Abstract
Histocompatibility leukocyte antigen (HLA)-A2 is used as a restricting element to present several melanoma-associated antigen (MAA)-derived peptides to cytotoxic T lymphocytes (CTLs). HLA-A2 antigen is selectively lost in primary melanoma lesions and more frequently in metastases. Only scanty information is available about the molecular mechanisms underlying this abnormality, in spite of its potentially negative impact on the clinical course of the disease and on the outcome of T cell–based immunotherapy. Therefore, in this study we have shown that the selective HLA-A2 antigen loss in melanoma cells 624MEL28 is caused by a splicing defect of HLA-A2 pre-mRNA because of a base substitution at the 5′ splice donor site of intron 2 of the HLA-A2 gene. As a result, HLA-A2 transcripts are spliced to two aberrant forms, one with exon 2 skipping and the other with intron 2 retention. The latter is not translated because of an early premature stop codon in the retained intron. In contrast, the transcript with exon 2 skipping is translated to a truncated HLA-A2 heavy chain without the α1 domain. Such a polypeptide is synthesized in vitro but is not detectable in cells, probably because of the low steady state level of the corresponding mRNA and the low translation efficiency. These results indicate that a single mutational event in an HLA class I gene is sufficient for loss of the corresponding allele. This may account, at least in part, for the high frequency of selective HLA class I allele loss in melanoma cells. Our conclusion emphasizes the need to implement active specific immunotherapy with a combination of peptides presented by various HLA class I alleles. This strategy may counteract the ability of melanoma cells with selective HLA class I allele loss to escape from immune recognition.
Keywords: histocompatibility leukocyte antigen class I, splicing defect, truncated heavy chain, melanoma
Immunohistochemical staining with mAbs has convincingly documented abnormalities in HLA class I antigen expression in primary melanoma lesions and more frequently in metastases 1. These defects range from total HLA class I antigen loss to selective loss of one of the HLA class I allospecificities encoded within a melanoma cell 2 3 4 5 6. The clinical significance of HLA class I antigen downregulation in melanoma cells is suggested by its association with a poor clinical course of the disease 7 and by its negative impact on the outcome of increasingly applied T cell–based immunotherapy 8 9. These findings have stimulated interest in the characterization of the molecular lesions underlying abnormalities in HLA class I antigen expression by melanoma cells and their effects on the interactions of melanoma cells with immune cells. Information derived from these studies contributes to our understanding of the molecular mechanism(s) used by melanoma cells to escape from immune surveillance and may eventually suggest strategies to correct these defects.
The molecular lesions causing total HLA class I antigen loss have been characterized in several melanoma cell lines 6 10 11 12. Mutations have been identified in β2-microglobulin (β2-μ)1 gene(s) which inhibit its translation in most cases and its transcription in rare cases 6 10 11 12. These mutations, which appear to represent an early event in the progression of the malignant phenotype 12, range from single base substitutions to partial gene deletion 10 11 12. Total HLA class I antigen loss by melanoma cells has marked effects on their in vitro interactions with cytotoxic lymphocytes. It causes resistance to lysis by melanoma-associated antigen (MAA)-specific, HLA class I antigen–restricted CTLs 6 12, and enhances their susceptibility to lysis by NK cells 13.
The molecular mechanism(s) underlying the spontaneous selective loss of an HLA class I allele by melanoma cells has not yet been investigated. To the best of our knowledge, the only available information in this regard derives from the analysis of the melanoma cell line SK-MEL-29.1.22, which had selectively lost HLA-A2 antigens in vitro after γ-irradiation and selection with MAA-specific, HLA-A2–restricted CTLs 14 15. The functional significance of the selective HLA class I allele loss has been investigated in a few cases. Loss of an HLA class I allele causes in vitro resistance of melanoma cells to lysis by MAA-specific CTLs, which use the lost allele as a restricting element 14 16 17.
To broaden our knowledge of the molecular mechanisms underlying the selective HLA class I allele loss by melanoma cells, in this study we have characterized the molecular lesion responsible for selective HLA-A2 antigen loss in the melanoma cell line 624MEL28. We have selected the HLA-A2 allele for our studies, since this allele has the highest frequency in patients with melanoma as well as in the control population, and has been found to be selectively lost in primary melanoma lesions and more frequently in metastases 3 4. Furthermore, this allele has been found to present the largest number of MAA-derived peptides to CTLs 18 19 and to be the restricting element for a large array of MAA-derived peptides that have been effectively used to implement active specific immunotherapy in patients with melanoma 20 21.
Materials and Methods
Cytokine.
Recombinant human IFN-γ was obtained from Hoffmann-La Roche, Inc.
Cells.
The human melanoma cell lines 624MEL28 and 624MEL38 17 and the B lymphoid cell line Raji were cultured at 37°C in a 5% CO2 atmosphere in RPMI 1640 medium (GIBCO BRL) supplemented with 10% Serum plus (Hazelton Biologics, Inc.).
mAbs and Conventional Antisera.
The mAb W6/32, which recognizes a monomorphic determinant expressed on β2-μ–associated HLA-A,B,C heavy chains; the mAb LGIII-220.6, which recognizes a determinant preferentially expressed on β2-μ–associated HLA-A heavy chains; the mAb H2-89.1, which recognizes a determinant preferentially expressed on β2-μ–associated HLA-B heavy chains; the anti–HLA-A2,A28 mAbs CR11-351, HO-4, and HO-5; the anti–HLA-A2,B17 mAbs HO-2 and MA2.1; the anti–HLA-B7 cross-reacting group mAb KS4; and the mAb HC-A2, which recognizes a determinant restricted to β2-μ–free HLA-A heavy chains, were developed and characterized as described elsewhere 22 23 24 25 26 27. The rabbit anti–β2-μ–free HLA class I heavy chain serum R5996-4 28 and the rabbit anti-HLA class I heavy chain cytoplasmic tail serum were obtained from Dr. N. Tanigaki (Roswell Park Cancer Institute) and Dr. H.L. Ploegh (Harvard Medical School, Boston, MA), respectively. Purified rabbit anti–mouse Ig antibodies and FITC-conjugated goat anti–mouse Ig antibodies (FITC-GAM) were purchased from Jackson ImmunoResearch Laboratories.
Synthetic Oligonucleotide Primers and Probes, and HLA-A2 Gene.
The HLA class I–specific synthetic oligonucleotide primers and probes listed in Table were synthesized on a BioSearch Cyclone DNA synthesizer (MilliGen/BioSearch), with the exception of those obtained from Dr. S.Y. Yang (Memorial Sloan-Kettering Cancer Center, New York). Oligonucleotide probes were radiolabeled with [γ-32P]ATP (>5,000 Ci/mmol; Nycomed Amersham plc) in the presence of T4 polynucleotide kinase 29.
Table 1.
HLA Class I–specific Oligonucleotide Primers and Probes
| Synthetic oligonucleotide | Sequence (5′–3′) | Exon | Location position | Specificity |
|---|---|---|---|---|
| 5′ primers | ||||
| 5pEh | CCCGAAGGCGGTGTATGGAT | −237–−217 | HLA class I | |
| 5pUT | CAGATTCTCCCCAGACGCCG | −24–−3 | HLA class I | |
| 5pE1A2 | TCCTGCTACTCTCGGGGGCT | 1 | 22–41 | HLA-A2 |
| AP31 | CGTCCCCAGGCTCTCACTCCAT | 2 | −9–13 | HLA class I |
| 5pE2A | GACGCCGCGAGCCAGAGGAT | 2 | 114–133 | HLA-A |
| 5pE3A2 | GCGGACATGGCATCTCAGAC | 3 | 135–155 | HLA-A2,A28 |
| 5P2 | GCGCGTCGACCCCAGACGCCGAG | |||
| GATGGCC | 1 | −13–6 | HLA class I | |
| 3′ primers | ||||
| AP2 | TCACTTTCCGTGCTCCCC | 2 | 186–203 | HLA-A2 |
| 3pE3A2 | CTCCCACTTGTGCTTGGTGG | 3 | 154–173 | HLA-A2,A28 |
| 3pE3 | TGCAGCGTCTCCTTCCCGTT | 3 | 248–267 | HLA class I |
| 3P2 | CCGCAAGCTTTCTCAGTCCCA | |||
| CACAAGGCAGCTGTC | 8 | 6–35 | HLA class I | |
| 3pE8A | AGTCACAAAGGGAAGGGCAGG | 8 | 46–66 | HLA class I |
| Probes | ||||
| 95V | CACACCGTCCAGAGG | 3 | 7–21 | HLA-A2,A69 |
| 161D | CTGGATGGCACGTGC | 3 | 207–222 | HLA-A3 |
The plasmid RSV.5neo-HLA-A2.1 was obtained from Dr. P. Cresswell (Yale University School of Medicine, New Haven, CT). The HLA-A2 cDNA for synthesis of RNA probe was constructed by cloning a 500-bp EagI/PstI fragment from plasmid RSV.5neo-HLA-A2.1 into pCR-Script™ SK(+) cloning vector in an antisense orientation. The EagI/PstI fragment contains most of the exon 2 and exon 3 of the HLA-A2 cDNA. The RNA probe was synthesized by in vitro transcription using MAXIscript™ T7 kit (Ambion, Inc.) with [α-32P]UTP (800 Ci/mmol; Amersham Pharmacia Biotech) following the manufacturer's instructions. The synthesized RNA probe was purified through a 5% polyacrylamide gel and eluted following the procedure recommended by the manufacturer. The HLA-A2 cDNA containing the whole coding region and the HLA-A2 cDNA lacking exon 2 to be used for in vitro translation were amplified by PCR from plasmid RSV5neo-HLA-A2.1 and melanoma 624MEL28 cells, respectively, using the primers 5P2 and 3P2. PCR products were cloned into SalI/HindIII sites of pCR-Script™ SK(+) cloning vector.
