Abstract
The control of CD4 expression is essential for proper T lymphocyte development. We have previously described a cis-acting silencer element required for repressing transcription of the CD4 gene. Here we report the cloning and characterization of a novel factor that binds to a critical functional site in the CD4 silencer. This factor, referred to as silencer-associated factor (SAF), is a member of the helix-turn-helix factor family and shares sequence similarity with the homeodomain class of transcriptional regulators. Introduction of a specific mutation into the SAF binding site in the CD4 silencer abrogates silencer activity in transgenic mice, supporting the hypothesis that SAF is important in mediating silencer function. Although SAF is expressed in all lymphocytes, immunofluorescence studies indicate that SAF is present primarily in the cytoplasm in T cells in which the endogenous silencer is nonfunctional, whereas it is present primarily in the nucleus in T cells in which the silencer is functional. We thus hypothesize that the subclass-specific subcellular compartmentalization of SAF plays an important role in mediating the specificity of function of the CD4 silencer during T cell development.
Keywords: transcriptional silencer, T cell development, subcellular localization
The control of CD4 coreceptor expression on developing T lymphocytes is tightly regulated and linked to the molecular events that drive repertoire selection 1 2 3. Expression of the CD4 gene is controlled by at least five distinct transcriptional control elements, including the promoter, three enhancers, and a silencer 4 5 6 7 8 9 10 11 12 13 14 15. The silencer is the critical controlling element that downregulates CD4 transcription at several stages of T cell development 7 8 11. During thymopoiesis, the CD4 silencer initially represses CD4 transcription in the CD4−CD8− double negative (DN)1 thymocyte, the most immature T cell precursor, and ceases function as the DN thymocyte continues to develop and expresses CD4. CD8, a similar coreceptor molecule, is also expressed at this stage; the resulting CD4+CD8+ double positive (DP) thymocyte then undergoes the T cell repertoire selection processes 16 17. Surviving T cells can either maintain expression of CD4 and become CD4+CD8− MHC class II–restricted T cells, which are primarily Th cells, or they can downregulate CD4 and become CD4− CD8+ T cells, which are primarily cytotoxic T (Tc) cells. The latter fate decision requires the reinitiation of function of the CD4 silencer and the repression of CD4 transcription. Because the function of the silencer is tightly linked to these cell fate decisions, a study of the nuclear factors that bind to the silencer and mediate its function will provide insight into the molecular mechanisms that drive these processes.
We have previously identified three factor binding sites in the CD4 silencer, which we refer to as S1, S2, and S3 11. Although all three sites are important for full silencer function, there is significant redundancy; silencer activity is only abrogated when S2 is deleted in combination with deletions in either S1 or S3. Using biochemical techniques, we determined that nuclear factors bind specifically to each of these sites 11. We have recently reported that the lin12/Notch pathway intermediate HES-1 binds to S1 18 and the hematopoietic-specific transcription factor c-Myb binds to S2 (reference 19 and Adlam, M., R.D. Allen, and G. Siu, manuscript submitted for publication); however, the factor(s) that bind to S3 and help mediate silencer function is (are) unknown. Here we report the identification and characterization of a novel homeodomain-like transcription factor, silencer-associated factor (SAF), which binds to the S3 region. Using transgenic reporter assays, we determined that the specific SAF binding site within the S3 region is important in silencer function, indicating that SAF may indeed mediate CD4 silencer function. Although SAF is expressed in all lymphocytes, immunofluorescence studies indicate that in cells in which the CD4 silencer is nonfunctional, such as CD4+CD8+ DP and CD4 single positive (SP) T cells, endogenous SAF is preferentially localized in the cytoplasm. Conversely, in cells in which the CD4 silencer is functional, such as CD4−CD8− DN and CD8 SP T cells, endogenous SAF is present in the nucleus. These observations indicate that the selective transport of SAF from the cytoplasm to the nucleus may be one mechanism for generating the functional specificity of the CD4 silencer.
Materials and Methods
Electrophoretic Mobility Shift Assays and Competitions.
Nuclear and whole-cell extracts were purified from the different cell lines and electrophoretic mobility shift assay (EMSA) analyses were conducted as previously described 11. For the cold competition experiments, 100- or 300-fold molar excess of nonradioactive oligonucleotides were added to the binding mix without the radioactive probe and incubated at room temperature for 20 min. The radioactive probe was added, and then the EMSA was carried out. For production of the glutathione S-transferase (GST)–SAF fusion protein, we used the pGEX2T system (Amersham Pharmacia Biotech). The pGEX2TSAF plasmid contains a 0.7-kb fragment containing the SAF cDNA obtained from our library screen cloned into the pGEX2T GST vector inserted in frame with the GST coding region. Purification of the GST–SAF fusion protein in the DH5α bacterial strain was conducted according to the manufacturer's instructions (Amersham Pharmacia Biotech). For antibody ablations, the extracts were preincubated for 30 min on ice with 0.5 μg of the appropriate antibody before completing the EMSA reaction.
Yeast One-hybrid Screening.
The basic protocol used for yeast one-hybrid screening has been described previously 20 21 22. Because of the large number of false positives obtained with this procedure, we designed a screen–counterscreen procedure to more easily identify the true positives. The lacZ reporter plasmids, pWK151, pWK153, and pWK154, were constructed by inserting fourfold multimerizations of the 56bp S3 region, the 18bp S2 region, or the pKs linker, respectively, into pJL638 (reference 22; gift of Dr. Joachim Li, University of California, San Francisco). These plasmids were integrated into the genome of yeast strain YJL 321 (reference 22; gift of Dr. Joachim Li), forming the YWK 101, YWK 102, and YWK 103 yeast strains, respectively. The plasmid pWK 152 was constructed by inserting the S3 four-mer into pJDM 373, which contains the HIS3 gene under the transcriptional control of a TATA box (reference 23; gift of Dr. Randall Reed, Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, Baltimore, MD). A dual reporter yeast strain was then constructed by transforming pWK 152 into yeast strain YWK 101; this strain thus contains both the HIS3 and lacZ reporter genes under the transcriptional control of a TATA box and the four-fold multimerization of S3. The yeast strains YJL 365 and YJL 363 containing lacZ plasmids under the transcriptional control of multimerizations of either wild-type or mutant ACS sites, respectively, were obtained from Dr. Joachim Li 22. A WEHI-3 cDNA library constructed into the pGADNOT vector was transformed into the YWK 101 yeast strain as previously described 24. 3 × 106 transformants were grown on LEU− HIS−TRP− 3-AT plates and subjected to an XGAL screen. Survival of transformants in the absence of leucine and tryptophan is due to the LEU2 gene present in the library vector and the TRP1 gene present in pWK 151; clones encoding putative S3 binding factors were identified using 3-AT and XGAL selection. The library plasmid DNA from these initial positives was then purified and subsequently transformed into yeast strains YWK 101, YWK 102, YWK 103, YJL 365, and YJL363, and an XGAL screen was performed. Library vectors that encode for a true S3 binding factor turn blue only when transformed into YWK 101, and remain white when transformed into YWK 102, 103, YJL 363, and YJL 365.
Antisera Preparation and Western Blot Analyses.
The rabbit anti-SAF antisera was generated against the GST–SAF fusion protein by BABCO. Antibodies against the GST moiety were removed by batch purification with GST-coupled beads and the serum was subsequently purified using Protein A–Sepharose (Amersham Pharmacia Biotech). For the immunofluorescence studies, the anti-SAF antisera was further purified by passing the GST-adsorbed SAF antisera through a GST–SAF-coupled Hi TrapR-N-hydroxysuccinimide activated column (Amersham Pharmacia Biotech) according to the manufacturer's instructions. Bound antisera was eluted by high and low pH cycles, and fractions containing antibody were pooled, dialyzed against PBS, and concentrated by running on a protein A–Sepharose column (Amersham Pharmacia Biotech). To determine the levels of SAF expressed in the cell lines, whole cell extracts were generated by lysing 5 × 107 cells in NP-40 lysis buffer for 60 min on ice. The extracts were then pelleted, and supernatant was collected. 20 μg of whole cell extracts from each cell line were loaded on a 12.5% SDS polyacrylamide gel and run at 15 mA for 4 h. Transfer to nitrocellulose membrane was conducted according to manufacturer's instructions (Bio-Rad). For detection, anti-SAF serum was used at 1:500 dilution and anti–β-Actin (Sigma Chemical Co.) at 1:1,000; blots were developed with the BM Chemiluminescence kit (Boehringer Mannheim) as described by the manufacturer.
