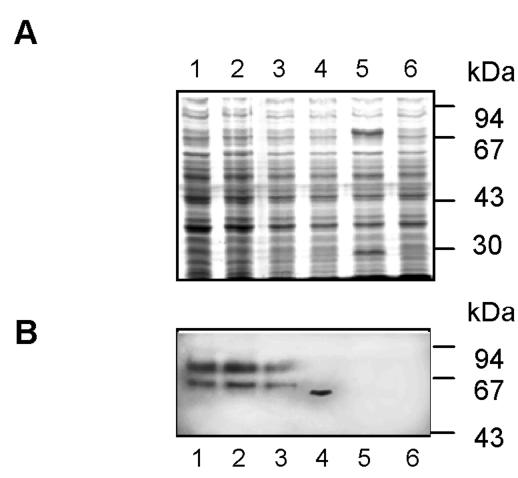
In a recent study, Lucas et al. identified three peptides of the UreA protein that contain T-cell epitopes recognized by protective UreA-specific CD4+ T cells (35). In the present study, we fused these peptides to AIDA-I to evaluate its potential as a T-cell epitope expression platform. All peptide fusion proteins were similarly expressed by the attenuated Salmonella aroA strain (Fig. 6), and the colonization capabilities of the constructs were comparable.
FIG. 6.
Expression of UreA CD4+ T-cell epitopes by Salmonella vaccine strains. Whole-cell lysates from cells grown overnight on LB agar plates at 37°C were prepared and subjected to SDS-PAGE. (A) Coomassie brilliant blue-stained SDS-PAGE gel. (B) Immunoblot with an anti-cholera toxin antibody. Lane 1, Crea1294(psdUreA74-95); lane 2, Crea1294(psdUreA27-53); lane 3, Crea1294(psdUreA209-230); lane 4, Crea1294(pCTB-AIDA); lane 5, SL3261(pYZ97); lane 6, Crea1283.
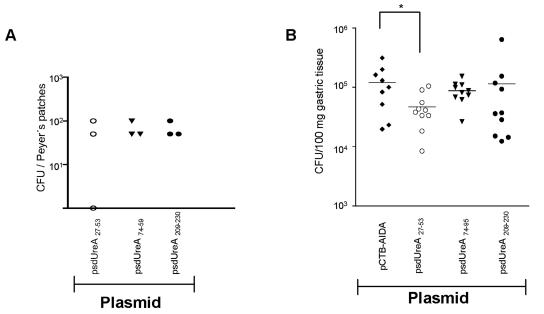
Mice were vaccinated with the three constructs or a negative control strain expressing AIDA-I without a fused UreA peptide and challenged with H. pylori as described above. While Salmonella strains expressing UreA74-95 and UreA209-230 had no protective effect, the UreA27-53 construct induced a significant reduction in the H. pylori burden compared to the burden observed with the Salmonella negative control strain (Fig. 7) (P = 0.03, as determined by the Student t test).
FIG. 7.
In vivo colonization and protective efficacy of Crea1294 expressing UreA CD4+ T-cell epitopes on the cell surface. Groups of three BALB/c mice were immunized orally with 1 × 109 cells of recombinant Salmonella vaccine strains. At day 7 postimmunization mice were killed, and the Peyer's patches were harvested, homogenized, and plated on selective LB agar plates to determine Salmonella colonization (A). (B) Analysis of H. pylori burden in mice orally vaccinated with Salmonella strains expressing UreA CD4+ T-cell epitopes after challenge. The results of one representative experiment are shown (10 mice per group) (two experiments were performed). Crea1294(psdUreA27-53), displaying the UreA27-53 peptide, conferred significant protection against H. pylori compared to the protection conferred by Crea1294(pCTB-AIDA) expressing only the carrier protein. An asterisk indicates a P value of 0.03, as determined by an unpaired t test. The horizontal lines indicate means.
