Abstract
Major histocompatibility complex (MHC) class Ib molecules have been implicated in CD8+ T cell–mediated defenses against intracellular bacterial infection, but the relative importance of MHC class Ib–restricted T cells in antimicrobial immunity is unknown. In this report, we use MHC tetramers to characterize T cell responses restricted by H2-M3, an MHC class Ib molecule that selectively presents N-formyl peptides. We find that sizeable H2-M3–restricted T cell responses, occurring earlier than MHC class Ia–restricted T cell responses, are mounted after primary infection with the intracellular bacterium Listeria monocytogenes. These H2-M3–restricted T cells are cytolytic and produce interferon γ. However, after a second L. monocytogenes infection, H2-M3–restricted memory T cell responses are minor in comparison to the much larger MHC class Ia–restricted responses. This first direct characterization of an MHC class Ib–restricted T cell response indicates that CD8+ T cells responding to L. monocytogenes infection can be divided into two groups: H2-M3–restricted responses, which provide rapid and quantitatively substantial effector function during primary infections but contribute relatively little to memory responses, and MHC class Ia–restricted responses, which expand later during primary infection but form memory T cells that respond rapidly and dramatically in response to subsequent infections by the same pathogen.
Keywords: H2-M3, major histocompatibility complex class Ib molecules, cytotoxic T lymphocytes, Listeria monocytogenes, bacterial infection
The mammalian immune system, molded by perpetual battle with diverse and evolving pathogens, can be divided into rapid, innate responses and somewhat delayed adaptive responses. Innate immunity targets microbial products that have been conserved through evolution and are shared by many microorganisms 1. The innate immune system uses germline-encoded molecules to quickly respond to a wide variety of pathogens. Many mechanisms have evolved to specifically identify bacterial products: LPS, a product of all gram-negative bacteria, is recognized by several molecules of the immune system, including LPS binding protein 2, CD14 3, and CD11c/CD18 4; macrophage scavenger receptors detect components of both gram-positive and gram-negative bacterial cell walls as well as the intact pathogens 5 6; and the N-formyl peptide receptor, found on neutrophils and macrophages, binds peptides starting with N-formyl methionine, the amino acid that initiates prokaryotic protein synthesis 7. These and perhaps many other molecules provide early detection of invading bacteria, thereby initiating antimicrobial defense and providing important cues for the developing adaptive immune response.
The adaptive, or specific immune system targets antigens expressed by a vast array of pathogens by generating B and T cell clones that each bear a rearranged specific antigen receptor 8. CD8+ T cells constitute an important arm of the adaptive immune response, recognizing pathogen-derived peptides that are presented by MHC class I molecules 9. MHC class I molecules have generally been divided into two groups: members of the highly polymorphic class Ia family present many different peptides to CD8+ T cells and have well-defined roles in immune defense against infection, whereas the function of molecules belonging to the more conserved MHC class Ib family has remained somewhat less clear 10 11 12. However, two different nonpolymorphic class Ib molecules appear specially suited for presentation of bacterial products. The divergent class Ib molecule CD1 is found outside the MHC locus in both humans and mice but is similar to MHC-encoded class I molecules in its structure and association with β2-microglobulin (β2m)1 13 14. Human CD1b presents mycobacterial glycolipids, such as Mycobacterium tuberculosis–derived mycolic acids 15 and Mycobacterium leprae–derived lipoarabinomannan 16, to unique subsets of T cells. Murine CD1d1 has a deep, hydrophobic binding groove 14; it is known to bind hydrophobic peptides of varying lengths and appears ideal to bind lipids and glycolipids as well.
The other murine class Ib molecule involved in antibacterial immunity, H2-M3, shows a strong preference for binding N-formyl peptides 12. N-formyl methionine is the initiating amino acid for bacterial protein synthesis, while in mammals only mitochondria use this amino acid. Thus, like CD1, H2-M3 seems ideally suited for the presentation of bacterial antigen to T cells. Unlike CD1, H2-M3 is encoded within the MHC and is believed to present only peptide antigen. Crystallographic study of H2-M3 revealed that the molecule has a structure very similar to that of MHC class Ia molecules, although the peptide groove is more hydrophobic, and the A pocket, which holds the first peptide residue in most class I molecules, is blocked 17. Binding of NH2-terminal formyl methionine occurs in the B pocket, and, as a consequence, peptides bound to H2-M3 are relatively short in comparison to the 8- to 10-mers that associate with class Ia molecules.