Transgenic Experiments.
The pTGΔ2, pTGΔ3, and pTGΔ2-3 transgenic constructs and mice were described previously 11. The pTGΔ2-3m4 construct is identical to pTGΔ2 except the M4 site specific mutation was placed in the S3 region as described previously (reference 11 and see Fig. 1 for sequence). Founder mice were generated by the Columbia-Presbyterian Cancer Center Transgenic/Chimeric Mouse Facility. The following antibodies were used to stain peripheral blood: FITC-conjugated GK1.5 (CD4), allophycocyanin-conjugated 53-6.7 (CD8) and ME1 (mouse IgG1 anti–HLA-B7) followed by PE-conjugated goat anti–mouse IgG1 (Caltag Labs.). Dead cells were excluded from analysis using propidium iodide. Multiple founders were generated and analyzed; representative data are shown. Cells were analyzed on a FACStarPLUS ™ flow cytometer (Becton Dickinson) using the Flo-Jo and CELLQuest™ data analysis software at the Flow Cytometry Facility at the Cancer Center of Columbia University.
Figure 1.
Sequence of CD4 silencer S3. Sequence of the complete probe used in the EMSA experiments is shown in the top line. SAF recognition sites are indicated by boxes. Sequence of mutant oligonucleotides used in EMSA competition analyses are shown below; substitutions from the wild-type sequence are indicated in bold.
Immunofluorescence Studies.
Immunofluorescence studies on the CD4 SP Th clone D10, the CD4+CD8+ thymoma AKR1G1, the CD4−CD8− DN thymoma S49, and the CD8 SP Tc clone L3 were carried out as described previously 25. 107 cells were cooled on ice, washed once in cold PBS, and fixed by resuspension for 30 min on ice in 1 ml of −20°C methanol. Fixed cells were then washed two times in cold PBS and incubated on ice with either affinity-purified SAF antisera or preimmune sera at a concentration of 2 μg/ml in 250 μl of PBS/5% sheep serum for 1 h. After incubation with the primary antibody was completed, cells were washed three times in cold PBS and incubated with Cy3-conjugated sheep anti–rabbit IgG (Sigma Chemical Co.) at a dilution of 1:300 in 250 μl of PBS/5% sheep serum for 30 min. Cells were then washed three times in cold PBS before mounting on poly-l-lysine–coated slides (Sigma Chemical Co.). Nuclear staining was performed by adding DAPI to one of the final washing steps. Microscopy was performed on a Leitz fluorescent microscope.
Results
Sequence Specificity of the S3 Binding Factor.
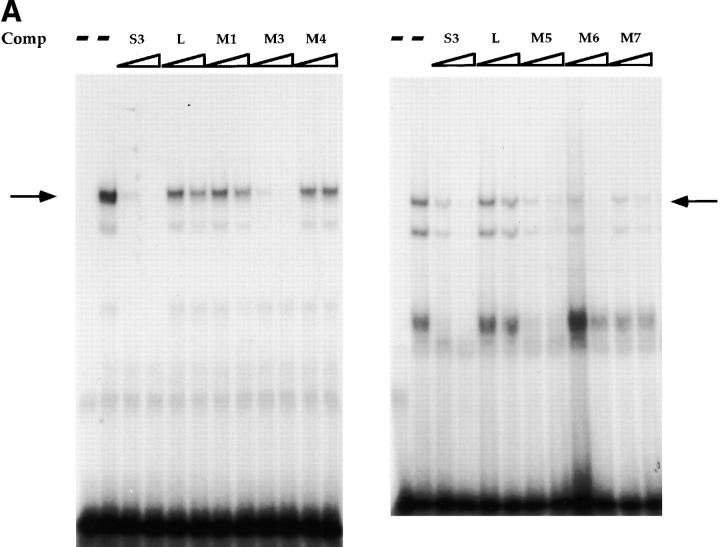
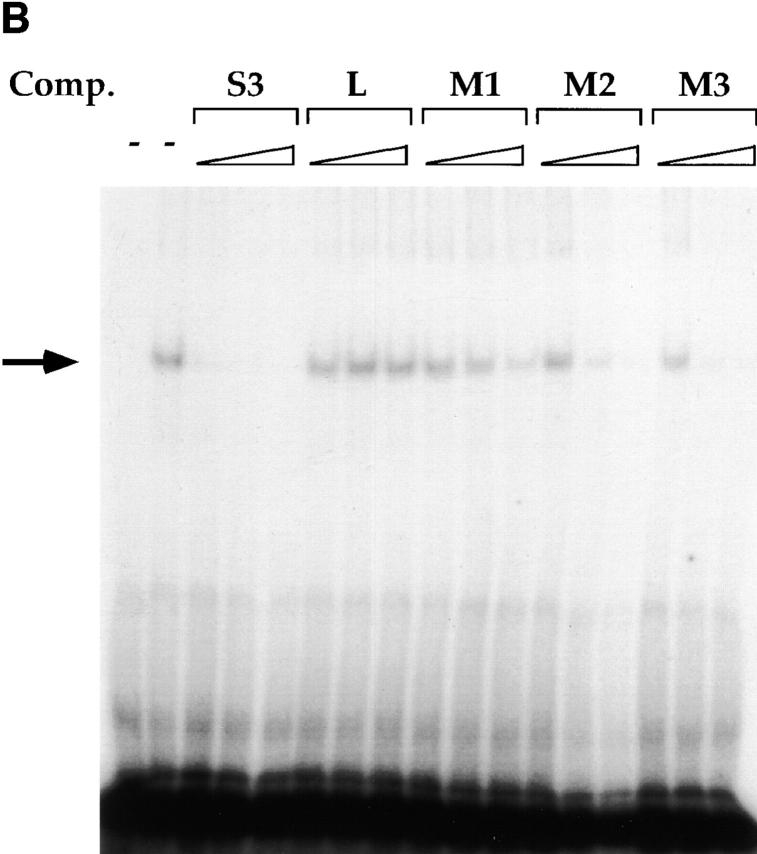
In previous studies, we identified three factor binding sites in the CD4 silencer using DNAse footprinting. Additional EMSA analyses identified one major and several minor factor–DNA complexes when the footprinted S3 region was used as a radioactive probe; we have concentrated our further analyses on the major S3 binding complex 11. Using o-phenanthroline copper footprinting, we narrowed the recognition site of the major S3 binding factor to a 16-bp region (reference 11 and Fig. 1). This region contains a 5-bp direct repeat (CTGTG) separated by 6 bp. A comparison of this 16-bp region with known binding site motifs revealed consensus LEF-1 26 27 28 29 and ETS 30 recognition sites; however, we were unable to demonstrate that either LEF-1 or an ETS family protein binds to S3 using biochemical approaches (data not shown). To identify a more precise recognition site for the major endogenous S3 binding factor, we designed a series of mutant S3 oligonucleotides to be used as competitors in EMSAs (Fig. 1 A). As we have reported previously 11, we can detect a major DNA–protein complex with the 16-bp S3 probe using nuclear extracts isolated from CD4−CD8+ Tc cells; complex formation can be completely inhibited by the addition of nonradioactive S3 probe but not linker, indicating that the S3 binding factor binds specifically to the S3 probe (Fig. 2 A). Oligonucleotides that contain mutations in both CTGTG repeats failed to compete away the major S3 binding complex (Fig. 1 and Fig. 2, M1 and M4). However, those containing mutations in either of the CTGTG repeats are still capable of competing for S3 complex formation (Fig. 1 and Fig. 2, M5 and M6). Oligonucleotides that contain mutations in sequences between the two CTGTG repeats also compete for complex formation efficiently (Fig. 1 and Fig. 2, M3; Fig. 1, M2; and data not shown); oligonucleotides that contain mutations in the spacer sequence directly adjacent to the CTGTG repeats also compete for complex formation, albeit somewhat less efficiently than do oligonucleotides with central mutations (Fig. 1 and Fig. 2; compare M7 to M2 and M3). We can detect three complexes with the S3 probe; the two slower mobility complexes appear to have similar sequence specificities and thus may represent modified versions of the same factor, whereas the fastest mobility complex does not appear to be reproducible from experiment to experiment (Fig. 2 A and data not shown). We can also detect S3 binding complexes in whole cell extracts purified from the D10 (CD4+CD8− Th), S49 (DN thymoma), AKR1G1 (DP thymoma), and the L3 and B18 (CD4−CD8+ SP Tc) T cell clones (Fig. 2b and Fig. c, and data not shown). Our data thus indicate that S3 binding proteins are present in T cells of all developmental phenotypes and bind to one of the two CTGTG direct repeats in the S3 probe.