The plasmid pHLA-A2, which carries the entire HLA-A2 gene including its regulatory sequences, and the neomycin resistance gene 30 were obtained from Dr. J.L. Strominger (Harvard University, Cambridge, MA).
Indirect Immunofluorescence.
Indirect immunofluorescence (IIF) staining was performed as described elsewhere 4. After staining, cells were analyzed by cytofluorometry on a FACS® analyzer (Becton Dickinson). Results are expressed as log fluorescence intensity.
Immunochemical Methods.
Radiolabeling of cells, indirect immunoprecipitation, SDS-PAGE under reducing conditions, isoelectric focusing (IEF), and fluorography were performed as described elsewhere 10 31.
Western blotting was performed as described elsewhere 32, with the modification that antibodies bound to proteins transferred to filters were detected using the ECL™ Western blotting detection reagents kit (Amersham Pharmacia Biotech) following the manufacturer's instructions.
Transfection of Cells with a Wild-type HLA-A2 Gene.
Plasmid pHLA-A2 was transfected into cells using the electroporation method as described 10. After transfection, cells were cultured for 2 wk in medium supplemented with G418-sulfate at the final concentration of 0.4 mg/ml. Cell colonies were picked up and expanded in medium supplemented with G418-sulfate at the final concentration of 0.2 mg/ml.
Amplification by PCR of cDNA and Genomic DNA.
cDNA was reverse transcribed from total RNA as described 33. Genomic DNA was prepared from cells using the cell lysis method 34. Amplification of cDNA and genomic DNA was performed as described elsewhere 15 using a DNA Thermal Cycler (Perkin-Elmer Cetus).
DNA Hybridization Analysis.
PCR products were size fractionated, transferred to nylon membranes, and hybridized with probes as described elsewhere 15.
DNA Sequencing.
DNA sequencing was performed as described elsewhere 35, using the Sequenase version 2.0 kit (United States Biochemical Corp.). PCR products were directly sequenced using products recovered from an agarose gel using the Geneclean II kit (Bio 101 Inc.). After denaturation at 98°C for 10 min in the presence of 10% DMSO, PCR products were cooled in ethanol/dry ice and then sequenced.
RNase Protection Assay.
The RNase protection assay was performed using the RPA II™ Ribonuclease protection assay kit (Ambion, Inc.) following the manufacturer's instructions, except for increasing the hybridization temperature to 45°C.
In Vitro Translation of HLA-A2 mRNA.
HLA-A2 mRNA was translated in vitro using the TNT® T7 coupled reticulocyte lysate system (Promega Corp.) following the manufacturer's instructions. The translation products were analyzed by SDS-PAGE under reducing conditions.
Results
Lack of HLA-A2 Antigen Cell Surface Expression by 624MEL28 Cells.
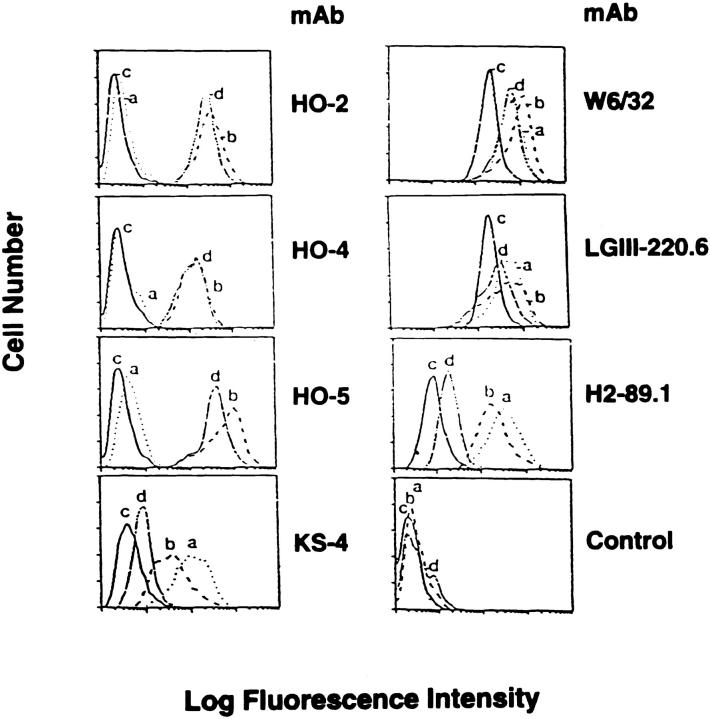
The clone 624MEL28 was isolated by limiting dilution from the melanoma cell line 624MEL (HLA-A2, -A3, -B7, -B14, -Cw7, -Cw8), which had been established from a metastatic lesion 17. This cell line was subcloned at the 35th passage, since the broad profile of the IIF staining with mAbs suggested marked heterogeneity in HLA-A2 antigen expression. The clone 624MEL28 was not stained in IIF by anti–HLA-A2,B17 mAb HO-2 and anti–HLA-A2,A28 mAbs HO-4 and HO-5, all of which stained the autologous HLA-A2 antigen–bearing clone 624MEL38 (Fig. 1). The lack of staining of the 624MEL28 cells by anti–HLA-A2 mAb is not due to loss of an antigenic determinant, since the three mAbs used recognize distinct determinants on HLA-A2 heavy chains (Ferrone, S., unpublished results). The HLA-A2 antigen loss is selective, since, like the autologous clone 624MEL38, 624MEL28 cells were stained strongly by anti–HLA-A,B,C mAb W6/32 and by anti–HLA-A mAb LGIII-220.6, and weakly by anti–HLA-B mAb H2-89.1 and anti–HLA-B7 cross-reacting group mAb KS4 (Fig. 1). It is noteworthy that the intensity of staining by the latter six mAbs of the clone 624MEL28 is comparable to that of the autologous clone 624MEL38. Furthermore, IFN-γ did not induce HLA-A2 antigen expression by 624MEL28 cells, since they were not stained by anti–HLA-A2 mAbs after a 72-h incubation at 37°C with IFN-γ (100 U/ml; Fig. 1). These findings indicate that 624MEL28 cells have selectively lost HLA-A2 antigens.
Figure 1.
Selective lack of cell surface HLA-A2 antigen expression by 624MEL28 cells. 624MEL28 (a) and 624MEL38 (b) cells were incubated at 37°C for 72 h with IFN-γ (100 U/ml). 624MEL28 (c) and 624MEL38 (d) cells incubated under the same experimental conditions but without IFN-γ were used as controls. At the end of the incubation, cells were harvested, washed with PBS, and incubated with anti–HLA-A2,B17 mAb HO-2, anti–HLA-A2,A28 mAbs HO-4 and HO-5, anti-HLA class I mAb W6/32, anti–HLA-A mAb LGIII-220.6, anti–HLA-B mAb H2-89.1, and anti–HLA-B7 cross-reacting group mAb KS4. After washings with PBS, cells were stained with FITC-GAM. Fluorescence intensity was determined on a FACS® analyzer (Becton Dickinson). Background was determined by incubating cells with FITC-GAM alone (control).
Lack of Wild-type HLA-A2 Heavy Chain Synthesis by 624MEL28 Cells.