Early studies of T cell responses to infection with the gram-positive bacterium Listeria monocytogenes demonstrated the prominent role of CD8+ T cells in adaptive immunity to the pathogen 18, and multiple Listeria-derived antigens recognized by MHC class Ia–restricted CD8+ T cells have been identified 19. Bacterial peptides are also presented in the context of the MHC class Ib molecule, H2-M3 20 21, a finding which explained the interesting observation that some CD8+ T cells recognize L. monocytogenes–infected cells in the absence of classical MHC restriction 22. Three N-formylated peptides derived from L. monocytogenes and presented to CD8+ T cells by H2-M3 have been identified 23 24 25. Despite the potential importance of H2-M3–restricted CD8+ T cells in immunity to bacterial infection, their contribution relative to MHC class Ia–restricted responses remains unclear.
In this study, we investigate the H2-M3–restricted T cell response specific for one of the L. monocytogenes–derived N-formyl peptides, f-MIGWIIA 23, and compare it to the well-characterized H2-Kd–restricted, listeriolysin O (LLO)91–99 specific T cell response. We show that H2-M3–restricted responses to L. monocytogenes are kinetically distinct from class Ia–restricted responses; functional CD8+ CTLs specific for f-MIGWIIA become apparent and reach a climax earlier than LLO91–99 specific T cells. Memory responses of H2-M3–restricted T cells are also distinctive; while LLO91–99 specific (class Ia–restricted) T cells undergo very dramatic, rapid expansion upon reinfection with L. monocytogenes, relatively few f-MIGWIIA–specific cells are detected during the recall response. These findings demonstrate that T cell responses restricted by H2-M3 are distinct from those restricted by class Ia molecules and suggest that H2-M3–restricted T cells may have a unique, early role in immune responses against primary bacterial infection while playing a relatively minor role in memory responses.
Materials and Methods
Mice, Bacteria, and Listeria Infection.
Female CB6/F1, BALB/c, C57BL/6, and C3H/HeJ mice were purchased from The Jackson Laboratory. L. monocytogenes strain 10403s was obtained from Daniel Portnoy (University of California Berkeley, Berkeley, CA) and grown in brain–heart infusion broth. Mice 8–12 wk old were infected by tail vein injection with 2 × 103 L. monocytogenes for primary infection. Recall infection with 105 bacteria was performed 7 wk after primary infection. Splenocytes were harvested at different time points during the course of infection as described previously 26.
Generation of H2-M3 Tetramers.
H2-M3 tetramers were generated following the approach described previously 26 27. The cDNA for mouse β2m was provided by Pamela Bjorkman (California Institute of Technology, Pasadena, CA) and amplified with the following primers: 5′-GGCATATGATCCAGAAAACCCCTCAA-3′ and 5′-CCGGATCCTACATGTCTCGATCCCAGTA-3′. The full-length cDNA clone for H2-M3 was obtained from Chyung-Ru Wang (University of Chicago, Chicago, IL) and modified and amplified by PCR. The following primers were used: 5′-GGCATATGGGTTCACATTCATTACGTTATTTCCACACTGCG-3′ and 5′-CCGGATCCTAATCGCGCAGCTCCATCTTCATAGCCTCGAAGATACCACCCAGAC-CACCACCCCATTTCAGGGCAAGGGG-3′. The former, NH2-terminal primer, removed the leader sequence. The latter, COOH-terminal primer, replaced the transmembrane and cytoplasmic domains with a biotinylation sequence. The initial H2-M3 sequence was modified to replace four C or G nucleotides at the NH2 terminus with A or T, after the construct failed to produce significant protein in Escherichia coli (see Fig. 1 A). This approach, used to increase expression of eukaryotic proteins in bacteria, was described previously by Garboczi et al. 28. Four nucleotides were changed as follows (changes underlined): ATG GGC TCA CAT TCA CTG CGC to ATG GGT TCA CAT TCA TTA CGT. The changes did not alter the amino acid sequence. PCR products were cloned into the pET3a isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible vector (Novagen, Inc.) and expressed as recombinant proteins in the E. coli expression host BL21(DE3). H2-M3 heavy chain and β2m were purified from inclusion bodies as described previously 26, dissolved in 8 M urea, and refolded in the presence (or absence) of ∼60 μg/ml f-MIGWIIA or f-MFINRW (cytochrome c oxidase subunit I [COI]) peptide (Research Genetics Inc.) and the protease inhibitors pepstatin (1 μg/ml), leupeptin (1 μg/ml), and PMSF (0.4 mM). Soluble monomeric H2-M3/β2m/peptide complexes were purified by gel filtration over a Superdex 200HR column (Amersham Pharmacia Biotech). Purified monomeric complexes were biotinylated in vitro at 20°C for 12 h in the presence of 15 μg BirA enzyme (Avidity), 80 μM biotin, 10 mM ATP, 10 mM MgOAc, 20 mM bicine, and 10 mM Tris-HCl (pH 8.3). Complexes were purified again by gel filtration to remove excess biotin and tetramerized with PE-conjugated streptavidin (Molecular Probes, Inc.) at a 4:1 molar ratio. Finally, tetramers were purified by gel filtration and stored at a concentration of ∼5 mg/ml at 4°C in PBS (pH 8.0) with 0.02% sodium azide, 1 μg/ml pepstatin, 1 μg/ml leupeptin, and 0.5 mM EDTA. H2-Kd/LLO91–99 tetrameric complexes were produced as described previously 26 27.