Figure 2.
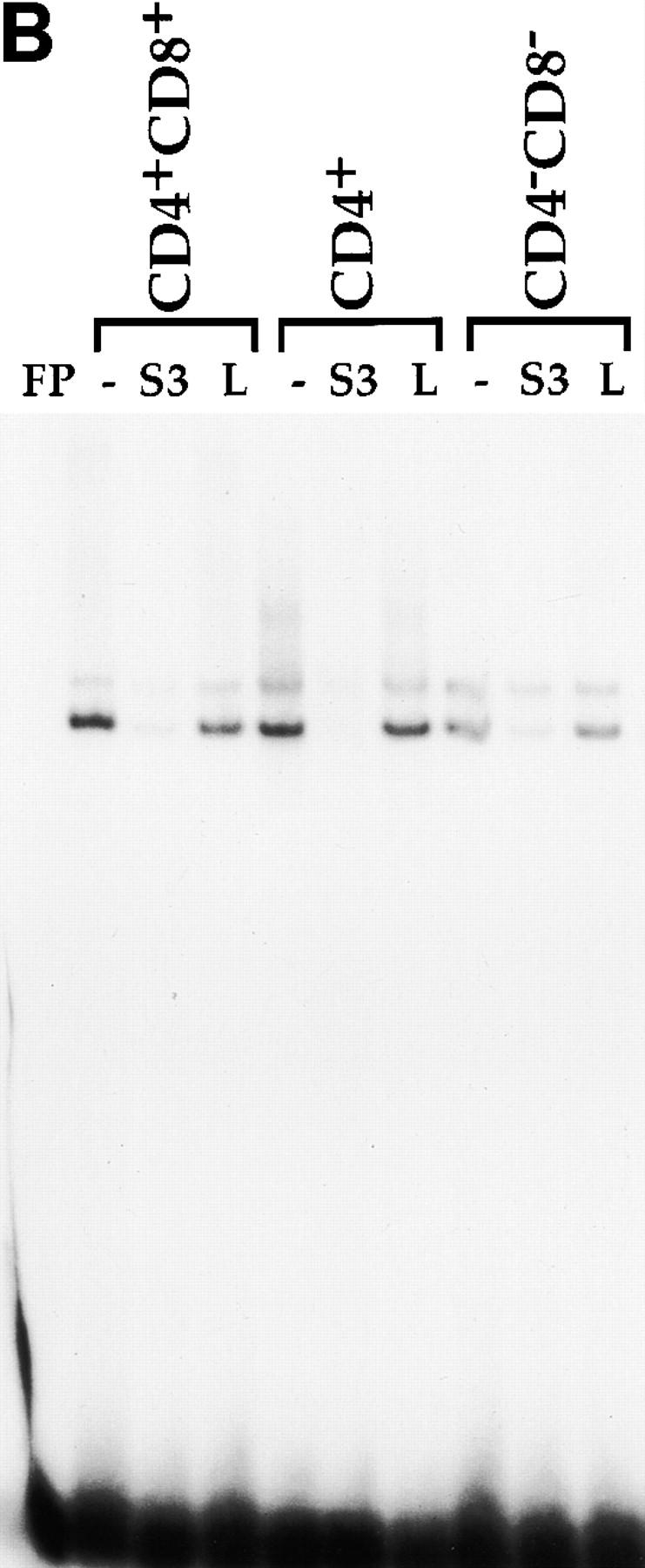
EMSAs using the S3 probe and CD8 SP Tc L3 nuclear extracts (A), whole cell extracts from the CD4 SP Th clone D10, the DP thymoma AKR1G1, and the DN thymoma S49 (B), or whole cell extracts from D10 and L3 (C). Arrows indicate putative SAF-containing complex; addition of increasing amounts of different nonradioactive competitor oligonucleotides are indicated above each lane.
Cloning of a Novel S3 Binding Factor.
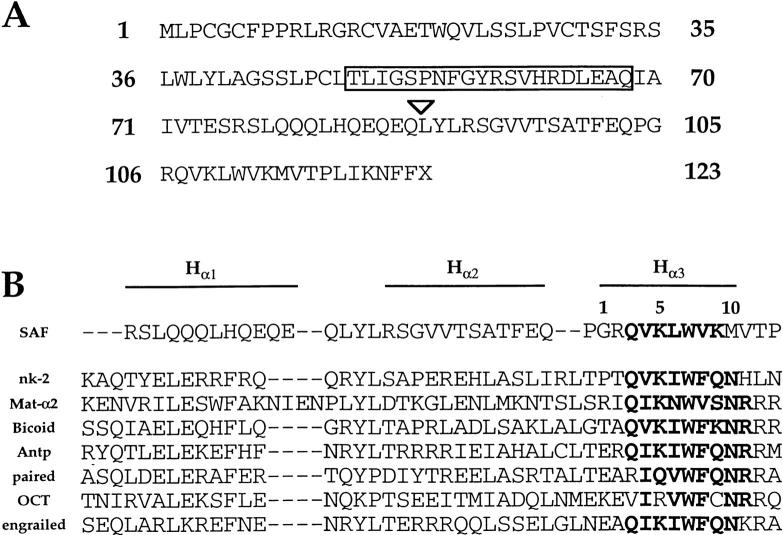
We next attempted to clone a cDNA encoding the S3 binding factor using a modified protocol of the previously published yeast one-hybrid technique (see Materials and Methods for details). We screened 3 × 106 colonies and identified one positive clone encoding a 0.7-kb cDNA. Using Northern blot analyses with the cDNA as a radioactive probe, we can detect a single 3.1-kb mRNA species in a wide variety of different tissues (data not shown). We isolated a full-length 3.1-kb cDNA clone by using the 0.7-kb cDNA as a radioactive probe to screen a λ phage thymus cDNA library. The cDNA contains a 2,123-bp 5′ untranslated region and a 623-bp 3′ untranslated region, as well as a 369-bp open reading frame (ORF) that potentially encodes for a 123-amino acid protein with a mol mass of 14 kD (Fig. 3 A and data not shown). We believe this ORF to be the one used in vivo for five reasons. First, this ORF is used in the GAL4 fusion protein originally isolated from the one-hybrid screen; second, GST fusion proteins containing this ORF are able to bind S3 specifically in EMSA analyses (see below); third, an antisera raised against the same GST fusion protein is able to recognize the S3 binding factor in T cell extracts (see below); fourth, Western blot analyses on both cell lines and tissues using this antisera identify one prominent 14-kD species (see below); and fifth, using databank searches we have identified a putative Caenorhabditis elegans homologue (see below). In addition, in vitro transcription/translation assays with the full-length cDNA produce a single 14-kD species that can be recognized by this antisera on Western blot analysis (data not shown). We refer to the novel transcription factor encoded by the cDNA as silencer-associated factor, or SAF.
Figure 3.
(A) Protein translation of the SAF ORF. Arrowhead indicates the beginning of the sequence of the original yeast one-hybrid clone isolated in the screen. (B) Sequence alignment of the putative HTH domain of SAF with the HTH regions of different homeodomain proteins. The three putative α-helical structures are indicated above; sequence similarity in the DNA recognition helix (HR) is indicated in bold.
We conducted BLAST homology searches using the translated protein sequence and could not identify significant sequence similarity with known proteins. Expressed sequence tagged cDNA sequences encoding portions of SAF were identified in several libraries, including those constructed from RNA purified from diverse tissues, such as thymus, 2-d embryo, and brain, further indicating that SAF is expressed in a wide variety of tissues at different stages of development. Interestingly, we have identified a putative homologue for SAF in C. elegans using databank searches. SAF shares an overall amino acid sequence identity of 32%; however, the similarity is highest in the COOH-terminal region, where a 21-amino acid domain located just NH2-terminal to the HHTH (helix-helix-turn-helix) domain shares 86% identity at the amino acid level (18 out of 21), with 96% similarity (20 out of 21) with its putative C. elegans homologue (boxed sequence, Fig. 3 A). However the exact function of this C. elegans ORF is unknown. These data indicate that SAF may be conserved throughout evolution, further supporting the hypothesis that SAF is an important factor.
SAF Shares Sequence Similarity with Homeodomain Proteins.