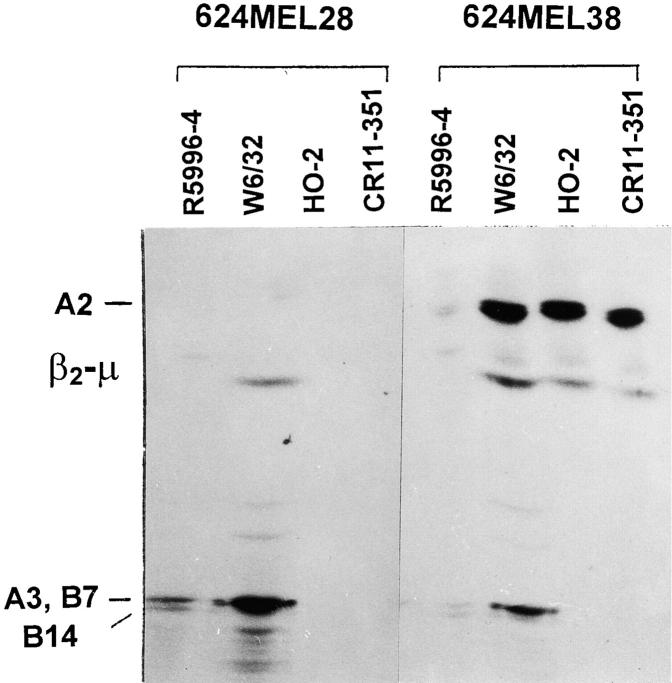
624MEL28 cells used as a source of HLA class I antigens for immunoprecipitation experiments were treated with IFN-γ in order to increase the level of HLA class I antigen expression. No component was detected by IEF analysis in the immunoprecipitates with anti–HLA-A2, A28 mAb CR11-351 and with anti–HLA-A2,B17 mAb MA2.1 from intrinsically radiolabeled 624MEL28 cells (Fig. 2). Furthermore, HLA-A2 heavy chains were not detected by IEF in the immunoprecipitates with mAb W6/32 and with rabbit antiserum R5996-4. However, the latter immunoprecipitates contain HLA-A3, -B7, and -B14 heavy chains. These immunochemical findings corroborate the results of binding assays and indicate that 624MEL28 cells do not synthesize wild-type HLA-A2 heavy chains.
Figure 2.
Lack of HLA-A2 heavy chain synthesis by 624MEL28 cells. 624MEL28 and 624MEL38 cells were incubated at 37°C for 72 h with IFN-γ (100 U/ml). Cells were metabolically radiolabeled with [32S]methionine and lysed with 1% NP-40. After indirect immunoprecipitation with anti–β2-μ–free HLA class I heavy chain rabbit serum R5996-4, anti-HLA class I mAb W6/32, anti–HLA-A2,B17 mAb HO-2, and anti–HLA-A2,A28 mAb CR11-351, antigens were analyzed by IEF on a slab gel apparatus. Gels were then processed for fluorography.
Induction of HLA-A2 Antigen Expression by 624MEL28 Cells after Transfection with a Wild-type HLA-A2 Gene.
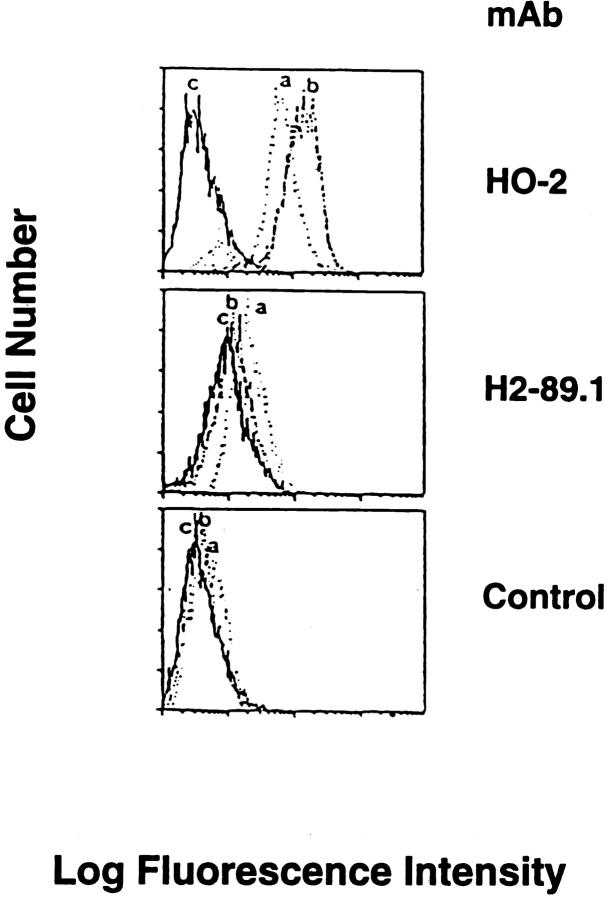
To investigate whether HLA-A2 antigen expression by 624MEL28 cells could be reconstituted by transferring a wild-type HLA-A2 gene, cells were transfected with a plasmid containing both a wild-type HLA-A2 gene and a neomycin resistance gene. Two clones selected in medium supplemented with G418 were both stained in IIF by anti–HLA-A2,B17 mAb HO-2 (Fig. 3). The staining intensity was increased when cells were incubated for 72 h at 37°C with IFN-γ (100 U/ml; data not shown). These results indicate that the lack of HLA-A2 antigen expression by 624MEL28 cells is caused by structural abnormality(ies) in the HLA-A2 gene.
Figure 3.
Induction of HLA-A2 antigen expression by 624MEL28 cells after transfection with a wild-type HLA-A2 gene. HLA-A2 gene–transfected 624MEL28 clones 2 (a) and 3 (b) were incubated with anti–HLA-A2,B17 mAb HO-2. After washings with PBS, cells were stained with FITC-GAM. Fluorescence intensity was determined on a FACS® analyzer (Becton Dickinson). Nonspecific fluorescence was determined by incubating cells with FITC-GAM alone. 624MEL28 cells that were not transfected with HLA-A2 gene (c) were used as an HLA-A2–negative control. The anti–HLA-B mAb H2-89.1 was used for comparison.
Detection of Two Alternative Forms of HLA-A2 mRNA in 624MEL28 Cells.
