Abstract
We have examined the question of whether there is an additional checkpoint in T cell development that regulates T cell receptor (TCR)-β expression in CD25+44− thymocytes by mechanisms that are independent of the pre-TCR. Our analysis in various mutant mice indicates that all changes in cytoplasmic TCR-β expression can be accounted for by pre-TCR–dependent signal mediation, putting into question the function of a putative pro-TCR.
Keywords: lymphocyte development, pre-T cell receptor, cytoplasmic T cell receptor β, clonotype-independent CD3 complex
Several years ago, it was found that on the surface of immature T cells CD3∈ could be expressed in the apparent absence of TCR chains for antigen, and that signals could be transduced by cross-linking this complex with CD3 antibodies 1 2 3. Since then, it has been discussed whether CD3∈ surface expression in apparent association with calnexin 4 represents a biological accident without implication for physiological T cell maturation 5, or whether it is perhaps indicative of a pro-TCR complex that controls development. This would represent yet another checkpoint, in addition to those controlled by the pre-TCR 6 and the α/β TCR 7. In fact, it was recently argued that the second possibility was likely to be correct, since impaired signal transduction in p56lck and CD3ζ-deficient mice appeared to be associated with reduced expression of productively rearranged TCR-β genes 8 9. However, it could not be excluded from these studies that the deficiency in TCR-β gene expression was dependent on defective signaling by the pre-TCR or the α/β TCR, since the analysis included thymocyte subsets whose generation depended either on the pre-TCR (some CD25+44− cells 10) or the α/β TCR (CD25−44+ NK T cells [11]). For that reason, we have analyzed TCR-β expression by intracellular staining in a variety of mutant mouse strains in small CD25+44− cells that are independent of the pre-TCR and the α/β TCR. Our data indicate that all differences that exist in TCR-β gene expression among various mouse strains can be attributed to signal transduction by the pre-TCR, since small CD25+44− thymocytes of pre-TCR α chain–deficient mice exhibited the same phenotype observed in CD3-deficient mice, thus arguing against a distinct checkpoint in development controlled by the CD3 complex in the absence of the pre-TCR.
Materials and Methods
Animals.
CD3∈−/−, pTα−/−, and γc−/− mice have been described 6 12 13 and were bred in the specific pathogen–free animal facilities of the Necker Institute, Paris. C57BL/6 and Rag2−/− mice were purchased from Iffa Credo. Animals were analyzed at 6–8 wk of age. Animal care was in accordance with institutional guidelines.
Antibodies and Immunofluorescence Analysis.
The following mAbs were used: CD25 (PC-61) conjugated to FITC, CD44 (Pgp-1)–PE, CD25-PE, CD4 (L3T4)-CyChrome, CD8 (Ly-2)–CyChrome (PharMingen), and anti-pan TCR-β (H57-597). Biotinylated mAbs were revealed with either streptavidin-PE (Southern Biotechnology) or streptavidin-allophycocyanin (APC; Molecular Probes Europe). Cells were stained in microtiter plates (106 cells/well in 50 μl) using combinations of directly conjugated mAbs. Simultaneous four-color cell analyses were performed on a FACSCalibur™ flow cytometer (Becton Dickinson). Dead cells were excluded by gating based on forward and side scatter characteristics.
Intracellular Staining for TCR-β Chains.
Thymocytes were enriched for the CD4−8− subset by negative depletion of CD4/CD8+ cells using Dynabeads (Dynal). For extracellular/intracellular double staining, cells were first incubated with culture supernatant of mAb 2.4G2 to block FcRII/III receptors. Cells were then stained with FITC-conjugated anti-CD25, PE-conjugated anti-CD44, and CyChrome-conjugated anti-CD4 and anti-CD8 at optimal concentration. After washing in PBS, cells were fixed in PBS plus 0.5% paraformaldehyde for 15 min at room temperature, followed by two washing steps in PBS. Cells were then permeabilized in 0.5% saponin for 10 min at room temperature and washed in PBS. Intracellular staining with biotinylated anti–pan TCR-β (H57-597) diluted in PBS/2% FCS plus 0.5% saponin was performed for 30 min at 4°C, washed twice in PBS/2% FCS, and revealed for 30 min at 4°C by streptavidin-APC diluted in PBS/2% FCS plus 0.5% saponin. Cytoplasmic staining was followed by two washing steps in PBS and 15 min on a rocking platform in PBS/2% FCS plus 0.5% saponin on ice. Finally, cells were washed in PBS/2% FCS.