Motif analyses indicated that the COOH-terminal portion of SAF has a predicted HHTH structure. The helix-turn-helix (HTH) motif is a common DNA binding domain that has been characterized for many different transcription factors 31 32. Based on the sequence similarity in the DNA binding helix, there are six major families of HTH proteins; SAF has the highest sequence similarity with members of the homeodomain HTH family (Fig. 3 B). The homeodomain proteins consist of a wide variety of different transcription factors that are important in the control of gene expression during development. All members of this family have the same general structure: a three-α helix bundle with each helix separated by short amino acid turns, and an NH2-terminal arm that stretches into the minor groove (Fig. 3 B) 32 33. Computer structural analysis of SAF indicates α-helical structures encompassing the glutamine- and glutamic acid–rich domain representing helix α1 as well as the helix α2 region (Fig. 3 B). The greatest sequence similarity is within the putative DNA recognition helix (helix α3), including a stretch of amino acids QVKLWVK that are seen often in the same position in homeodomain proteins (Fig. 3 B, bold sequence). Although SAF shows the greatest sequence similarity to the homeodomain proteins, there is a major difference between SAF and the homeodomain protein family: all homeodomain proteins have an asparagine at position 10 of α3, whereas SAF has a methionine (Fig. 3 B). As this region is believed to be important in DNA sequence recognition, these observations indicate that SAF is likely to bind to DNA in a manner distinct from the classical homeodomain–DNA interaction.
Endogenous SAF Binds to S3 of the CD4 Silencer.
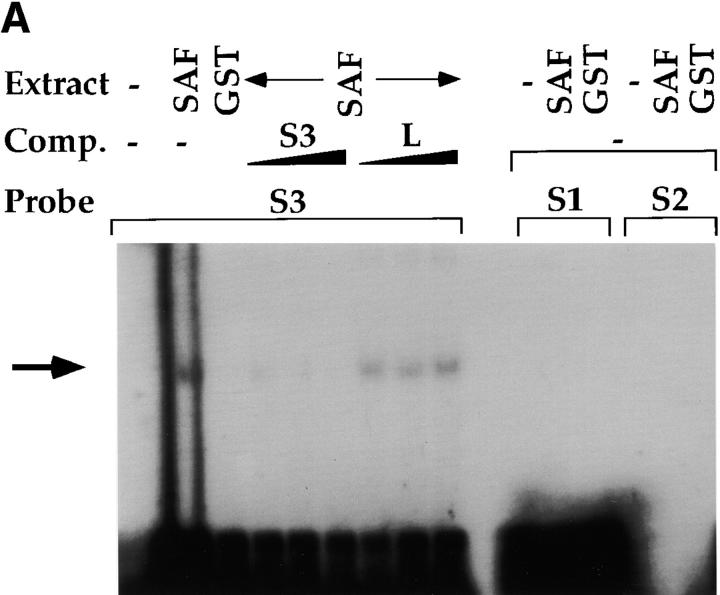
To confirm that the SAF cDNA isolated from the yeast one-hybrid screen binds S3, we generated a GST–SAF fusion protein to be used in EMSAs. The SAF-encoding cDNA that was isolated in the yeast one-hybrid screen was excised from the yeast one-hybrid vector and subcloned into the pGEX2T GST vector (Amersham Pharmacia Biotech); the cDNA encompassed the COOH-terminal 34 amino acids that contain the putative helix α2 and helix α3 of the HTH domain (referred to as SAF89–123). The GST–SAF89–123 fusion protein was then used in EMSA analyses with different radioactive probes. We can detect a single DNA–protein complex when we conduct EMSA analyses with GST–SAF89–123 and the S3 probe (Fig. 4). This complex formation is not contingent upon the GST moiety, since purified GST alone cannot bind S3 (Fig. 4 A). The complex formation is specific for the S3 sequence as it can be inhibited with addition of nonradioactive S3 but not with linker, and it cannot bind to oligonucleotide probes taken from S1 or S2 of the CD4 silencer (Fig. 4 A). These data indicate that SAF can bind to S3 in a sequence-specific fashion. In addition, these data indicate that the DNA binding domain of SAF is in the COOH-terminal in the same region as the HTH motif, thus providing additional evidence that HTH motif contains at least a portion of the DNA binding domain. To determine if SAF has the same fine specificity of binding as the endogenous S3 binding factor, we conducted competition EMSAs with the S3 probe and the mutant oligonucleotide probes used above (Fig. 1, Fig. 2, and Fig. 4). Similar to what we observed for the endogenous S3 binding factor, an oligonucleotide that contains mutation in both CTGTG repeats (M1) does not compete for SAF binding to S3, whereas mutations in the spacer sequence (M2 and M3) or only in one CTGTG repeat (M5 and M6) compete effectively for complex formation (Fig. 4 and data not shown). Thus, SAF has the same DNA binding fine specificity as the endogenous S3 binding factor, supporting the hypothesis that SAF is the endogenous S3 binding factor.
Figure 4.
Sequence specificity of the SAF DNA binding domain. (A) EMSAs using the GST–SAF89–123 fusion protein and CD4 silencer S1, S2, and S3 probes. Probe, titration of nonradioactive competitor oligonucleotides, and protein extract used are indicated above lanes. Arrow indicates SAF–DNA complex. (B) Competition EMSAs using the GST–SAF89–123 fusion protein, the S3 probe, and S3 mutant variants. Titrations of different nonradioactive competitor mutant S3 oligonucleotides are shown above lanes. Arrow indicates SAF–DNA complex.
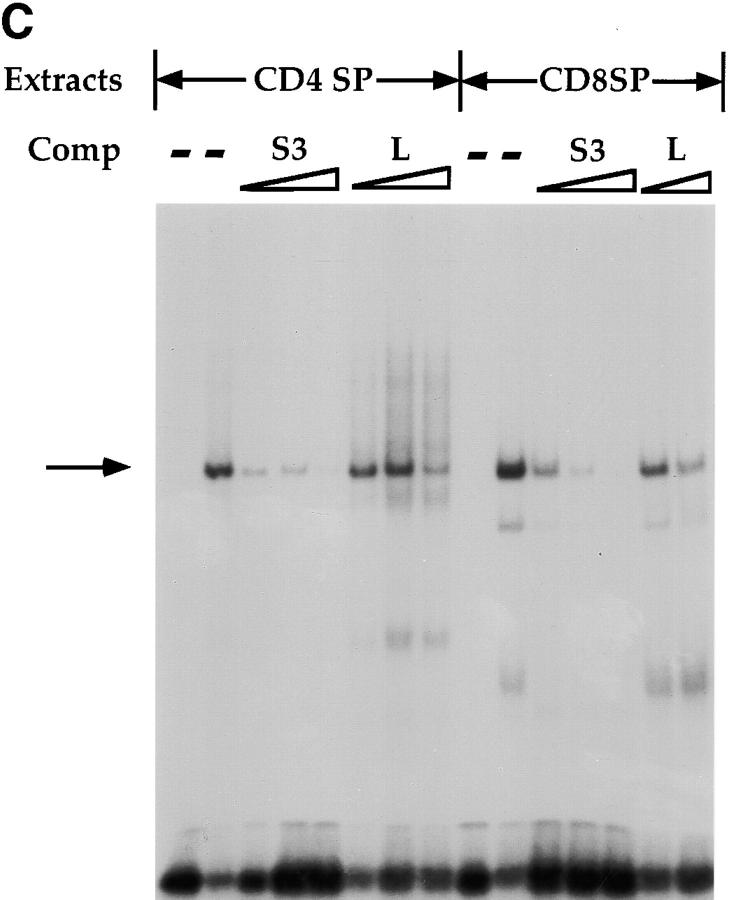
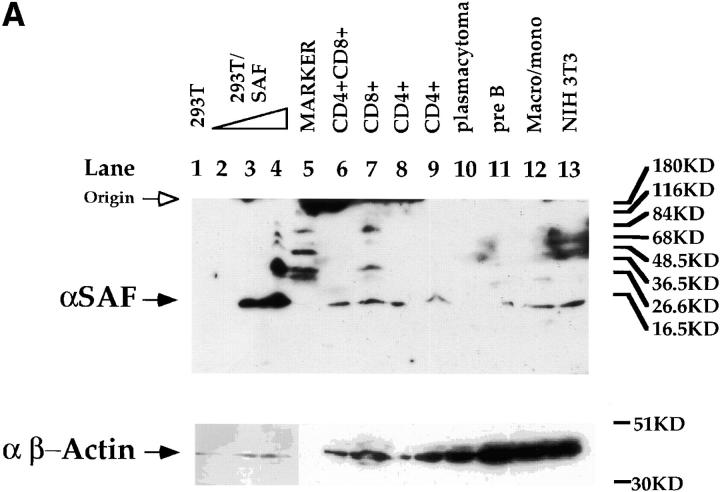
To characterize SAF in greater detail, we generated a rabbit polyclonal antisera against SAF using the GST–SAF89–123 fusion protein as antigen. The specificity of the antisera was tested by Western blot analyses using whole cell extracts purified from a variety of cells of different phenotypes (Fig. 5 A). We can detect the induction of expression of a 14-kD species in 293T cells transfected with a CMV expression vector containing the full-length SAF cDNA, supporting the hypothesis that the 369-bp ORF is indeed the reading frame used in vivo (Fig. 5 A). In addition, we can detect the same 14-kD species in all T cell subclasses, B cells, macrophages, and fibroblasts (Fig. 5 A), indicating a wide tissue distribution of expression of SAF protein, consistent with the EMSA data discussed above. To prove that the endogenous S3 binding protein is indeed SAF, we tested whether the SAF antisera would affect endogenous S3 binding factor–DNA complex formation in EMSAs. As can be seen in Fig. 5 B, we can ablate S3–protein complex formation completely in both CD4 SP Th and CD8 SP Tc cell extracts using the SAF antisera. We cannot inhibit S3–protein complex formation significantly using either the preimmune sera or an antibody directed against Elf-1, indicating that this ablation is specific for the endogenous S3 binding protein. These data indicate that the endogenous S3 binding protein shares antigenic epitopes with SAF, supporting the hypothesis that SAF is the endogenous S3 binding factor.