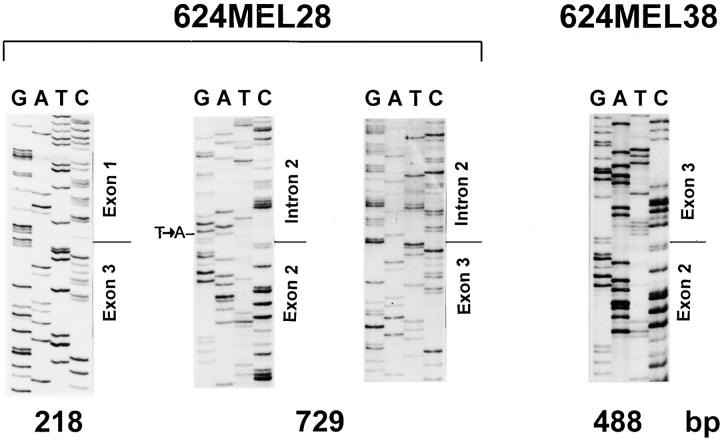
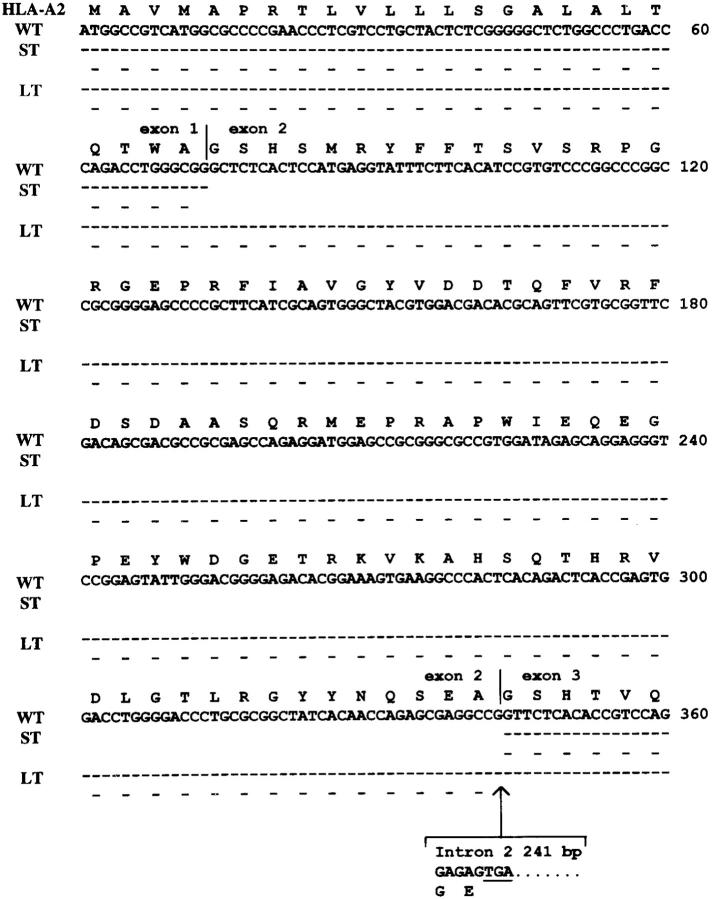
Reverse transcription (RT)-PCR amplification of the mRNA corresponding to a region spanning from the middle of exon 1 to exon 3 of the HLA-A2 gene yielded a 218- and a 729-bp product from 624MEL28 cells. The latter product is 241 bp larger than the one amplified from 624MEL38 cells with the expected size of 488 bp, whereas the former product is 270 bp smaller. Both RT-PCR products are HLA-A2 specific, since they hybridized with the HLA-A2,A69–specific probe 95V (Fig. 4 A). These results indicate that there are two forms of the HLA-A2 mRNA in 624MEL28 cells, one with a 270-base deletion and the other with a 241-base fragment inclusion. The size of deleted and inserted fragments is compatible with that of exon 2 and intron 2 of the HLA-A2 gene, respectively, suggesting the lack of the sequence corresponding to exon 2 or insertion of intron 2 in HLA-A2 mRNA. These possibilities were corroborated by the generation of a 571-bp product, but not of a 330-bp product when cDNA corresponding to exons 2 and 3 of the HLA-A2 gene was isolated from 624MEL28 cells and amplified by PCR (Fig. 4 B). Exon 2 skipping and insertion of intron 2 in the two aberrant forms of HLA-A2 mRNA identified in 624MEL28 cells were conclusively proven by DNA sequencing of the 729-, 571-, and 218-bp RT-PCR products. Only the sequences of exons 1 and 3 of the HLA-A2 gene were found in the 218-bp RT-PCR product (Fig. 5). The sequence of intron 2 of the HLA-A2 gene was found in both the 729- and the 571-bp RT-PCR products. It is noteworthy that the sequence of intron 1 was not detected in the 729-bp RT-PCR product, indicating that the latter was derived from the mature HLA-A2 mRNA with intron 2 retention. A T to A base substitution was found at position 2 in intron 2. This mutation results in a splicing defect, since the U at this position in pre-mRNA is required for spliceosome recognition for pre-mRNA splicing. As shown in Fig. 6, the intron 2 retention causes a reading frameshift, which introduces the stop codon TGA at position 6–8 of the unspliced intron 2. Thus, the synthesis of the HLA-A2 polypeptide encoded by the large HLA-A2 transcript is blocked. In contrast, the small HLA-A2 transcript is in frame in spite of the exon 2 skipping. Therefore, a truncated HLA-A2 heavy chain lacking the α1 domain is expected to be synthesized by 624MEL28 cells. No additional mutation was found in the remaining sequence of HLA-A2 cDNA.
Figure 4.
Detection of two alternative forms of HLA-A2 cDNA amplified by RT-PCR from 624MEL28 cells. HLA class I heavy chain cDNA was synthesized from RNA isolated from 624MEL28 cells by reverse transcriptase reaction using HLA class I–specific 3′ primer 3pE3. HLA-A2 cDNA was amplified by PCR using the two pairs of HLA-A2–specific primer (A and B). HLA-A locus cDNA was amplified using an HLA-A locus–specific primer pair (C). The regions from which the primers are derived are indicated below each panel. The corresponding exon regions in the cDNA are indicated by numbers. RT-PCR products were subjected to Southern blotting and hybridized with HLA-A2,A69–specific probe 95V or HLA-A3–specific probe 161D as indicated. DNA amplified from HLA-A2–positive 624MEL38 cells and from HLA-A2–negative Raji cells were used as HLA-A2–positive and –negative controls, respectively. DNA amplified from HLA-A3–negative SKMEL-29.1 cells was used as a negative control for HLA-A3.
Figure 5.
Identification of exon 2 skipping and intron 2 retention in HLA-A2 cDNA from 624MEL28 cells. The 218- and 729-bp RT-PCR products amplified from RNA isolated from 624MEL28 cells using HLA-A2–specific primers 5pE1A2 and 3pE3A2 were purified from agarose gel using a Geneclean II (Bio 101 Inc.) kit. RT-PCR products were then sequenced using a Sequenase 2.0 kit (United States Biochemical Corp.). The autoradiography of the sequencing gels for these RT-PCR products is shown as indicated. The 488-bp RT-PCR products amplified from 624MEL38 cells were sequenced using the same primers as a normal control.
Figure 6.
Comparison of nucleotide and deduced amino acid sequences of the two forms of HLA-A2 cDNA from 624MEL28 cells with those of the wild-type HLA-A2.1 cDNA. The coding region and deduced amino acid sequences of HLA-A2 cDNA corresponding to exon 1, exon 2, and part of exon 3 of the wild-type HLA-A2 cDNA (WT) and of the HLA-A2 cDNA with exon 2 skipping (ST) and intron 2 retention (LT) isolated from 624MEL28 cells are compared. Dashes and blanks indicate sequence identity and deletion, respectively. Intron 2 retention is indicated by a large arrowhead. The stop codon is underlined.
Mutation at the 5′ Splice Donor Site in Intron 2 of the HLA-A2 Gene in 624MEL28 Cells.
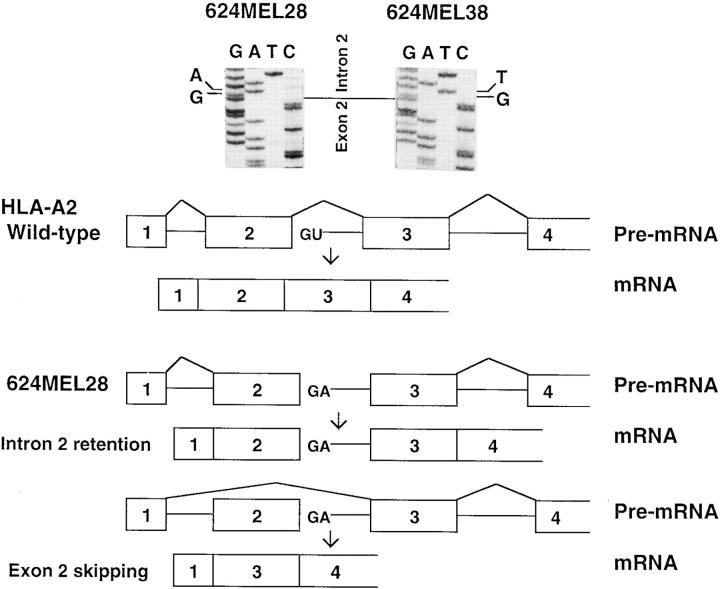
To define the molecular lesion responsible for exon 2 skipping in the HLA-A2 mRNA, the region from exon 1 to exon 3 of the HLA-A2 gene was amplified from genomic DNA of 624MEL28 cells using HLA-A2–specific primers. Only the PCR products with the expected size of 980 bp were amplified from 624MEL28 cells as well as from the autologous 624MEL38 cells, which express HLA-A2 antigens (data not shown). These results rule out the presence of additional HLA-A2 gene copies with exon 2 deletion in 624MEL28 cells. Furthermore, DNA sequencing of the four clones containing the 980-bp PCR product identified a T to A substitution at position 2 of the 5′ splice donor site in intron 2 (Fig. 7). This mutation is the same mutation found in the large mutated HLA-A2 cDNA. No mutation was found at the 3′ acceptor site in introns 1 and 2 of the HLA-A2 gene. Therefore, the mutation at position 2 of intron 2, which inactivates the 5′ splice donor site, is responsible for both exon 2 skipping and intron 2 retention in the HLA-A2 mRNA in 624MEL28 cells.
Figure 7.