DISCUSSION
Recombinant attenuated Salmonella strains are promising vaccine carriers for oral delivery of heterologous antigens. For example, we and other workers have previously shown that immunization with Salmonella strains expressing H. pylori ureases A and B can protect mice against a subsequent Helicobacter challenge infection (16).
In a number of experimental models, Salmonella vaccine efficacy can be enhanced by surface display or secretion of the foreign antigen. Autotransporter domains of gram-negative bacteria can be used as one such surface display system (27-29, 31, 36, 45). Autotransporters are widespread among gram-negative bacteria, and their main physiological function is to translocate virulence factors through the cell envelope to the surface (19). The fact that they are expressed as a single polypeptide chain containing all features necessary to translocate an N-terminal passenger domain to the cell surface (25) makes them attractive candidates for antigen display. Furthermore, functional B-cell epitopes (42) and T-cell epitopes (29, 30) have already been successfully expressed as autotransporter fusion proteins. In this study, we tested if Salmonella surface display of urease A or urease A fragments that contain T-cell epitopes (35) by using the AIDA-I autotransporter provides better anti-Helicobacter protection than an otherwise identical cytoplasmic UreA construct provides. A nearly full-length variant of UreA, as well as three different UreA peptides, could be expressed as AIDA-I fusion proteins and were displayed on the surface of attenuated Salmonella cells. The nearly full-length variant encoded on psdUreA27-238 lacks the N terminus (amino acids 1 to 26), which contains six lysine residues, to avoid potential inhibition of translocation across the inner membrane by the Sec pathway (1). Inhibition of translocation was noted when the full-length UreA was introduced into E. coli or Salmonella (Lattemann, unpublished observations).
In the murine H. pylori infection model a significant reduction in the H. pylori burden was detected in mice vaccinated with a Salmonella strain expressing either the almost complete UreA protein [Crea1294(psdUreA27-238)] or the T-cell epitope containing peptide UreA27-53 [Crea1294(psdUreA27-53)] as an AIDA-I fusion protein. The results obtained with the latter strain also provided independent functional confirmation that there is at least one protective epitope in UreA, which has been suggested previously by Lucas and coworkers, who identified the UreA fragments mentioned above as CD4+ T-cell epitope-containing regions (35). Additionally, this study demonstrated the feasibility of immunizing against H. pylori with Salmonella vaccine strains expressing CD4+ T-cell epitopes on the cell surface. In contrast, the H. pylori burdens in mice vaccinated with a Salmonella strain conventionally expressing UreA in the cytoplasm [Crea1294(pcUreA)] were not significantly reduced compared to the burdens in mice vaccinated with the carrier strain.
CD4+ T-cell induction is directly correlated with the number of Salmonella cells colonizing the Peyer's patches on the first few days after vaccination (4). For comparing various Salmonella vaccine constructs, it is thus important to determine the corresponding colonization capabilities. The two vaccine strains expressing cytoplasmic and surface-displayed urease A showed similar colonization levels in Peyer's patches of immunized mice. This suggests that other factors and probably the different localizations account for the different vaccination efficacies of the two strains. Superior antigen processing of surface antigens by antigen-presenting cells and/or an altered urease A conformation might be involved, but further studies are required to test these hypotheses. Improvement of the urease A-AIDA-I fusion protein-expressing strains is, however, still needed, and this should be possible by increasing Salmonella fitness in vivo. Our standard laboratory vaccine strain, strain SL3261(pYZ97), colonized at least 10-fold better and expressed UreA as well as UreB subunits. Both of these aspects, increased fitness and expression of two vaccine antigens, are likely to be responsible for the superior efficacy of this strain. Therefore, in addition to improvement of the fitness of carriers by modulating the level of expression of the AIDA-I fusions, addition of immunogenic epitopes from other antigens to the existing constructs and prime-boost regimens are potential ways to increase the efficacy of the new strains, which, as reported here, indicates that AIDA-I is an attractive candidate for the development of live vaccines against H. pylori.