Figure 5.
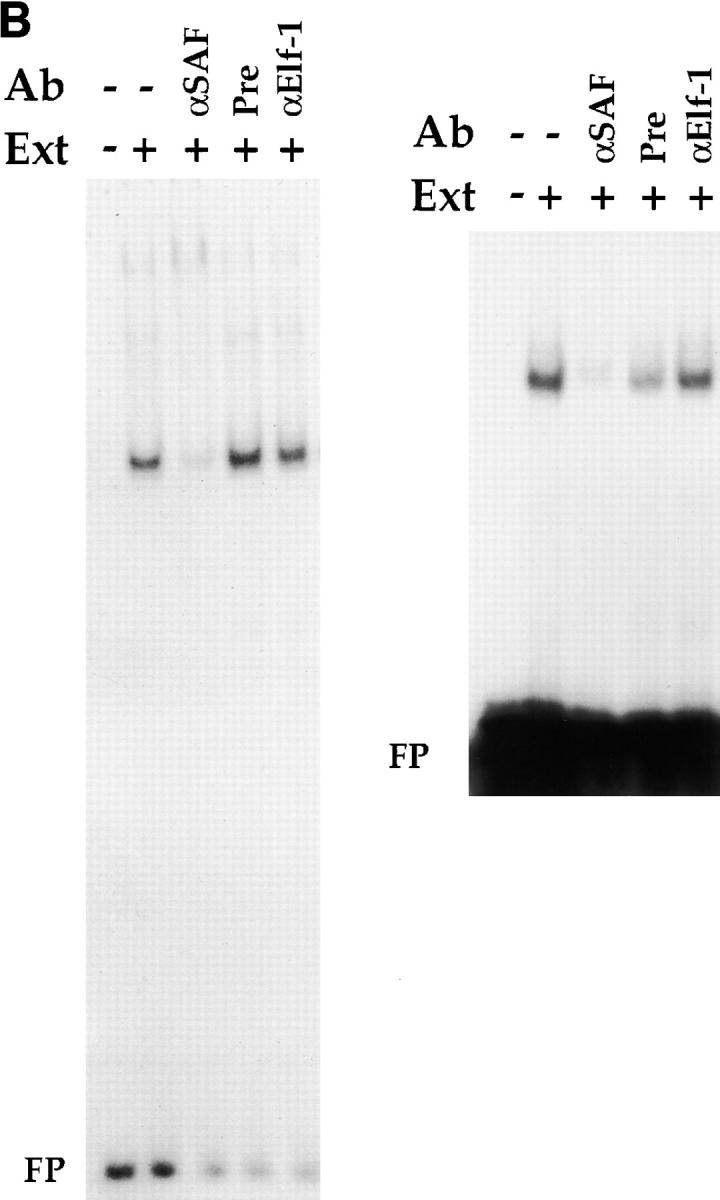
Expression and binding of endogenous SAF. (A) Western blot analysis with the SAF antisera (top) and the control anti–β-actin antibody (bottom). Western blot analysis on whole cell extracts from 293T cells (lane 1): 293T cells transfected with 5, 10, and 20 μg of the CMV–SAF expression construct (lanes 2–4); the CD4+CD8+ DP AKR1G1 thymoma (lane 6); the CD8+ SP Tc L3 cell clone (lane 7); the CD4+ SP Th D10 cell clone (lanes 8 and 9); the P3X63 plasmacytoma (lane 10); the 103 pre-B cell lymphoma (lane 11); the WEHI-3B macrophage/monocyte (lane 12); and the 3T3 fibroblast (lane 13). Molecular weight standards were loaded in lane 5. (B) Antibody ablation EMSA analyses with the SAF antisera and (left) CD4+ SP Th D10 or (right) CD8+ SP Tc L3 extracts. Lanes containing S3 probe only, S3 probe with extract, and S3 probe with extract and antibody are indicated above each lane. ‘Pre’ indicates preimmune sera.
The SAF Binding Site Is Important for CD4 Silencer Function.
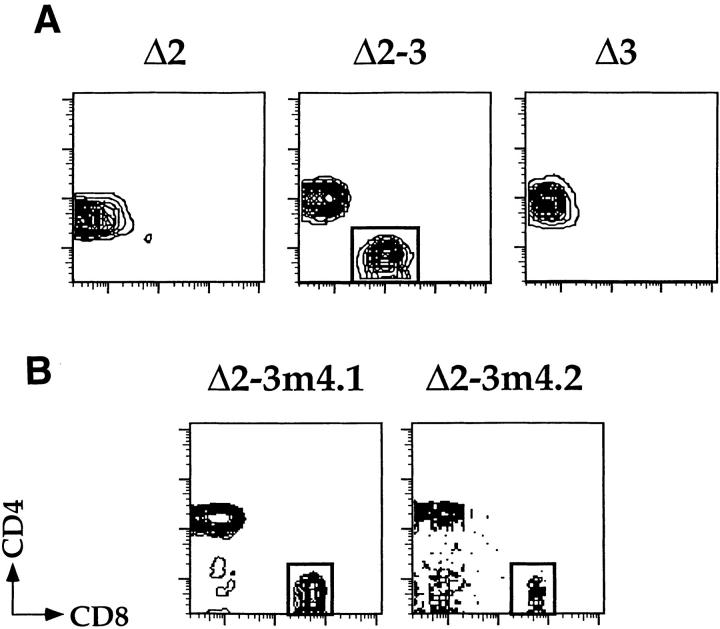
Our biochemical data indicate that SAF binds to an important functional region of the CD4 silencer, indicating that SAF is playing a role in silencer function. Should this be the case, we can predict that we would abrogate CD4 silencer function by making a site-specific mutation in the SAF binding site. To test this, we generated mutant silencers and tested them in transgenic assays (Fig. 6). We have previously shown that single deletions of any of the three factor binding sites in the CD4 silencer, referred to as S1, S2, and S3, do not affect silencer activity. In fact, silencer function is abrogated only when S2 is deleted in conjunction with deletions in either S1 or S3 11. We have generated a series of mice transgenic for constructs that contain silencers with different combinations of factor binding site deletions 11. The base pTG construct contains the HLA-B7 marker gene under the transcriptional control of the CD4 promoter and enhancers; the pTGSil series constructs also contain either the unmutated silencer or mutated silencers 11. The pTGSilΔ2 construct contains the silencer with an 18-bp deletion of the S2 region, the pTGSilΔ3 construct contains the silencer with a 56-bp deletion of the S3 region including the SAF binding site, and the pTGSilΔ2-3 construct contains the silencer with both deletions. As can be seen in Fig. 6 A, cells that express the HLA-B7 marker gene in the pTGSilΔ2 and pTGSilΔ3 transgenic mice are confined to the peripheral CD4 SP T cell population, as would be expected if silencer function is intact. In contrast, the pTGSilΔ2-3 transgenic mice express the marker gene in both CD4 SP and CD8 SP T cells, indicating that silencer function has been abrogated. As discussed above, these observations are consistent with our previous data 11.