Results and Discussion
Intracellular TCR-β Gene Expression in CD4−8− Subsets of Thymocytes.
We have analyzed thymocytes from wild-type, γc−/− 12, pTα−/− 6, CD3∈−/− 13, and Rag2−/− mice 14 in order to analyze the effect of each mutation on TCR-β gene expression in small CD25+44− cells. The subset distribution among CD4−8− cells according to CD44 and CD25 expression is shown in Fig. 1. Wild-type and γc−/− mice exhibit a similar phenotype except for an elevated proportion of CD44+25+ cells in the latter due to a partial block at this stage of development in γc−/− mice. pTα−/− mice look similar to CD3∈−/− and Rag2−/− mice, but due to their incomplete block at the CD44−25+ stage of development, contain more CD44−25− cells than the latter two strains. Of these, some 70% are γ/δ T cells 6. When intracellular TCR-β expression versus CD25 expression was analyzed in all CD4−8− cells, it became clear that wild-type and γc−/− thymocytes express TCR β chains in the majority of cells, but γc−/− thymocytes less so because of an early partial block before TCR-β rearrangement at the CD44+25+ stage 12. In these two strains, most TCR-β expression was present in CD25− cells. In contrast, in pTα−/− and CD3∈−/− mice, most TCR-β expression was found in CD25+ cells, although less completely so in pTα−/− mice because of a partial developmental block at the CD25+44− stage resulting in a population of CD25−44− cells, of which up to 70% are γ/δ T cells. Of these γ/δ T cells, up to 25% expressed cytoplasmic TCR β chains 15, which accounts for the cytoplasmic TCR-β staining in the CD25− cells in pTα−/− mice (Fig. 2 A). There is naturally no TCR-β expression in Rag2−/− mice (Fig. 2). However, this picture changed, to some extent, when the analysis was performed on smaller cells where the proportion of TCR-β+ cells among CD25+ cells was significantly decreased in wild-type and γc−/− mice but not at all or only marginally in pTα−/− and CD3∈−/− mice (Fig. 2 B). What is also apparent in Fig. 2 B is that the proportion of TCR-β1 cells among small CD25+ cells is significantly smaller in wild-type and γc−/− mice, while it is larger in pTα−/− and CD3∈−/− mice. This is due to the fact that in CD25+ cells from pTα−/− and CD3∈−/− mice, TCR-β rearrangement proceeds further than in normal mice 16 17. It is also clear from Fig. 2a and Fig. b, that CD25+ cells in wild-type and γc−/− mice express on average higher TCR-β levels than CD25+ cells from pTα−/− and CD3∈−/− mice, and that with regard to this parameter there is no significant difference between CD25+ cells from pTα−/− and CD3∈−/− cells. Actually, there is a continuous spectrum of TCR-β expression rather than a discrete peak, which would be expected from a population of cells that undergoes TCR-β rearrangement and begins to express productive genes. Nevertheless, there is no doubt that the staining is specific, since there is no staining in the same population of cells in Rag2−/− mice (Fig. 2), and also because an irrelevant control antibody of the same Ig class does not stain in all different mouse strains (data not shown). Thus, all differences that exist between wild-type and CD3∈−/− mice with regard to TCR-β expression in CD25+ cells can be attributed to defective signaling by the pre-TCR rather than to an independent control of TCR-β expression by the CD3 complex alone.
Figure 1.