Identification of a base substitution in intron 2 of the HLA-A2 gene and alternative splicing of HLA-A2 pre-mRNA in 624MEL28 cells. Exon 2 and intron 2 segments of the HLA-A2 gene were amplified by PCR from genomic DNA of 624MEL28 cells using the HLA-A2–specific primer pair 5pE1A2 and 3pE3. HLA-A2 DNA amplified from 624MEL38 cells was used as a control. DNA was sequenced using a Sequenase 2.0 kit. The exon 2–intron 2 boundary and the first two nucleotides of intron 2 are indicated in the DNA sequencing gel (top). The splicing of HLA-A2 pre-mRNA in 624MEL28 cells is compared diagrammatically with that of wild-type HLA-A2 pre-mRNA (bottom).
Marked Reduction of the Steady State Level of the Mutated HLA-A2 mRNA in 624MEL28 Cells.
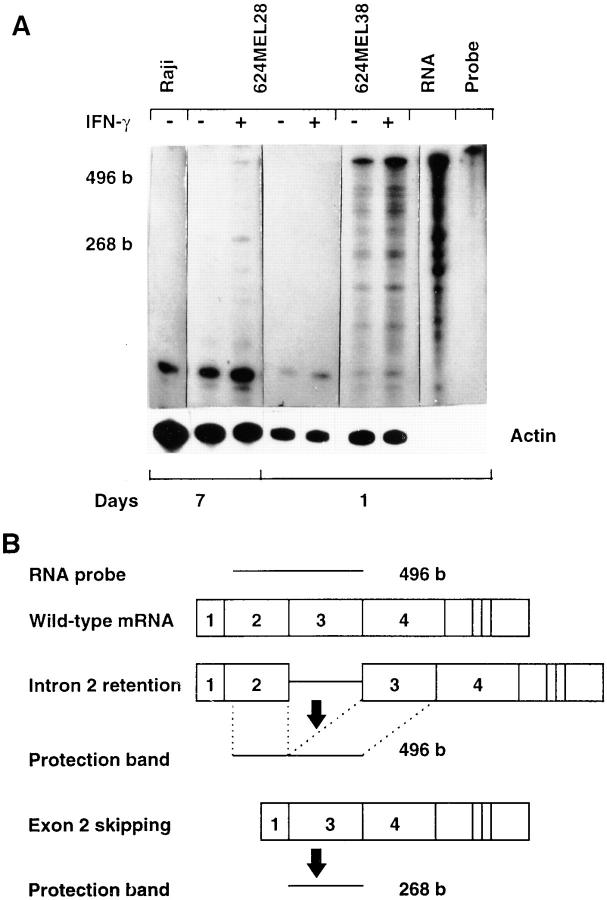
Although the HLA-A2 mRNA with exon 2 skipping in 624MEL28 cells was expected to synthesize a truncated polypeptide, such a polypeptide was not detected by SDS-PAGE analysis of antigens immunoprecipitated from intrinsically labeled 624MEL28 cells by the anti–β2-μ–free HLA class I heavy chain xenoantiserum R5996-4 or by an anti-HLA class I heavy chain cytoplasmic tail xenoantiserum. Furthermore, the truncated HLA-A2 polypeptide was not detected by testing a 624MEL28 cell extract with the two xenoantisera in Western blotting. To determine whether these results reflect a dramatic decrease in the level of the HLA-A2 mRNA lacking the exon 2 sequence because of the splicing defect, its steady state mRNA level in 624MEL28 cells was measured using the RNase protection assay. A 32P-labeled antisense HLA-A2 RNA complementary to 496 bp of the region spanning most of exons 2 and 3 of the HLA-A2 gene was used as a probe. As shown in Fig. 8, digestion with RNase of this probe protected by mRNA isolated from 624MEL28 cells yielded a 268- and a 496-base fragment. The 268-base fragment, which has the same size as exon 3 in the probe, is derived from the probe protected from RNase digestion by the HLA-A2 mRNA lacking the exon 2 sequence. The protection of the probe by mRNA from 624MEL38 cells did not yield this fragment. The 496-base fragment is likely to derive from the probe protected by the HLA-A2 mRNA with unspliced intron 2, since a fragment with the same size was generated from the probe protected by the RNA synthesized in vitro by an HLA-A2 genomic DNA fragment containing exon 2, intron 2, and exon 3. This fragment is unlikely to derive from protection by a wild-type HLA-A2 mRNA from 624MEL28 cells, since such an mRNA was not detected by RT-PCR in these cells. The level of the 268- and 496-base fragments protected by mRNA isolated from 624MEL28 cells is very low, at least 20-fold less than that of the 496-base fragment derived from the probe protected by the wild-type HLA-A2 mRNA isolated from control 624MEL38 cells. These results suggest that the steady state level of HLA-A2 mRNA with exon 2 skipping is too low to produce a detectable level of truncated HLA-A2 heavy chain.
Figure 8.
Reduced steady state level of the aberrant HLA-A2 mRNA in 624MEL28 melanoma cells. 624MEL28 cells were incubated at 37°C for 72 h with IFN-γ (100 U/ml). Control cells were incubated under the same experimental conditions, but without IFN-γ. At the end of the incubation, total cellular RNA was extracted from cells and hybridized with a 32P-labeled HLA-A2 riboprobe (496 bases) and with a 32P-labeled human β-actin riboprobe (250 bases). After hybridization, RNA was digested by RNase, and the generated fragments were size fractionated by 5% PAGE. Gels were autoradiographed for 1 or 7 d. RNA from 624MEL38 and Raji cells were used as HLA-A2–positive and –negative controls, respectively. (A) RNA synthesized in vitro by an HLA-A2 genomic fragment containing exon 2, intron 2, and exon 3 (RNA) was used to identify the fragments generated by RNase digestion from the probe protected by the HLA-A2 mRNA containing intron 2. (B) The fragments generated by RNase digestion from the probe protected by a wild-type HLA-A2 mRNA, by HLA-A2 mRNA with exon 2 skipping, and by HLA-A2 mRNA with intron 2 retention are shown diagrammatically.
In Vitro Translation of HLA-A2 mRNA with Deletion of the Segment Corresponding to Exon 2.
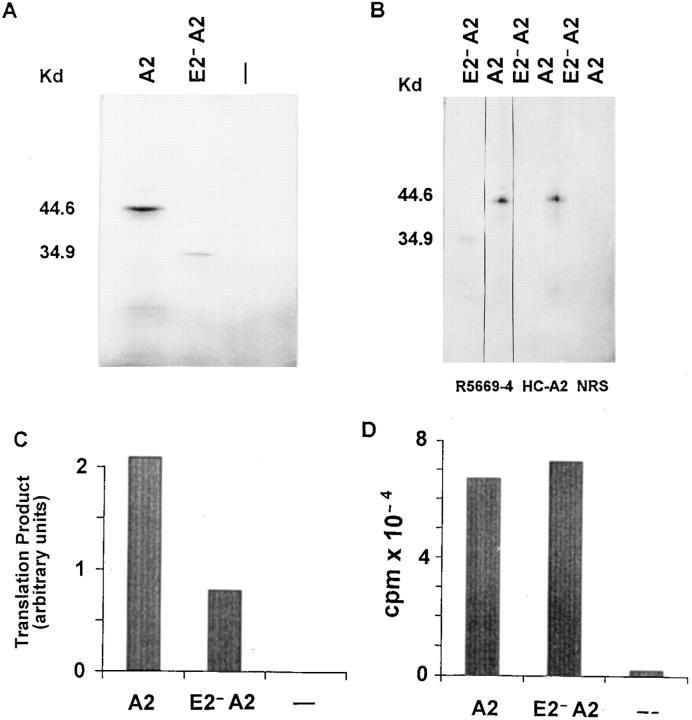
Translation experiments using a transcription/translation-coupled system tested whether the HLA-A2 mRNA lacking the exon 2 sequence can synthesize proteins in vitro. To this end, the mutated HLA-A2 cDNA containing the whole coding sequence, except the exon 2–encoded region, was isolated from 624MEL28 cells and used for in vitro mRNA synthesis. A 35-kD polypeptide, ∼10 kD smaller than the wild-type, was synthesized by mutated HLA-A2 mRNA (Fig. 9 A). The loss of the α1 domain, which is encoded by exon 2, accounts for the reduction in size of the synthesized polypeptide. This interpretation is consistent with the results of the SDS-PAGE analysis of the translation products immunoprecipitated by three xenoantibodies. The truncated polypeptide was not immunoprecipitated by mAb HC-A2, which recognizes a determinant expressed on the α1 domain of β2-μ–free HLA class I heavy chains 36, but was immunoprecipitated by the anti–β2-μ–free HLA class I heavy chain xenoantiserum R5996-4 (Fig. 9 B) and by the anti-HLA class I heavy chain cytoplasmic tail xenoantiserum (data not shown). These results indicate that the HLA-A2 mRNA with exon 2 skipping synthesizes in vitro a truncated HLA class I heavy chain lacking the α1 domain. It is noteworthy that the intensity of the 35-kD translation product is about twice as low as that of the 44.6-kD translation product (Fig. 9A and Fig. C), although the extent of RNA synthesis by the mutated and the wild-type HLA-A2 cDNA is similar, as measured by the level of [32P]UTP incorporation (Fig. 9 D). Therefore, the efficiency of in vitro translation of the HLA-A2 mRNA with exon 2 skipping is lower than that of the wild-type HLA-A2 mRNA.