Figure 6.
Site-specific mutation of the SAF binding site in S3 leads to abrogation of CD4 silencer function. Peripheral T cells were isolated from the (A) pTGΔ2, pTGΔ2-3, and pTGΔ3 and the (B) pTGΔ2-3m4 transgenic lines and stained with antibodies to CD4, CD8, and the HLA-B7 marker. The HLA-B7+ cells were then gated on and analyzed for expression of CD4 (y axis) and CD8 (x axis). Presence of transgenic marker–positive CD8 SP T cells in the pTGΔ2-3m4 and pTGΔ2-3 mice (box, lower right quadrant) indicate loss of silencer function in the marker construct. Five different pTGΔ2-3m4 founders were generated; data from representative high expressing (B, left) and low expressing (B, right) transgenic mice are shown.
Although the original S3 deletion removed the SAF binding site, this deletion encompassed 56 bp, so it is possible that the SAF binding site is irrelevant and that other sequences within S3 are required for silencer function. To determine if the SAF binding site itself is important in silencer function, we generated a mutant silencer, SilΔ2-3m4, that contains the 18-bp S2 deletion and the M4 mutation of the SAF binding site. The M4 mutation completely abrogates SAF binding as defined by our biochemical data (see above). This mutant silencer was cloned into the pTG reporter construct, transgenic mice were generated, and peripheral T cells were harvested from expressing founders and analyzed for expression of the HLA-B7 marker gene (Fig. 6 and data not shown). Similar to the pTGSilΔ2-3 transgenic mice, both CD4 SP and CD8 SP peripheral T cells in the pTGSilΔ2-3m4 transgenic mice express the marker gene, indicating that silencer function is abrogated (Fig. 6 B). These data indicate that the site-specific mutation of the SAF binding site is functionally similar to the original 56-bp deletion of S3, which in combination with a deletion in S2 leads to abrogation of silencer function. Therefore, we can correlate the loss of SAF binding to S3 with the loss of silencer function, supporting the hypothesis that SAF is indeed playing an important role in CD4 silencer function.
Subcellular Compartmentalization of SAF.
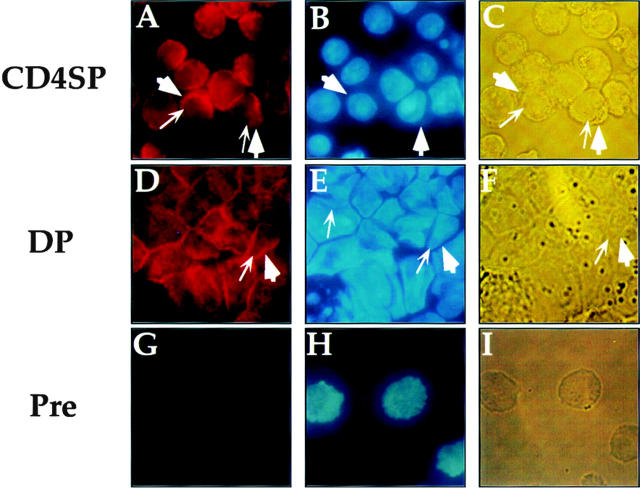
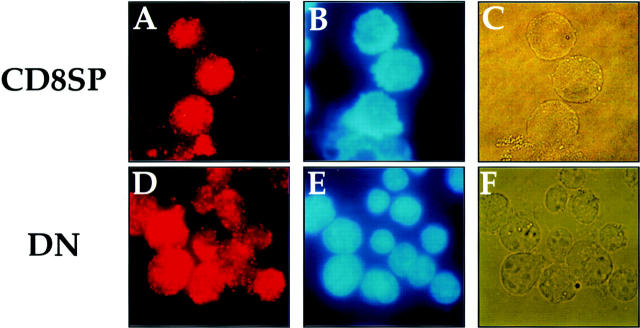
As mentioned above, SAF is expressed in T cells of all developmental phenotypes. The fact that the CD4 silencer functions in CD4− but not CD4+ cells indicates that the specificity of silencer function cannot be mediated by cell type–specific SAF expression. Therefore, if SAF plays a role in the specificity of silencer function, we predict that there are posttranslational events that permit SAF to function only in CD4− cells, thus conveying specificity of silencer function. One potential mechanism is the cell type–specific partitioning of the factor in different subcellular compartments. For example, it is possible that a ubiquitously expressed transcription factor is sequestered outside of the nucleus to prevent DNA binding. In the case of the CD4 silencer, cells that express CD4 would localize SAF to the cytoplasm, thus preventing it from binding to the silencer and inducing its function and therefore allowing CD4 transcription. In cells that do not express CD4, SAF is transported to the nucleus, thus allowing SAF to bind to the silencer and induce its function leading to the repression of CD4 transcription. This hypothesis predicts that we would detect SAF protein primarily in the nucleus in T cells that do not express CD4 and primarily in the cytoplasm in T cells that express CD4. To test this hypothesis, we used affinity-purified anti-SAF antisera in immunofluorescence experiments with T cells of different developmental phenotypes (Fig. 7 and Fig. 8). Interestingly, the anti-SAF antisera stains the cytoplasm of both the CD4 SP and the CD4+ CD8+ cells most intensely. As can be seen in Fig. 7A and Fig. D, the nucleus of each of these cells are present as a shadow with only faint staining (nuclear membrane indicated with thick arrowheads; compare panels A with B, and D with E), whereas the ring of cytoplasm surrounding the nucleus stains intensely (cell membrane indicated with thin arrows; compare panels A with B and C, and D with E and F). These observations correlate with the expression of CD4 in these two T cell developmental subclasses; SAF is predominantly in the cytoplasm and thus cannot access its S3 binding site and help mediate silencer function. The preimmune sera yields only low levels of staining, indicating that the signal detected is specific (Fig. 7G–I).
Figure 7.
Subcellular localization of SAF in CD4+ SP and CD4+CD8+ T cells. Staining with the affinity-purified anti-SAF antisera (A and D), DAPI (B and E) and the light-phase field (C and F) of the D10 CD4 SP and the AKR1G1 CD4+CD8+ T cells are indicated. Each set of fields is from the same experiment. G–I show staining with the preimmune serum, DAPI, and light-phase field for a CD8 SP T cell; all other preimmune stainings for the other cell types showed similar patterns (data not shown). Thick arrows indicate cell membrane, and thin arrows indicate nuclear membrane.
Figure 8.
Subcellular localization of SAF in CD8+ SP and CD4−CD8− T cells. Staining with the affinity-purified anti-SAF antisera (A and D), DAPI (B and E), and the light-phase field (C and F) of the L3 CD8 SP and the S49 DN T cells are indicated. Each set of fields is from the same experiment.
In contrast, for the DN and CD8 SP T cells, anti-SAF staining colocalizes with the nuclear DAPI stain, indicating that for these CD4− T cell subclasses SAF is present at high levels in the nucleus (Fig. 8; compare panels A with B, and D with E). The localization of SAF to the nucleus in CD4− T cells correlates both with the expression of CD4 and endogenous silencer function; in these cells, SAF is in the nucleus and thus presumably has access to its cognate binding site in the CD4 silencer, which will thus allow it to mediate silencer function. Taken together, our data indicate that SAF protein, although synthesized in T cells of all developmental phenotypes, is preferentially localized in different subcellular compartments depending on the expression of CD4 and are consistent with the hypothesis that the developmental stage-specific subcellular localization of SAF plays a role in the specificity of function of the CD4 silencer. In contrast, immunofluorescence with antisera against both c-Myb and HES-1, the other CD4 silencer binding factors, indicate that both of these factors are nuclear in T cells of all developmental phenotypes (data not shown), suggesting that SAF may play a unique role in the control of CD4 silencer function and CD4 transcription.
Discussion
SAF: A Novel CD4 Silencer Binding Protein.