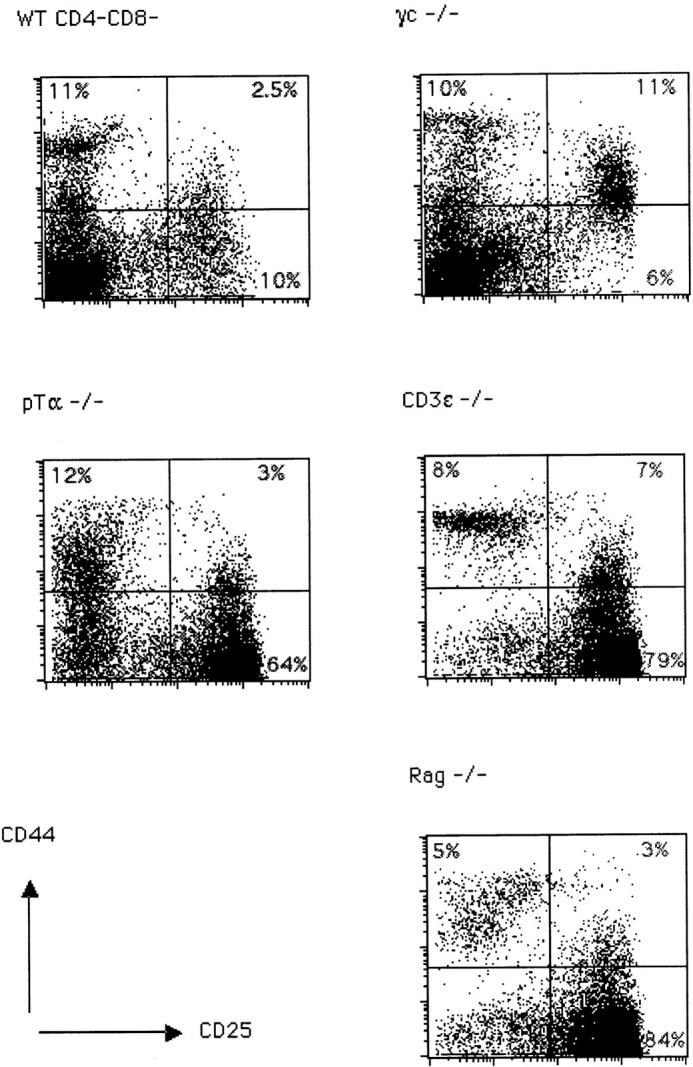
Representative FACS® staining profile of CD4−8− thymocytes from C57BL/6 (WT), γc−/−, pTα−/−, CD3∈−/−, and Rag2−/− mice. Thymocytes were double stained for CD25 and CD44 surface antigens as described. The percentages of cells in each quadrant are indicated.
Figure 2.
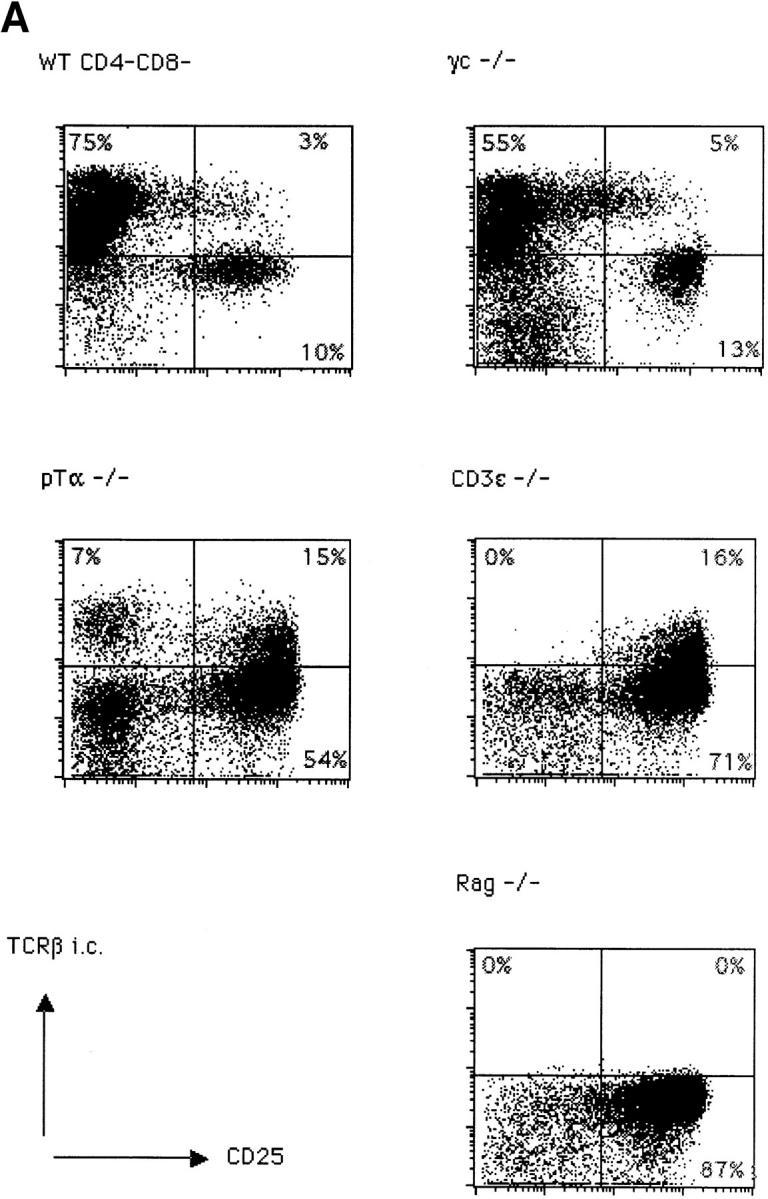
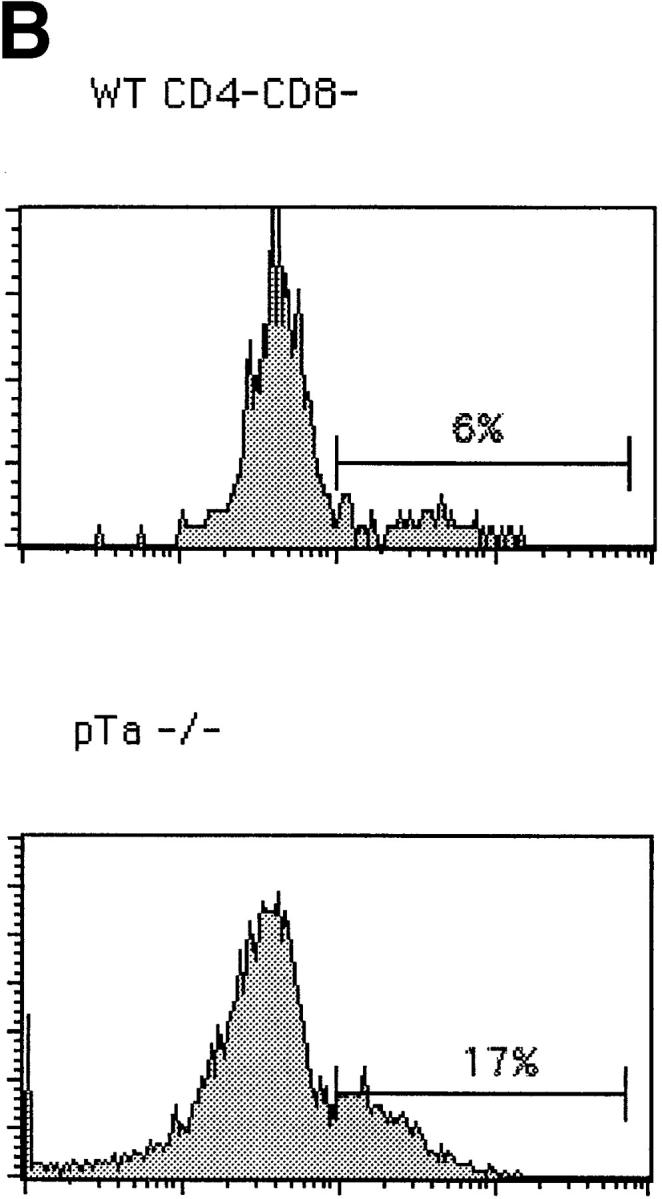
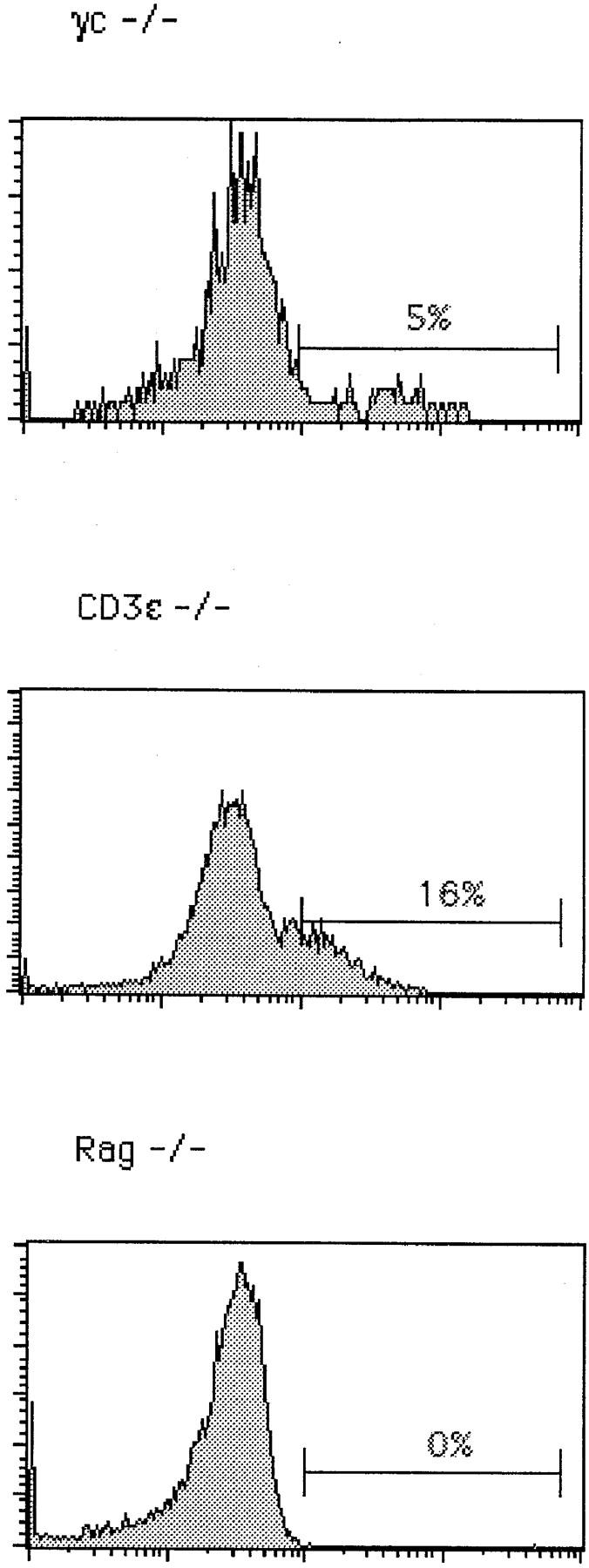
Intracellular staining for TCR-β in CD4−8− thymocytes from C57BL/6 (WT), γc−/−, pTα−/−, CD3∈−/−, and Rag2−/− mice. (A) Total CD4−8− cells were surface stained with anti-CD25; cytoplasmic staining was performed with anti–pan TCR-β (H57) antibodies. (B) Intracellular TCR-β expression in gated small CD25+4−8− thymocytes.
We have focused on TCR-β expression in small CD25+44− cells only, and it is clear that in this thymocyte subset the proportion of cells expressing TCR-β in their cytoplasm is much smaller than in a population that contains CD25+ as well as CD44+ cells, due to the presence of CD44+ NK T