Several other approaches have been used to develop expression systems that are alternatives to the conventional somatic expression of antigens for the induction of cellular immune responses (13, 20, 44). These include secretion of antigens into the surrounding environment (13, 20) or into the cytoplasm of the host cell (44). Salmonella vaccine strains endowed with the α-hemolysin secretion apparatus of E. coli in trans were able to secrete full-length antigens and were efficacious in several animal models (15). The type III secretion system encoded on the SPI-1 pathogenicity island of S. enterica serovar Typhimurium has also been successfully used in Salmonella live vaccine carriers to translocate major histocompatibility complex I restricted epitopes fused to the N terminus of YopE, a secreted effector protein of Yersinia enterocolitica, into host cells, resulting in protective immune responses (43, 44).
Humoral immune responses have been observed after vaccination of mice with attenuated Salmonella strains with immunogenic determinants exposed on their surfaces by means of outer membrane proteins from E. coli, like OmpA (17), PhoE (24, 47), LamB (6, 18, 32), and P87 fimbriae (7), the ice-nucleating protein from Pseudomonas aeruginosa (33), the main flagellar component FliC from Salmonella (9, 40, 40), or the Salmonella autotransporter MisL (42). Recently, use of E. coli outer membrane protein TolC for surface display of protective listerial B- and T-cell epitopes in a Salmonella vaccine strain has been shown to be efficacious in vivo (46).
In summary, autotransporters facilitate effective surface display of antigenic determinants on live Salmonella vaccine carriers, leading to humoral (42) and cellular immune responses in vivo (29). This study demonstrated that surface display of a Helicobacter antigen via the AIDA-I autotransporter can induce protective immune responses.
Acknowledgments
We thank Elke Gerland, Kirstin Hoffmann, and Kerstin Burmeister for excellent technical assistance.
This work was supported in part by grants from the BMBF (grants PTJ-BIO/0312178 and G01III1-1/3-VIIZV-17) and from DFG Novel Vaccination Strategies priority program (grant Bu 971/4-2).
Editor: D. L. Burns
REFERENCES
- 1.Andersson, H., and G. von Heijne. 1994. Membrane protein topology: effects of delta mu H+ on the translocation of charged residues explain the ′positive inside' rule. EMBO J. 13:2267-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angelakopoulos, H., and E. L. Hohmann. 2000. Pilot study of phoP/phoQ-deleted Salmonella enterica serovar Typhimurium expressing Helicobacter pylori urease in adult volunteers. Infect. Immun. 68:2135-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benz, I., and M. A. Schmidt. 1989. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect. Immun. 57:1506-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bumann, D. 2001. Regulated antigen expression in live recombinant Salmonella enterica serovar Typhimurium strongly affects colonization capabilities and specific CD4+ T-cell responses. Infect. Immun. 69:7493-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bumann, D., W. G. Metzger, E. Mansouri, O. Palme, M. Wendland, R. Hurwitz, G. Haas, T. Aebischer, B. U. von Specht, and T. F. Meyer. Safety and immunogenicity of live recombinant Salmonella enterica serovar Typhi Ty21a expressing urease A and B from Helicobacter pylori in human volunteers. Vaccine 20:645-652. [DOI] [PubMed]