We have identified a novel factor, which we refer to as SAF, that binds to a critical functional site of the CD4 silencer. We have determined that endogenous SAF binds to S3 and probably mediates silencer function; we draw this conclusion on the basis of several different experimental approaches. First, we have established that the fine DNA binding specificity of SAF is identical to that of the endogenous S3 binding factor. In addition, antisera raised against SAF specifically ablate the formation of the endogenous S3 binding complex. Finally, in our transgenic reporter assay system, a site-specific mutation in S3 that abrogates both SAF and endogenous S3 factor binding also breaks CD4 silencer function in conjunction with the S2 deletion. These functional results are consistent with our earlier data, which indicated that a large deletion in the S3 region, in combination with a deletion in the S2 region, abrogates silencer function 11. Taken together, our data provide strong evidence supporting the hypothesis that endogenous SAF binds to the CD4 silencer at S3 and mediates its function.
SAF has several interesting structural features. The function of the NH2-terminal domain of SAF is unknown. However, SAF shares some sequence similarity with the homeodomain class of transcription factors that are known to be important both in transcriptional repression and in the control of developmentally-regulated genes 31 32. In particular, the DNA binding domain of SAF contains an HHTH motif similar to that of the homeodomain class of proteins. There is significant sequence similarity between SAF and homeodomain proteins in the DNA recognition helix, although SAF lacks an important conserved asparagine at position 10 of helix α3. It is interesting to note that the SAF binding site (CTGTGNNNNNNCTGTG) differs significantly from the consensus homeodomain recognition sequence (TCAATTAAAT) 34 35. The asparagine at position 10 of helix α3 interacts via hydrogen bonds and van der Waals interactions with a central adenine, a base that is not present in the SAF recognition site 36. Thus, this asparagine-to-methionine change may reflect a difference in the consensus recognition sequences between the two factors. Despite these sequence similarities, a complete structure analysis of SAF is required before we can draw definitive conclusions about its relationship with the homeodomain proteins.
We have been unable to reproduce CD4 silencer function in transfection assays by transfecting silencer-containing reporter constructs into CD4 SP and CD8 SP T cell clones. Although the reasons for this are not known, several possibilities exist. Perhaps the most likely is that silencer function cannot be recapitulated using transfected reporters because it requires higher order chromatin structure that is normally lacking in transfection systems. The only reliable assay of silencer function is the transgenic reporter system 11, as this requires the insertion of the reporter construct into the genome, this observation is consistent with this hypothesis. Alternatively, it may be that SAF requires a non-DNA binding corepressor to mediate transcriptional repression. For example, the Hairy family of DNA binding factors require the non-DNA binding cofactor groucho to mediate transcriptional repression 37; similarly, the Ssn6-Tup1 complex serves as a general repressor of transcription in yeast that is recruited to target promoters by many different sequence-specific DNA binding proteins 38. Finally, levels of SAF expressed during transient transfection may exceed endogenous levels of post-translational modification enzymes that might be required for its activity. In this latter case, most of the overexpressed SAF in transfected T cells would be unable to mediate repression due to lack of modification. In any case, the lack of a good transfection system to study silencer function indicates that further characterization of the role of SAF in mediating CD4 silencer function will most likely require more complicated mouse genetic experiments. For example, it may be possible to generate altered specificity binding mutants of SAF to study its function; similar experiments have been conducted to characterize other mammalian transcription factors 39 40. In addition, generation of the targeted-disruption of the endogenous SAF gene will probably provide useful information on the role of SAF in T cell development and CD4 silencer function.
SAF and the Specificity of CD4 Silencer Function.
Although in genetic experiments the CD4 silencer is the critical controlling element that mediates the specificity of CD4 gene expression during T cell development, all of the silencer binding factors we have identified, including SAF, are expressed in T cells of all developmental stages 11 18 19. However, our data indicate that although SAF is expressed in all T cells, its subcellular localization differs in a T cell subclass–specific manner. In cells that express CD4, the CD4 SP, and the DP T cells, SAF localizes to the cytoplasm; whereas in cells that do not express CD4 (the DN and CD8 SP T cells) SAF localizes to the nucleus. This is the first report of biochemical subclass specificity of the factors that mediate CD4 gene expression and provide a potential mechanism for the control of CD4 silencer function during T cell development. We hypothesize that in cells in which the CD4 silencer is functioning, such as the CD8 SP and DN T cells, SAF is transported to the nucleus, where it binds to S3 of the CD4 silencer and helps mediate its repression of CD4 gene expression. In cells in which the silencer is nonfunctional, such as the CD4 SP and DP T cells, SAF is specifically excluded from the nucleus. Because of this, SAF cannot bind to the CD4 silencer, and thus does not mediate its function. This model indicates that the mechanisms controlling SAF nuclear transport may be linked to the processes that transmit the differentiation signal from the surface of the thymocyte during the selection process.
The nature of the mechanism that restricts SAF localization in specific cell types is unclear. Although SAF does not have a consensus nuclear localization signal, its small size should in principle permit it to migrate freely through the nuclear pore, and thus some mechanism to compartmentalize it must be hypothesized. It is possible that the posttranslational modification of SAF either allows or blocks its nuclear transport; a similar mechanism is used to initiate the translocation of the cytoplasmic component of nuclear factor of activated T cells to the nucleus 41. Alternatively, SAF may be bound to another factor, which causes SAF to be sequestered in the cytoplasm; the posttranslational modification of the binding partner would then release SAF, allowing it to be transported to the nucleus. Such a mechanism is used to regulate the nuclear localization of nuclear factor κB 42. Because SAF is a homeodomain-like protein, one particularly interesting example of transcription factor translocation involves the Drosophila melanogaster factor Extradenticle (EXD). EXD is a homeodomain transcription factor that mediates cell fate during embryonic development 43. EXD is often found in the cytoplasm; however, at specific stages of development, EXD binds to a second homeodomain protein, Homothorax, which causes the heterodimer to translocate to the nucleus 44 45. Once in the nucleus, the EXD–Homothorax heterodimer binds to adjacent DNA recognition sites in the promoters of target genes and induces their expression. It is possible that SAF is translocated specifically to the nucleus during T cell development by a member of the Meis family, the mammalian Homothorax homologues. Interestingly, there is a consensus Meis recognition site within the S3 region, directly adjacent to the SAF binding site; preliminary biochemical experiments indicate that a nuclear factor in CD4− T cells recognizes the Meis recognition site specifically (Sarafova, S., and G. Siu, unpublished data). It is thus possible that, similar to EXD–Homothorax, Meis is shuttling SAF to the nucleus and binding as a dimer to their respective recognition sites in the CD4 silencer. It is important to note that for many promoters EXD requires its Homothorax partner to mediate element function; thus, should SAF require a partner for nuclear localization, it is likely that it will also require this partner to mediate silencer function. We are currently conducting experiments to address these issues directly.
Acknowledgments
We thank Drs. Joachim Li and Randall Reed for the yeast one-hybrid plasmids; Aneel Aggarwal, Richard Axel, Ronald Liem, Richard Mann, Alessandra Pernis, Christy Rohani, Ben Shykind, and Bob Soliman for helpful discussions and assistance; and Kathryn Calame, Nika Danial, Andrew Henderson, Paul Rothman, and Chris Schindler for comments on the manuscript.
This work was supported by a predoctoral training grant from the Integrated Program to W.W.S. Kim, and grants from the National Institutes of Health (AI34925) and the Irma T. Hirschl–Monique Caulier-Weill Charitable Trust to G. Siu.