Figure 1.
Expression and peptide-dependent refolding of recombinant H2-M3. (A) H2-M3 cDNA was modified by PCR (as described in Materials and Methods), cloned into the pET3a vector, and expressed in E. coli strain BL21(DE3). Samples taken before and 1 and 3 h after IPTG induction were subjected to SDS-PAGE, followed by staining with Coomassie blue. The number of hours of IPTG induction before collection of each sample is indicated above the gel. (B) The H2-M3 construct containing the biotinylation sequence was further mutagenized by PCR to replace four C/G nucleotides in the first seven codons of the 5′ coding region with A/T nucleotides (see Materials and Methods). These changes greatly increased H2-M3 expression in E. coli after IPTG induction. Samples taken before and after IPTG induction were treated as in A. H2-M3 and β2m protein were refolded in the presence of the Listeria-derived f-MIGWIIA peptide (C) or the mitochondrial COI self-peptide (D) as described in Materials and Methods, concentrated, and subjected to size-exclusion chromatography on a Superdex 200HR column. Absorbance at 280 nm was measured and plotted. (E) Refolding of H2-M3/β2m complexes was also attempted in the absence of exogenous formyl peptide.
In Vitro Restimulation of Splenocytes.
Splenocytes were harvested from BALB/c or C57BL/6 mice 7 d after infection. After lysis of erythrocytes, 3–4 × 107 splenocytes were resuspended in 5 ml RP10+ (RP 1640 [GIBCO BRL] supplemented with 10% FCS, l-glutamine [0.2 μg/ml], Hepes [1.2 μg/ml, pH 7.5], β-ME [50 μm], penicillin [100 U/ml], streptomycin [100 μg/ml], and gentamicin [50 μg/ml]). Stimulator cells were prepared from syngeneic naive mice. Splenocytes were irradiated (3,000 rads) and then pulsed with 10−6 M f-MIGWIIA or LLO91–99 peptide for 1 h at 37°C. Cells were washed to remove unbound peptide, and 3 × 107 cells resuspended in 5 ml RP10+ were added to 5 ml immune splenocytes in a T 25 cell culture flask. Cultures were stimulated every week with 3 × 107 peptide-coated splenocytes to generate T cell lines. RP10+ medium was supplemented with 5% T-STIM Culture Supplement (Collaborative Biomedical Products) after the second restimulation. Three BALB/c T cell clones were generated from a f-MIGWIIA–specific CTL line after 3 wk of restimulation in vitro. Limiting dilution was performed in a 96-well plate, and medium was replaced weekly until expansion was sufficient to allow restimulation in a T 25 culture flask.
Enrichment, Staining, and Analysis of CD8+ T Cells.