- 6.Charbit, A., A. Molla, W. Saurin, and M. Hofnung. 1988. Versatility of a vector for expressing foreign polypeptides at the surface of gram-negative bacteria. Gene 70:181-189. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H., and D. M. Schifferli. 2000. Mucosal and systemic immune responses to chimeric fimbriae expressed by Salmonella enterica serovar typhimurium vaccine strains. Infect. Immun. 69:3129-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corthesy-Theulaz, I. E., S. Hopkins, D. Bachmann, P. F. Saldinger, N. Porta, R. Haas, Y. Zheng-Xin, T. F. Meyer, H. Bouzourene, A. L. Blum, and J. P. Kraehenbuhl. 1998. Mice are protected from Helicobacter pylori infection by nasal immunization with attenuated Salmonella typhimurium phoPc expressing urease A and B subunits. Infect. Immun. 66:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Almeida, M. E., S. M. Newton, and L. C. Ferreira. 1999. Antibody responses against flagellin in mice orally immunized with attenuated Salmonella vaccine strains. Arch. Microbiol. 172:102-108. [DOI] [PubMed] [Google Scholar]
- 10.DiPetrillo, M. D., T. Tibbetts, H. Kleanthous, K. P. Killeen, and E. L. Hohmann. 1999. Safety and immunogenicity of phoP/phoQ-deleted Salmonella typhi expressing Helicobacter pylori urease in adult volunteers. Vaccine 18:449-459. [DOI] [PubMed] [Google Scholar]
- 11.Ermak, T. H., P. J. Giannasca, R. Nichols, G. A. Myers, J. Nedrud, R. Weltzin, C. K. Lee, H. Kleanthous, and T. P. Monath. 1998. Immunization of mice with urease vaccine affords protection against Helicobacter pylori infection in the absence of antibodies and is mediated by MHC class II-restricted responses. J. Exp Med. 188:2277-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrero, R. L., J. M. Thiberge, M. Huerre, and A. Labigne. 1994. Recombinant antigens prepared from the urease subunits of Helicobacter spp.: evidence of protection in a mouse model of gastric infection. Infect. Immun. 62:4981-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galen, J. E., and M. M. Levine. 2001. Can a ‘flawless’ live vector vaccine strain be engineered? Trends Microbiol. 9:372-376. [DOI] [PubMed] [Google Scholar]
- 14.Garmory, H. S., K. A. Brown, and R. W. Titball. 2002. Salmonella vaccines for use in humans: present and future perspectives. FEMS Microbiol. Rev. 26:339-353. [DOI] [PubMed] [Google Scholar]
- 15.Gentschev, I., G. Dietrich, S. Spreng, A. Kolb-Maurer, J. Daniels, J. Hess, S. H. Kaufmann, and W. Goebel. 2000. Delivery of protein antigens and DNA by virulence-attenuated strains of Salmonella typhimurium and Listeria monocytogenes. J. Biotechnol. 83:19-26. [DOI] [PubMed] [Google Scholar]
- 16.Gomez-Duarte, O. G., B. Lucas, Z. X. Yan, K. Panthel, R. Haas, and T. F. Meyer. 1998. Protection of mice against gastric colonization by Helicobacter pylori by single oral dose immunization with attenuated Salmonella typhimurium producing urease subunits A and B. Vaccine 16:460-471. [DOI] [PubMed] [Google Scholar]
- 17.Haddad, D., S. Liljeqvist, S. Kumar, M. Hansson, S. Stahl, H. Perlmann, P. Perlmann, and K. Berzins. 1995. Surface display compared to periplasmic expression of a malarial antigen in Salmonella typhimurium and its implications for immunogenicity. FEMS Immunol. Med. Microbiol. 12:175-186. [DOI] [PubMed] [Google Scholar]
- 18.Hayes, L. J., J. W. Conlan, J. S. Everson, M. E. Ward, and I. N. Clarke. 1991. Chlamydia trachomatis major outer membrane protein epitopes expressed as fusions with LamB in an attenuated aroA strain of Salmonella typhimurium; their application as potential immunogens. J. Gen. Microbiol. 137:1557-1564. [DOI] [PubMed] [Google Scholar]