Footnotes
1used in this paper: DN, double negative; DP, double positive; EMSA, electrophoretic mobility shift assay; EXD, Extradenticle; GST, glutathione S-transferase; HHTH, helix-helix-turn-helix; HTH, helix-turn-helix; ORF, open reading frame; SAF, silencer-associated factor; SP, single positive; Tc, cytotoxic T cell
References
- Perlmutter R.M., Marth J.D., Ziegler S.F., Garvin A.M., Pawar S., Cooke M.P., Abraham K.M. Specialized protein tyrosine kinase proto-oncogenes in hematopoietic cells. Biochim. Biophys. Acta. 1988;948:245–262. doi: 10.1016/0304-419x(89)90001-2. [DOI] [PubMed] [Google Scholar]
- Fowlkes B.J., Pardoll D. Molecular and cellular events of T cell development. Adv. Immunol. 1989;44:207–264. doi: 10.1016/s0065-2776(08)60643-4. [DOI] [PubMed] [Google Scholar]
- Littman D.R., Davis C.B., Killeen N., Xu H. Signal transduction during T cell development. Adv. Exp. Med. Biol. 1994;365:63–71. doi: 10.1007/978-1-4899-0987-9_7. [DOI] [PubMed] [Google Scholar]
- Siu G., Wurster A.L., Lipsick J.S., Hedrick S.M. Expression of the CD4 gene requires a Myb transcription factor. Mol. Cell. Biol. 1992;12:1592–1604. doi: 10.1128/mcb.12.4.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S., Littman D.R. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol. Cell. Biol. 1993;13:5620–5628. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon P., Giovane A., Wasylyk B., Klatzmann D. Characterization of the human CD4 gene promotertranscription from the CD4 gene core promoter is tissue-specific and is activated by Ets proteins. Proc. Natl. Acad. Sci. USA. 1993;90:7739–7743. doi: 10.1073/pnas.90.16.7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S., Scarborough J.D., Killeen N., Littman D.R. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- Siu G., Wurster A.L., Duncan D.D., Soliman T.M., Hedrick S.M. A transcriptional silencer controls the developmental expression of the CD4 gene. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:3570–3579. doi: 10.1002/j.1460-2075.1994.tb06664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurster A.L., Siu G., Leiden J., Hedrick S.M. Elf-1 binds to a critical element in a second CD4 enhancer. Mol. Cell. Biol. 1994;14:6452–6463. doi: 10.1128/mcb.14.10.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan D.D., Stupakoff A., Hedrick S.M., Marcu K.B., Siu G. A Myc-associated zinc-finger protein (MAZ) binding site is one of four important functional regions in the CD4 promoter. Mol. Cell. Biol. 1995;15:3179–3186. doi: 10.1128/mcb.15.6.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan D.D., Adlam M., Siu G. Asymmetric redundancy in CD4 silencer function. Immunity. 1996;4:301–311. doi: 10.1016/s1074-7613(00)80438-0. [DOI] [PubMed] [Google Scholar]
- Salmon P., Boyer O., Lores P., Jami J., Klatzmann D. Characterization of an intronless CD4 minigene expressed in mature CD4 and CD8 T cells, but not expressed in immature thymocytes. J. Immunol. 1996;156:1873–1879. [PubMed] [Google Scholar]
- Adlam M., Duncan D.D., Ng D.K., Siu G. Positive selection induces CD4 promoter and enhancer function. Int. Immunol. 1997;9:877–887. doi: 10.1093/intimm/9.6.877. [DOI] [PubMed] [Google Scholar]
- Donda A., Schulz M., Burki K., De Libero G., Uematsu Y. Identification and characterization of a human CD4 silencer. Eur. J. Immunol. 1996;26:493–500. doi: 10.1002/eji.1830260232. [DOI] [PubMed] [Google Scholar]
- Uematsu Y., Donda A., De Libero G. Thymocytes control the CD4 gene differently from mature T lymphocytes. Int. Immunol. 1997;9:179–187. doi: 10.1093/intimm/9.1.179. [DOI] [PubMed] [Google Scholar]
- Fink P.J., Bevan M.J. Positive selection of thymocytes. Adv. Immunol. 1995;59:99–133. doi: 10.1016/s0065-2776(08)60630-6. [DOI] [PubMed] [Google Scholar]
- Nossal G.J.V. Negative selection of lymphocytes. Cell. 1994;76:229–239. doi: 10.1016/0092-8674(94)90331-x. [DOI] [PubMed] [Google Scholar]
- Kim H.K., Siu G. The Notch pathway intermediate HES-1 silences CD4 gene expression. Mol. Cell. Biol. 1998;18:7166–7175. doi: 10.1128/mcb.18.12.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R.D., Bender T.P., Siu G. c-Myb is essential for early T cell development. Genes Dev. 1999;13:1073–1078. doi: 10.1101/gad.13.9.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye C., Remondelli P., Karin M., Elledge S. Isolation of a cDNA encoding a metal response element binding protein using a novel expression cloning procedurethe one hybrid system. DNA Cell Biol. 1994;13:731–742. doi: 10.1089/dna.1994.13.731. [DOI] [PubMed] [Google Scholar]
- Wang C.Y., Petryniak B., Thompson C.B., Kaelin W.G., Leiden J.M. Regulation of the Ets-related transcription factor Elf-1 by binding to the retinoblastoma protein. Science. 1993;260:1330–1335. doi: 10.1126/science.8493578. [DOI] [PubMed] [Google Scholar]
- Li J.J., Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- Wang M.M., Reed R.R. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- Durfee T., Becherer K., Chen P.L., Yeh S.H., Yang Y., Kilburn A.E., Lee W.H., Elledge S.J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; New York, NY: 1995. [Google Scholar]
- Giese K., Amsterdam A., Grosschedl R. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 1991;5:2567–2578. doi: 10.1101/gad.5.12b.2567. [DOI] [PubMed] [Google Scholar]
- Travis A., Amsterdam A., Belanger C., Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- Waterman M.L., Jones K.A. Purification of TCF-1α, a T-cell-specific transcription factor that activates the T-cell receptor Cα gene enhancer in a context-dependent manner. New Biol. 1990;2:621–636. [PubMed] [Google Scholar]
- Waterman M.L., Fischer W.H., Jones K.A. A thymus-specific member of the HMG protein family regulates the human T cell receptor Cα enhancer. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- Karim F.D., Urness L.D., Thummel C.S., Klemsz M.J., McKercher S.R., Celada A., Van Beveran C., Maki R.A., Gunther C.V., Nye J.A., Graves B.J. The ETS-domaina new DNA-binding motif that recognizes a purine-rich core DNA sequence. Genes Dev. 1990;4:1451–1453. doi: 10.1101/gad.4.9.1451. [DOI] [PubMed] [Google Scholar]
- Scott M.P., Tamkun J.W., Hartzell G.W., III. The structure and function of the homeodomain. Biochim. Biophys. Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Wintjens R., Rooman M. Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J. Mol. Biol. 1996;262:294–313. doi: 10.1006/jmbi.1996.0514. [DOI] [PubMed] [Google Scholar]
- Burglin T. A comprehensive classification of homeobox genes. In: Duboule D., editor. Guidebook to the Homeobox Genes. Oxford University Press; Oxford, UK: 1994. pp. 27–71. [Google Scholar]
- Desplan C., Theis J., O'Farrell P.H. The sequence specificity of homeodomain-DNA interaction. Cell. 1988;54:1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T., Levine M. Divergent homeobox proteins recognize similar DNA sequences in Drosophila . Nature. 1988;332:858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Hirsch J.A., Aggarwal A.K. Structure of the even-skipped homeodomain complexed to AT-rich DNAnew perspectives on homeodomain specificity. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:6280–6291. doi: 10.1002/j.1460-2075.1995.tb00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroush Z., Finley R.L., Jr., Kidd T., Wainwright S.M., Ingham P.W., Brent R., Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Keleher C.A., Redd M.J., Schultz J., Carlson M., Johnson A.D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Shah P.C., Bertolino E., Singh H. Using altered specificity Oct-1 and Oct-2 mutants to analyze the regulation of immunoglobulin gene transcription. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:7105–7117. doi: 10.1093/emboj/16.23.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillemans N., Tewari R., Lindeboom F., Rottier R., de Wit T., Wijgerde M., Grosveld F., Philipsen S. Altered DNA-binding specificity mutants of EKLF and Sp1 show that EKLF is an activator of the β-globin locus control region in vivo. Genes Dev. 1998;12:2863–2873. doi: 10.1101/gad.12.18.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A. Signaling mechanisms in T cells. Crit. Rev. Immunol. 1991;10:495–519. [PubMed] [Google Scholar]
- Grilli M., Chiu J.-S., Lenardo M.J. NF-κB and relparticipants in a multiform transcriptional regulatory system. Int. Rev. Cytol. 1991;143:1–63. doi: 10.1016/s0074-7696(08)61873-2. [DOI] [PubMed] [Google Scholar]
- Mann R.S., Chan S.K. Extra specificity from Extradenticlethe partnership between HOX and PBX/EXD homeodomain proteins Trends Genet. 12 1996. 258 262(See published erratum 12:328). [DOI] [PubMed] [Google Scholar]
- Rieckhof G., Casares F., Ryoo H.D., Abu-Shaar M., Mann R.S. Nuclear translocation of Extradenticle requires homothorax, which encodes an Extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Pai C.-Y., Kuo T.-S., Jaw T.J., Kurant E., Chen C.-T., Bessarab D.A., Salzberg A., Sun Y.H. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, Extradenticle, and suppresses eye development in Drosophila . Genes Dev. 1998;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]