cells in the latter population that express α/β TCRs on their cell surface 11 and were included in previous analyses 8. The NK T cell population is likely to be absent in lck−/−, ζ−/− mice because it is a highly selected population that requires signaling through the CD3 complex. Thus, this population would depend on signaling by the α/β TCR rather than just by p56lck and ζ chains, as suggested by Wurch et al. 8. The NK T cell population, for reasons so far unknown, is absent in the pTα−/− mice 18. Moreover, the CD25+44− subset contains a population of pre-TCR–dependent large cells that apparently was included in previous studies 8. If one eliminates these populations of cells from analysis, one nevertheless finds differences in the proportion of CD25+ cells expressing TCR-β, as well as in the level of TCR-β per cell between wild-type and CD3∈−/− mice: in the former, fewer CD25+ cells express TCR-β and at higher levels compared with CD3∈−/− mice. However, the same difference is also noted between wild-type and pTα−/− mice, indicating that this difference is dependent on signal transduction by the pre-TCR rather than signal transduction by p56lck and CD3ζ in the absence of the pre-TCR, as suggested previously 8 9.
In summary, analysis of TCR-β expression in CD25+ cells argues against the notion that levels of TCR-β are upregulated by a pro-TCR rather than the pre-TCR. Rather, they demonstrate that it is the pre-TCR complex in which CD3 signal transducing molecules exert their biological function for the first time in the development of α/β lineage cells.
Acknowledgments
I. Aifantis is the recipient of a Biotechnology Grant from the European Commission, H. von Boehmer is supported by the Institut Universitaire de France, the Juvenile Diabetes Foundation (USA), and by the Körber Foundation (Germany). Supported in part by the Institut National de la Santé et de la Recherche Médicale, Paris, and by the Faculté Necker-Enfants Malades, Descartes Université, Paris. The Basel Institute for Immunology was founded and is supported by F. Hoffmann-La Roche.