- 19.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hess, J., I. Gentschev, D. Miko, M. Welzel, C. Ladel, W. Goebel, and S. H. Kaufmann. 1996. Superior efficacy of secreted over somatic antigen display in recombinant Salmonella vaccine induced protection against listeriosis. Proc. Natl. Acad. Sci. USA 93:1458-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hohmann, E. L., C. A. Oletta, W. P. Loomis, and S. I. Miller. 1995. Macrophage-inducible expression of a model antigen in Salmonella typhimurium enhances immunogenicity. Proc. Natl. Acad. Sci. USA 92:2904-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 23.Hussell, T., P. G. Isaacson, J. E. Crabtree, and J. Spencer. 1993. The response of cells from low-grade B-cell gastric lymphomas of mucosa-associated lymphoid tissue to Helicobacter pylori. Lancet 342:571-574. [DOI] [PubMed] [Google Scholar]
- 24.Janssen, R., and J. Tommassen. 1994. PhoE protein as a carrier for foreign epitopes. Int. Rev. Immunol. 11:113-121. [DOI] [PubMed] [Google Scholar]
- 25.Jose, J., F. Jahnig, and T. F. Meyer. 1995. Common structural features of IgA1 protease-like outer membrane protein autotransporters. Mol. Microbiol. 18:378-380. [DOI] [PubMed] [Google Scholar]
- 26.Jose, J., J. Kramer, T. Klauser, J. Pohlner, and T. F. Meyer. 1996. Absence of periplasmic DsbA oxidoreductase facilitates export of cysteine-containing passenger proteins to the Escherichia coli cell surface via the IgA beta autotransporter pathway. Gene 178:107-110. [DOI] [PubMed] [Google Scholar]
- 27.Kjaergaard, K., H. Hasman, M. A. Schembri, and P. Klemm. 2002. Antigen 43-mediated autotransporter display, a versatile bacterial cell surface presentation system. J. Bacteriol. 184:4197-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klauser, T., J. Pohlner, and T. F. Meyer. 1990. Extracellular transport of cholera toxin B subunit using Neisseria IgA protease beta-domain: conformation-dependent outer membrane translocation. EMBO J. 9:1991-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konieczny, M. P., M. Suhr, A. Noll, I. B. Autenrieth, and M. A. Schmidt. 2000. Cell surface presentation of recombinant (poly-) peptides including functional T-cell epitopes by the AIDA autotransporter system. FEMS Immunol. Med. Microbiol. 27:321-332. [DOI] [PubMed] [Google Scholar]
- 30.Kramer, U., K. Rizos, H. Apfel, I. B. Autenrieth, and C. T. Lattemann. 2003. Autodisplay: development of an efficacious system for surface display of antigenic determinants in Salmonella vaccine strains. Infect. Immun. 71:1944-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lattemann, C. T., J. Maurer, E. Gerland, and T. F. Meyer. 2000. Autodisplay: functional display of active beta-lactamase on the surface of Escherichia coli by the AIDA-I autotransporter. J. Bacteriol. 182:3726-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leclerc, C., A. Charbit, A. Molla, and M. Hofnung. 1989. Antibody response to a foreign epitope expressed at the surface of recombinant bacteria: importance of the route of immunization. Vaccine 7:242-248. [DOI] [PubMed] [Google Scholar]
- 33.Lee, J. S., K. S. Shin, J. G. Pan, and C. J. Kim. 2000. Surface-displayed viral antigens on Salmonella carrier vaccine. Nat. Biotechnol. 18:645-648. [DOI] [PubMed] [Google Scholar]