Figure 9.
In vitro synthesis of a truncated HLA class I heavy chain by HLA-A2 mRNA with exon 2 skipping isolated from 624MEL28 cells. The cloned HLA-A2 cDNA lacking exon 2 sequence (E2− A2) was used as a template for mRNA synthesis. The synthesized mRNA was translated in the presence of [35S]methionine in a coupled transcription/translation system driven by T7 promoter (Promega Corp.). A wild-type HLA-A2 cDNA (A2) was used as a control. Translation products were analyzed by SDS-PAGE (A) or were immunoprecipitated by the rabbit anti–β2-μ–free HLA class I heavy chain serum R5996-4 and by the anti–β2-μ–free HLA heavy chain mAb HC-A2 before analysis by SDS-PAGE (B). Normal rabbit serum (NRS) was used as a negative control. The degree of translation was quantitated by scanning densitometry of the intrinsically radiolabeled proteins in the gel (C). The level of RNA synthesis was measured by [32P]UTP incorporation obtained from TCA precipitation (D).
Discussion
This study has characterized for the first time the molecular lesion underlying the spontaneous selective loss of an HLA class I allele by melanoma cells. HLA-A2 antigen has been selectively lost by melanoma cells 624MEL28 because of a defect in the splicing of HLA-A2 pre-mRNA. Pre-mRNA is spliced into two aberrant forms of mature mRNA, one with exon 2 skipping and the other with intron 2 retention. The latter mRNA form is distinct from pre-mRNA since it does not contain the sequences of introns 1, 3, 4, 5, 6, and 7. The mRNA with intron 2 retention cannot be translated into a wild-type HLA-A2 heavy chain, since the intron 2 retention introduces a premature stop codon at position 6–8 of the unspliced intron. The mRNA form with exon 2 skipping is expected to be translated into a truncated HLA-A2 heavy chain lacking the α1 domain. Although detected in in vitro translation experiments, such a polypeptide was not detected in 624MEL28 cells by testing cell extracts with xenoantibodies to distinct domains of HLA class I antigens in indirect immunoprecipitation and Western blotting assays. The latter finding may reflect the very low steady state level of the mRNA form with exon 2 skipping, as measured by the RNase protection assay, and the low translation efficiency, as suggested by the results of in vitro translation experiments.
The abnormal splicing of the HLA-A2 mRNA in 624MEL28 cells is caused by a mutation at position 2 of the 5′ splice donor site in intron 2, which is highly conserved in mammalian cells 37. This mutation inactivates the 5′ splice donor site, since pre-mRNA splicing requires the U at this position for spliceosome recognition 38. The importance of the conserved nucleotide at this position has been proven with site-directed mutagenesis experiments in which replacement of the T with a purine prevented in vivo splicing of the 12S mRNA 39 and in vitro splicing of β-globin mRNA 40. The base substitution at the 5′ splice donor site in intron 2 of the HLA-A2 gene results either in exon 2 skipping or in intron 2 retention in the two aberrant transcripts of the HLA-A2 gene in 624MEL28 cells, since no HLA-A2 gene copy with exon 2 deletion or with normal sequence at the 5′ splice donor site of intron 2 was detected in 624MEL28 cells.
The occurrence of either exon skipping or intron retention in pre-mRNA splicing caused by the same mutation at the 5′ splice donor site of an intron is an uncommon phenomenon. To the best of our knowledge, this abnormality has been detected only in the type III procollagen gene in members of a family with aortic aneurysms and easy bruisability 41 and in the β-hexoaminidase α chain gene in an Ashkenazi Jewish patient with Tay-Sachs disease 42. In both cases, as in our own, exon skipping and intron retention were caused by a mutation in the splice site at the 5′ end of the mutated intron. Exon skipping is a predominant phenotype caused by mutations at the 5′ splice donor site of an intron in mammalian cells 43. The mutation of a 5′ splice donor site inhibits the interaction of spliceosome with the 3′ and 5′ splice sites across the exon so that it blocks exon definition 44. If no cryptic site is activated, splicing of the exon leads directly to exon skipping. In contrast, intron retention is a rare phenomenon, found in only ∼6% of the cases with alternative splicing in mammalian cells 43. In the splicing of a small intron, the spliceosome uses the intron as the initial mode for selection of splice sites. A mutation of a 5′ splice donor site in a small intron leads to intron retention 45. The mutated intron 2 of the HLA-A2 gene in 624MEL28 cells is a small intron, as it is ∼241 bp in size. Thus, it is likely that “exon and intron definition,” two recognition mechanisms, are used for selection of splice sites in the splicing of HLA-A2 pre-mRNA.
The aberrant splicing of the HLA-A2 mRNA in the melanoma cell line 624MEL28 is different in several respects from the alternative splicing that results in skipping of exon 5 in the mRNA for HLA class I heavy chains 46 and of exon 3 or of both exons 3 and 4 in the mRNA for HLA-G heavy chains 47. First, exon 5 skipping may occur in the mRNA for both HLA-A and -B locus gene products in the same cell line. Second, more than one exon may be skipped in the mRNA for HLA-G heavy chain. Finally, normally spliced mRNA encoding the various antigens is more abundant than the corresponding mRNA resulting from alternative splicing. In contrast, in 624MEL28 cells exon 2 is skipped only in the mRNA transcribed by a mutated HLA class I heavy chain gene. No normally spliced HLA-A2 mRNA was detected in 624MEL28 cells.
To the best of our knowledge, the molecular lesion of the HLA-A2 gene identified in 624MEL28 cells is the first to have been characterized in a melanoma cell line with a spontaneous selective HLA class I allele loss. This lesion is distinct from that found in the γ-irradiation–induced HLA-A2 loss mutant melanoma cell line SK-MEL-29.1.22 in which a partial deletion of the HLA-A2 gene results in its transcriptional blockade 15. Furthermore, the molecular defect in 624MEL28 cells is different from the lesions underlying HLA class I allelic losses in other malignant cells. Deletion and transcriptional downregulation of the HLA-A11 gene cause selective loss of this allele in a colon carcinoma 48 and in a Burkitt's lymphoma 49 cell line, respectively. Furthermore, mutations in the HLA class I gene itself or in the upstream promoter region are likely to underlie the selective loss of HLA class I alleles described in two colon and two cervical carcinoma cell lines 50 51. It is noteworthy that one single mutational event is sufficient to cause the selective loss of an HLA class I allospecificity. In contrast, at least two mutational events that inactivate the two B2m genes present in a cell are required to cause total HLA class I antigen loss. This difference may account for the higher frequency of a selective HLA class I allele loss than of total HLA class I antigen loss in melanoma cells 52.
Preliminary results suggest that the HLA-A2 antigen loss 624MEL28 melanoma cells do not induce an MAA-specific CTL response restricted by the expressed HLA class I alleles. These findings, which parallel similar data in mouse model systems 53 54, could account for the escape of HLA-A2 antigen loss melanoma cells from CTL-mediated recognition and for the high frequency of selective HLA-A2 loss in metastatic melanoma lesions 1 4. The negative impact of these variants on the outcome of T cell–based immunotherapy emphasizes the need to design strategies to induce MAA-specific CTL responses restricted not only by HLA-A2 antigens, but also by the other HLA class I alleles present in HLA-A2–positive patients with melanoma. An alternative, although not exclusive, strategy is to combine T cell–based immunotherapy with immunotherapeutic modalities that are not negatively affected by HLA class I antigen loss by melanoma cells.