References
- Levelt C.N., Ehrfeld A., Eichmann K. Regulation of thymocyte development through CD3. I. Timepoint of ligation of CD3 determines clonal deletion or induction of developmental program. J. Exp. Med. 1993;177:707–716. doi: 10.1084/jem.177.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H., Vandeputte D., Tolkamp L., De Vries E., Borst J., Berns A. CD3 components at the surface of pre-T cells can mediate pre-T cell development in vivo. Eur. J. Immunol. 1994;24:934–939. doi: 10.1002/eji.1830240423. [DOI] [PubMed] [Google Scholar]
- Shinkai Y., Alt F.W. CD3-mediated signals rescue the development of CD4+CD8+ thymocytes in RAG-2−/− mice in the absence of TCR-chain expression. Int. Immunol. 1994;6:995–1001. doi: 10.1093/intimm/6.7.995. [DOI] [PubMed] [Google Scholar]
- Wiest D.L., Burgess W.H., McKean D., Kearse K.P., Singer A. The molecular chaperone calnexin is expressed on the surface of immature thymocytes in association with clonotype-independent CD3 complexes. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:3425–3433. doi: 10.1002/j.1460-2075.1995.tb07348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer H., Fehling H.J. Structure and function of the pre-T cell receptor. Annu. Rev. Immunol. 1997;15:433–452. doi: 10.1146/annurev.immunol.15.1.433. [DOI] [PubMed] [Google Scholar]
- Fehling H.J., Krotkova A., Saint-Ruf C., von Boehmer H. Crucial role of the pre-T-cell receptor gene in the development of αβ but not γδ T cells. Nature. 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- von Boehmer H. Positive selection of lymphocytes. Cell. 1994;76:219–228. doi: 10.1016/0092-8674(94)90330-1. [DOI] [PubMed] [Google Scholar]
- Wurch A., Biro J., Potocnik A.J., Falk I., Mossmann H., Eichmann K. Requirement of CD3 complex–associated signaling functions for expression of rearranged T cell receptor VDJ genes in early thymic development. J. Exp. Med. 1998;188:1669–1678. doi: 10.1084/jem.188.9.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biro J., Wurch A., Potocnik A.J., Falk I., Mossmann H., Eichmann K. Regulation of T cell receptor (TCR) beta gene expression by CD3 complex signaling in immature thymocytesimplications for TCRbeta allelic exclusion. Proc. Natl. Acad. Sci. USA. 1999;96:3882–3887. doi: 10.1073/pnas.96.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman E.S., Passoni L., Crompton T., Leu T.M., Schatz D.G., Koff A., Owen M.J., Hayday A.C. Productive T-cell receptor beta-chain gene rearrangementcoincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 1996;10:948–962. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- Bendelac A., Rivera M.N., Park S.H., Roark J.H. Mouse CD1-specific NK1 T cellsdevelopment, specificity, and function. Annu. Rev. Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- Di Santo J.P., Muller W., Guy-Grand D., Fischer A., Rajewsky K. Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor gamma chain. Proc. Natl. Acad. Sci. USA. 1995;92:377–381. doi: 10.1073/pnas.92.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissen M., Gillet A., Ardouin L., Bouvier G., Trucy J., Ferrier P., Vivier E., Malissen B. Altered T cell development in mice with a targeted mutation of the CD3-epsilon gene. EMBO (Eur. Mol. Biol. Organ.) J. 1994;14:4641–4653. doi: 10.1002/j.1460-2075.1995.tb00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Steward V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M., Alt F. RAG-2 deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- Aifantis I., Azogui O., Feinberg J., Saint-Ruf C., Buer J., von Boehmer H. On the role of the pre-T cell receptor in αβ versus γδ T lineage commitment. Immunity. 1998;9:649–655. doi: 10.1016/s1074-7613(00)80662-7. [DOI] [PubMed] [Google Scholar]
- Aifantis I., Buer J., von Boehmer H., Azogui O. Essential role of the pre-T cell receptor in allelic exclusion. Immunity. 1997;7:601–608. doi: 10.1016/s1074-7613(00)80381-7. [DOI] [PubMed] [Google Scholar]
- Ardouin L., Ismaili J., Malissen B., Malissen M. The CD3-γδ∈ and CD3-ζ/η modules are each essential for allelic exclusion at the T cell receptor β locus but are both dispensable for the initiation of V to (D)J recombination at the T cell receptor-β, -γ, and -δ loci. J. Exp. Med. 1998;87:105–116. doi: 10.1084/jem.187.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert G., Fehling H.J., von Boehmer H., MacDonald H.R. Absolute requirement for the pre-T cell receptor α chain during NK1.1+ TCRαβ cell development. Eur. J. Immunol. 1999;In press doi: 10.1002/(SICI)1521-4141(199906)29:06<1966::AID-IMMU1966>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]