- 34.Londono-Arcila, P., D. Freeman, H. Kleanthous, A. M. O'Dowd, S. Lewis, A. K. Turner, E. L. Rees, T. J. Tibbitts, J. Greenwood, T. P. Monath, and M. J. Darsley. 2002. Attenuated Salmonella enterica serovar Typhi expressing urease effectively immunizes mice against Helicobacter pylori challenge as part of a heterologous mucosal priming-parenteral boosting vaccination regimen. Infect. Immun. 70:5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucas, B., D. Bumann, A. Walduck, J. Koesling, L. Develioglu, T. F. Meyer, and T. Aebischer. 2001. Adoptive transfer of CD4+ T cells specific for subunit A of Helicobacter pylori urease reduces H. pylori stomach colonization in mice in the absence of interleukin-4 (IL-4)/IL-13 receptor signaling. Infect. Immun. 69:1714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurer, J., J. Jose, and T. F. Meyer. 1997. Autodisplay: one-component system for efficient surface display and release of soluble recombinant proteins from Escherichia coli. J. Bacteriol. 179:794-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maurer, J., J. Jose, and T. F. Meyer. 1999. Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J. Bacteriol. 181:7014-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michetti, P., I. Corthesy-Theulaz, C. Davin, R. Haas, A. C. Vaney, M. Heitz, J. Bille, J. P. Kraehenbuhl, E. Saraga, and A. L. Blum. 1994. Immunization of BALB/c mice against Helicobacter felis infection with Helicobacter pylori urease. Gastroenterology 107:1002-1011. [DOI] [PubMed] [Google Scholar]
- 39.Morona, R., J. Yeadon, A. Considine, J. K. Morona, and P. A. Manning. 1991. Construction of plasmid vectors with a non-antibiotic selection system based on the Escherichia coli thyA+ gene: application to cholera vaccine development. Gene 107:139-144. [DOI] [PubMed] [Google Scholar]
- 40.Newton, S. M., C. O. Jacob, and B. A. Stocker. 1989. Immune response to cholera toxin epitope inserted in Salmonella flagellin. Science 244:70-72. [DOI] [PubMed] [Google Scholar]
- 41.Parsonnet, J., S. Hansen, L. Rodriguez, A. B. Gelb, R. A. Warnke, E. Jellum, N. Orentreich, J. H. Vogelman, and G. D. Friedman. 1994. Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med. 330:1267-1271. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz-Perez, F., R. Leon-Kempis, A. Santiago-Machuca, G. Ortega-Pierres, E. Barry, M. Levine, and C. Gonzalez-Bonilla. 2002. Expression of the Plasmodium falciparum immunodominant epitope (NANP)(4) on the surface of Salmonella enterica using the autotransporter MisL. Infect. Immun. 70:3611-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russmann, H., E. I. Igwe, J. Sauer, W. D. Hardt, A. Bubert, and G. Geginat. 2001. Protection against murine listeriosis by oral vaccination with recombinant Salmonella expressing hybrid Yersinia type III proteins. J. Immunol. 167:357-365. [DOI] [PubMed] [Google Scholar]
- 44.Russmann, H., H. Shams, F. Poblete, Y. Fu, J. E. Galan, and R. O. Donis. 1998. Delivery of epitopes by the Salmonella type III secretion system for vaccine development. Science 281:565-568. [DOI] [PubMed] [Google Scholar]
- 45.Shimada, K., Y. Ohnishi, S. Horinouchi, and T. Beppu. 1994. Extracellular transport of pseudoazurin of Alcaligenes faecalis in Escherichia coli using the COOH-terminal domain of Serratia marcescens serine protease. J. Biochem. (Tokyo) 116:327-334. [DOI] [PubMed] [Google Scholar]
- 46.Spreng, S., G. Dietrich, W. Goebel, and I. Gentschev. 2003. Protection against murine listeriosis by oral vaccination with recombinant Salmonella expressing protective listerial epitopes within a surface-exposed loop of the TolC-protein. Vaccine 21:746-752. [DOI] [PubMed] [Google Scholar]
- 47.Tommassen, J., M. Agterberg, R. Janssen, and G. Spierings. 1993. Use of the enterobacterial outer membrane protein PhoE in the development of new vaccines and DNA probes. Zentralbl. Bakteriol. 278:396-406. [DOI] [PubMed] [Google Scholar]
- 48.Wotherspoon, A. C., C. Doglioni, T. C. Diss, L. Pan, A. Moschini, M. de Boni, and P. G. Isaacson. 1993. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet 342:575-577. [DOI] [PubMed] [Google Scholar]