Acknowledgments
The authors acknowledge the secretarial assistance of Ms. Harriett V. Harrison, Ms. Donna James, Ms. Elba Osorio, and Ms. Derina McDonald. The authors also thank Dr. H.L. Ploegh, Harvard Medical School, Boston, MA; Dr. J.L. Strominger, Harvard University, Cambridge, MA; Dr. N. Tanigaki, Roswell Park Cancer Institute; Dr. P. Cresswell, Yale University School of Medicine, New Haven, CT; and Dr. S.Y. Yang, Memorial Sloan-Kettering Cancer Center, New York, NY, for providing antibodies, oligonucleotide primers, and probes.
This work was supported by U.S. Public Health Service grant CA67108 awarded by the National Cancer Institute, Department of Health and Human Services.
Footnotes
1used in this paper: β2-μ, β2-microglobulin; FITC-GAM, FITC-conjugated goat anti–mouse Ig antibodies; IEF, isoelectric focusing; IIF, indirect immunofluorescence; MAA, melanoma-associated antigen(s); RT, reverse transcription
References
- Ferrone S., Marincola F.M. Loss of HLA class I antigens by melanoma cellsmolecular mechanisms, functional significance and clinical relevance. Immunol. Today. 1995;16:487–494. doi: 10.1016/0167-5699(95)80033-6. [DOI] [PubMed] [Google Scholar]
- Versteeg R., Noordermeer I.A., Kruse-Wolters M., Ruiter D.J., Schrier P.I. c-myc down-regulates class I HLA expression in human melanomas. EMBO (Eur. Mol. Biol. Organ.) J. 1988;7:1023–1029. doi: 10.1002/j.1460-2075.1988.tb02909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natali P.G., Nicotra M.R., Bilgotti A., Venturo I., Marcenaro L., Giacomini P., Russo C. Selective changes in expression of HLA class I polymorphic determinants in human solid tumors. Proc. Natl. Acad. Sci. USA. 1989;86:6719–6723. doi: 10.1073/pnas.86.17.6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageshita T., Wang Z., Calorini L., Yoshii A., Kimura T., Ono T., Gattoni-Celli S., Ferrone S. Selective loss of human leukocyte class I allospecificities and staining of melanoma cells by monoclonal antibodies recognizing monomorphic determinants of class I human leukocyte antigens. Cancer Res. 1993;53:3349–3354. [PubMed] [Google Scholar]
- Marincola F.M., Shamamian P., Alexander R.B., Gnarra J.R., Turetskaya R.L., Nedospasov S.A., Simonis B., Taubenberger J.K., Yannelli J., Mixon A. Loss of HLA haplotype and B locus down-regulation in melanoma cell lines. J. Immunol. 1994;153:1225–1237. [PubMed] [Google Scholar]
- Wang Z., Margulies L., Hicklin D.J., Ferrone S. Molecular and functional phenotypes of melanoma cells with abnormalities in HLA class I antigen expression. Tissue Antigens. 1996;47:382–390. doi: 10.1111/j.1399-0039.1996.tb02573.x. [DOI] [PubMed] [Google Scholar]
- van Duinen S.G., Ruiter D.J., Broecker E.B., van der Velde E.A., Sorg C., Welvaart K., Ferrone S. Level of HLA antigens in locoregional metastases and clinical course of the disease in patients with melanoma. Cancer Res. 1988;48:1019–1025. [PubMed] [Google Scholar]
- Boon T., Cerottini J.-C., Van den Eynde B., van der Bruggen P., Van Pel A. Tumor antigens recognized by T lymphocytes. Annu. Rev. Immunol. 1994;12:337–365. doi: 10.1146/annurev.iy.12.040194.002005. [DOI] [PubMed] [Google Scholar]
- Restifo N.P., Marincola F.M., Kawakami Y., Taubenberger J., Yannelli J.R., Rosenberg S.A. Loss of functional β2-microglobulin in metastatic melanomas from five patients receiving immunotherapy. J. Natl. Cancer Inst. 1996;88:100–108. doi: 10.1093/jnci/88.2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Urso C.M., Wang Z., Cao Y., Tatake R., Zeff R.A., Ferrone S. Lack of HLA class I antigen expression by cultured melanoma cells FO-1 due to a defect in β2-microglobulin gene expression. J. Clin. Invest. 1991;87:284–292. doi: 10.1172/JCI114984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Cao Y., Albino A.P., Zeff R.A., Houghton A., Ferrone S. Lack of HLA class I antigen expression by melanoma cells SK-MEL-33 caused by a reading frameshift in β2-microglobulin messenger RNA. J. Clin. Invest. 1993;91:684–692. doi: 10.1172/JCI116249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicklin D.J., Wang Z., Arienti F., Rivoltini L., Parmiani G., Ferrone S. β2-microglobulin mutations, HLA class I antigen loss, and tumor progression in melanoma. J. Clin. Invest. 1998;101:2720–2729. doi: 10.1172/JCI498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maio M.M., Altomonte R., Tatake R., Zeff R.A., Ferrone S. Reduction in susceptibility to natural killer cell–mediated lysis of human FO-1 melanoma cells after induction of HLA class I antigen expression by transfection with B2-microglobulin gene. J. Clin. Invest. 1991;88:282–289. doi: 10.1172/JCI115289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfel T., Klehmann E., Muller C., Schutt K.-H., Meyer-sum-Buschenfelde K.-H., Knuth A. Lysis of human melanoma cells by autologous cytolytic T cell clones. Identification of human histocompatibility leukocyte antigen A2 as a restriction element for three different antigens. J. Exp. Med. 1989;170:797–810. doi: 10.1084/jem.170.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Seliger B., Mike N., Momburg F., Knuth A., Ferrone S. Molecular analysis of the HLA-A2 antigen loss by melanoma cells SK-MEL-29.1.22 and SK-MEL-29.1.29. Cancer Res. 1998;58:2149–2157. [PubMed] [Google Scholar]
- Lehmann F., Marchand M., Hainaut P., Pouillart P., Sastre X., Ikeda H., Boon T., Coulie P.G. Differences in the antigens recognized by cytolytic T cells on two successive metastases of a melanoma patient are consistent with immune selection. Eur. J. Immunol. 1995;25:340–347. doi: 10.1002/eji.1830250206. [DOI] [PubMed] [Google Scholar]
- Rivoltini L., Baracchini K.C., Viggiano V., Kawakami Y., Smith A., Mixon A., Restifo N.P., Topalian S.L., Simonis T.B., Rosenberg S.A., Marincola F.M. Quantitative correlation between HLA class I allele expression and recognition of melanoma cells by antigen-specific cytotoxic T lymphocytes. Cancer Res. 1995;55:3149–3157. [PMC free article] [PubMed] [Google Scholar]
- Anichini A., Mortarini R., Maccalli C., Squarcina P., Fleischhauer K., Mascheroni L., Parmiani G. Cytotoxic T cells directed to tumor antigens not expressed on normal melanocytes dominate HLA-A2.1-restricted immune repertoire to melanoma. J. Immunol. 1996;156:208–217. [PubMed] [Google Scholar]
- Kim C.J., Parkinson D.R., Marincola F. Immunodominance across HLA polymorphismimplications for cancer immunotherapy. J. Immunother. 1998;21:1–16. [PubMed] [Google Scholar]
- Rosenberg S.A., Yang J.C., Schwartzentruber D.J., Hwu P., Marincola F.M., Topalian S.L., Restifo N.P., Dudley M.E., Schwarz S.L., Spiess P.J. Immunologic and therapeutic evaluation of a synthetic peptide vaccine for the treatment of patients with metastatic melanoma. Nat. Med. 1998;4:321–327. doi: 10.1038/nm0398-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestle F.O., Alijagic S., Gilliet M., Sun Y., Grabbe S., Dummer R., Burg G., Schadendorf D. Vaccination of melanoma patients with peptide- or tumor lysate-pulsed dendritic cells. Nat. Med. 1998;4:328–332. doi: 10.1038/nm0398-328. [DOI] [PubMed] [Google Scholar]
- Barnstable C.J., Bodmer W.F., Brown G., Galfre G., Milstein C., Williams A.F., Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens—new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- Tanabe M., Sekimata M., Ferrone S., Takiguchi M. Structural and functional analysis of monomorphic determinants recognized by monoclonal antibodies reacting with the HLA class I α3 domain. J. Immunol. 1992;148:3202–3209. [PubMed] [Google Scholar]
- Russo C., Ng A.-K., Pellegrino M.A., Ferrone S. The monoclonal antibody CR11-351 discriminates HLA-A2 variants identified by T cells. Immunogenetics. 1983;18:23–35. doi: 10.1007/BF00401353. [DOI] [PubMed] [Google Scholar]
- Tsujisaki M., Sakaguchi K., Igarashi M., Richiardi P., Perosa F., Ferrone S. Fine specificity and idiotype diversity of the murine anti-HLA-A2, A28 monoclonal antibodies CR11-351 and KS-1. Transplantation. 1988;45:632–639. doi: 10.1097/00007890-198803000-00026. [DOI] [PubMed] [Google Scholar]
- McMichael A.J., Parham P., Rust N., Brodsky F. A monoclonal antibody that recognizes an antigenic determinant shared by HLA-A2 and B17. Hum. Immunol. 1980;1:121–129. doi: 10.1016/0198-8859(80)90099-3. [DOI] [PubMed] [Google Scholar]
- Stam N.J., Vroom T.M., Peters P.J., Pastoors E.B., Ploegh H.L. HLA-A- and HLA-B-specific monoclonal antibodies reactive with free heavy chains in Western blots, in formalin-fixed, paraffin-embedded tissue sections and in cryo-immuno-electron microscopy. Int. Immunol. 1990;2:113–125. doi: 10.1093/intimm/2.2.113. [DOI] [PubMed] [Google Scholar]
- Nakamuro K., Tanigaki N., Pressman D. Common antigenic structures of HL-A antigens. VI. Common antigenic determinants located on the 33,000-Dalton alloantigenic fragment portion of papain-solubilized HL-A molecules. Immunology. 1975;29:1119–1132. [PMC free article] [PubMed] [Google Scholar]
- Geliebter J., Zeff R.A., Melvold R.W., Nathenson S.G. Mitotic recombination in germ cells generated two major histocompatibility complex mutant genes shown to be identical by RNA sequence analysisKbm9 and Kbm6 . Proc. Natl. Acad. Sci. USA. 1986;83:3371–3375. doi: 10.1073/pnas.83.10.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega M.A., Strominger J.L. Constitutive endocytosis of HLA class I antigens requires a specific portion of the intracytoplasmic tail that shares structural features with other endocytosed molecules. Proc. Natl. Acad. Sci. USA. 1989;86:2688–2692. doi: 10.1073/pnas.86.8.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.Y. A standardized method for detection of HLA-A and HLA-B alleles by one-dimensional isoelectric focusing (IEF) gel electrophoresis. In: Dupont B., editor. Immunobiology of HLA. Vol. 1. Springer-Verlag Inc; New York: 1989. pp. 332–335. [Google Scholar]
- Burnette W.N. “Western blotting”electrophoretic transfer of proteins from sodium dodecyl sulfate–polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Loh E.Y., Elliott J.F., Cwirla S., Lanier L.L., Davis M.M. Polymerase chain reaction with single-sided specificityanalysis of T cell receptor δ chain. Science. 1989;243:217–220. doi: 10.1126/science.2463672. [DOI] [PubMed] [Google Scholar]
- Kawasaki E.S. Sample preparation from blood, cells and other fluids. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR ProtocolsA Guide to Methods and Applications. Academic Press, Inc.; San Diego: 1990. pp. 146–158. [Google Scholar]
- Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sernee M.F., Ploegh H.L., Schust D.J. Why certain antibodies cross-react with HLA-A and HLA-Gepitope mapping of two common MHC class I reagents. Mol. Immunol. 1998;35:177–188. doi: 10.1016/s0161-5890(98)00026-1. [DOI] [PubMed] [Google Scholar]
- Mount S.M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D.A., Steitz J.A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E.F., Caruthers M.H., Berk A.J. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature. 1982;295:380–384. doi: 10.1038/295380a0. [DOI] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Weissmann C. 5′ cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5′ splice region, not by the conserved 5′ GU. Cell. 1987;50:237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- Kontusaari S., Tromp G., Kuivaniemi H., Ladda R.L., Prockop D.J. Inheritance of an RNA splicing mutation (G+1 IVS20) in the type III procollagen gene (COL3A1) in a family having aortic aneurysms and easy bruisabilityphenotype overlap between familial arterial aneurysms and Ehlers-Danlos syndrome type IV. Am. J. Hum. Genet. 1990;47:112–120. [PMC free article] [PubMed] [Google Scholar]
- Ohno K., Suzuki K. Multiple abnormal β-hexosaminidase α chain mRNAs in a compound-heterozygous Ashkenazi Jewish patient with Tay-Sachs disease. J. Biol. Chem. 1988;263:18563–18567. [PubMed] [Google Scholar]
- Nakai K., Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene. 1994;141:171–177. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- Berget S.M. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Talerico M., Berget S.M. Intron definition in splicing of small Drosophila introns. Mol. Cell. Biol. 1994;14:3434–3445. doi: 10.1128/mcb.14.5.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krangel M.S. Secretion of HLA-A and -B antigens via an alternative RNA splicing pathway. J. Exp. Med. 1986;163:1173–1190. doi: 10.1084/jem.163.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani A., Geraghty D.E. Alternative splicing of HLA-G transcripts yields proteins with primary structures resembling both class I and class II antigens. Proc. Natl. Acad. Sci. USA. 1992;89:3947–3951. doi: 10.1073/pnas.89.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning M.J., Krausa P., Rowan A., Bicknell D.C., Bodmer J.G., Bodmer W.F. Tissue typing the HLA-A locus from genomic DNA by sequence-specific PCRcomparison of HLA genotype and surface expression on colorectal tumor cell lines. Proc. Natl. Acad. Sci. USA. 1993;90:2842–2845. doi: 10.1073/pnas.90.7.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imreh M.P., Zhang Q.-J., de Campos-Lima P.O., Imreh S., Krausa P., Browning M., Klein G., Masucci M.G. Mechanisms of allele-selective down-regulation of HLA class I in Burkitt's lymphoma. Int. J. Cancer. 1995;62:90–96. doi: 10.1002/ijc.2910620117. [DOI] [PubMed] [Google Scholar]
- Browning M.J., Krausa P., Rowan A., Hill A.B., Bicknell D.C., Bodmer J.G., Bodmer W.F. Loss of human leukocyte antigen expression on colorectal tumor cell linesimplications for anti-tumor immunity and immunotherapy. J. Immunother. 1993;14:163–168. doi: 10.1097/00002371-199310000-00001. [DOI] [PubMed] [Google Scholar]
- Koopman L.A., Mulder A., Corver W.E., Anholts J.D.H., Giphart M.J., Claas F.H.J., Fleuren G.J. HLA class I phenotype and genotype alterations in cervical carcinomas and derivative cell lines. Tissue Antigens. 1998;51:623–636. doi: 10.1111/j.1399-0039.1998.tb03005.x. [DOI] [PubMed] [Google Scholar]
- Hicklin D.J., Marincola F.M., Ferrone S. HLA class I antigen downregulation in human cancersT-cell immunotherapy revives an old story. Mol. Med. Today. 1999;5:178–186. doi: 10.1016/s1357-4310(99)01451-3. [DOI] [PubMed] [Google Scholar]
- Seung S., Urban J.L., Schreiber H. A tumor escape variant that has lost one major histocompatibility complex class I restriction element induces specific CD8+ T cells to an antigen that no longer serves as a target. J. Exp. Med. 1993;178:933–940. doi: 10.1084/jem.178.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Waes C., Monach P.A., Urban R.D., Wortzel R.D., Schreiber H. Immunodominance deters the response to other tumor antigens thereby favoring escapeprevention by vaccination with tumor variants selected with clone cytolytic T cells in vitro. Tissue Antigens. 1996;47:399–407. doi: 10.1111/j.1399-0039.1996.tb02575.x. [DOI] [PubMed] [Google Scholar]
- Fernandez-Vina M.A., Falco M., Sun Y., Stastny P. DNA typing for HLA class I allelesI Subsets of HLA-A 2and of -A28. Hum. Immunol. 331992. 163 173 [DOI] [PubMed] [Google Scholar]
- Oh S.-H., Fleischhauer K., Yang S.Y. Isoelectric focusing subtypes of HLA-A can be defined by oligonucleotide typing. Tissue Antigens. 1993;41:135–142. doi: 10.1111/j.1399-0039.1993.tb01992.x. [DOI] [PubMed] [Google Scholar]
- Ennis P.D., Zemmour J., Salter R.D., Parham P. Rapid cloning of HLA-A, B cDNA by using the polymerase chain reactionfrequency and nature of errors produced in amplification. Proc. Natl. Acad. Sci. USA. 1990;87:2833–2837. doi: 10.1073/pnas.87.7.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]