Splenocytes were enriched for CD8+ T cells by depletion using magnetically activated cell sorting (MACS®; Miltenyi Biotec). Cells were first incubated with antibodies against CD4 (GK1.5), MHC class II (TIB120), and MAC-1 (TIB128) for 20 min on ice in separation buffer (PBS, 0.5% BSA, and 2 mM EDTA, pH 7.45). After extensive washing, spleen cells were incubated with goat anti–rat IgG magnetic microbeads (Miltenyi Biotec) for 20 min at 4°C. Splenocytes were washed again, applied to a type LS column (Miltenyi Biotec), and separated using the MidiMACS® (Miltenyi Biotec) following the manufacturer's instructions. For staining, ∼3 × 105 CD8+-enriched splenocytes were blocked with unconjugated streptavidin (0.5 mg/ml; Molecular Probes, Inc.) and Fc-block (PharMingen) in staining buffer (SB; PBS, 0.5% BSA, 0.02% sodium azide, pH 7.45) in a 96-well plate for 20 min on ice. Cells were then triple stained with anti-CD62L–FITC (clone MEL-14; PharMingen); PE-conjugated H2-M3/f-MIGWIIA, H2-M3/COI, or H2-Kd/LLO91–99 tetrameric complexes (0.25–0.5 mg/ml); and anti-CD8α–CyChr (clone 53-6.7; PharMingen) in SB for 1 h on ice. After three washes in SB, cells were fixed in 1% paraformaldehyde/PBS (pH 7.45). Data were acquired using a FACSCalibur™ flow cytometer and analyzed using CELLQuest software (Becton Dickinson). For cell sorting, staining of enriched CD8+ T cells from each spleen was performed in a single tube using a staining buffer of PBS supplemented with 0.5% FCS, and cells were not fixed before sorting.
Chromium-release Assays.
P815 target cells (American Type Culture Collection) were labeled with 51Cr and washed. FACS®-sorted cells were incubated with 5 × 103 labeled cells alone or in the presence of 10−6 M f-MIGWIIA, f-MFINRW (COI self), or LLO91–99 peptide for 7.5 h at 37°C. The percent specific lysis was calculated as described previously 26 based on the amount of 51Cr release.
Enzyme-linked Immunospot Assay.
The enzyme-linked immunospot (ELISPOT) assay for detecting IFN-γ–producing T cells was performed as described previously 29. In brief, P815 target cells were irradiated (10,000 rads) and coated with 10−6 M f-MIGWIIA or LLO91–99 peptide, or incubated in the absence of peptide, for 1 h at 37°C. After washing, 105 target cells were added along with 105 immune splenocytes to each well of a 96-well nitrocellulose plate (Millipore, Inc.) coated with anti–IFN-γ antibody (clone R4-6A2; PharMingen). The plate was incubated for 24 h at 37°C in the presence of 30 U/ml IL-2. IFN-γ production was detected with biotin-labeled anti–IFN-γ antibody (clone XMG1.2; PharMingen), followed by addition of streptavidin peroxidase (Kirkegaard & Perry Labs., Inc.) and development with a substrate solution of 1 mg/ml DAB (HRP Color Development Reagent; Bio-Rad Laboratories), 50 mM Tris-HCl, and 0.5 μl/ml 30% hydrogen peroxide. Spots were counted under a dissecting microscope.
Results
Generation of H2-M3 Tetramers.
H2-M3 is an MHC class Ib molecule that binds N-formylated, hydrophobic peptides and presents antigen derived from intracellular bacteria to CD8+ T cells 12. To visualize H2-M3–restricted T cell responses to bacterial infection, we generated tetrameric complexes of H2-M3 with the L. monocytogenes–derived peptide, f-MIGWIIA 23. As a negative control, H2-M3 tetramers complexed with a mitochondrially derived self-peptide, COI (f-MFINRW 30), were generated. We PCR mutagenized H2-M3 to remove the leader, transmembrane, and cytosolic regions and to add a COOH-terminal biotinylation site 31 32, but expression of this construct in E. coli was poor (Fig. 1 A). We then modified the 5′ end of the coding sequence, changing four C/G into A/T nucleotides within the first seven codons without altering the amino acid sequence of the protein (see Materials and Methods, and reference 28). These changes greatly enhanced IPTG-inducible expression of recombinant H2-M3 in E. coli (Fig. 1 B), thereby allowing purification of high quantities of recombinant protein for in vitro refolding. Purified H2-M3 and β2m were solubilized in 8 M urea and refolded in the presence of synthetic f-MIGWIIA or COI peptide.
To ensure that refolding and stabilization of soluble H2-M3 complexes were peptide dependent, refolding reactions were performed in the absence of peptide (Fig. 1 E) or in the presence of f-MIGWIIA (Fig. 1 C) or COI (Fig. 1 D) peptide. Size-exclusion chromatography demonstrated that heavy chain and β2m form stable complexes in the presence (Fig. 1C and Fig. D) but not the absence (Fig. 1 E) of N-formylated peptide. Purified H2-M3 complexes were biotinylated and tetramerized with PE-conjugated streptavidin. Tetrameric complexes were further purified by gel filtration before concentration and used as staining reagents for flow cytometric analyses 27.
Staining and Specificity of H2-M3 Tetramers.
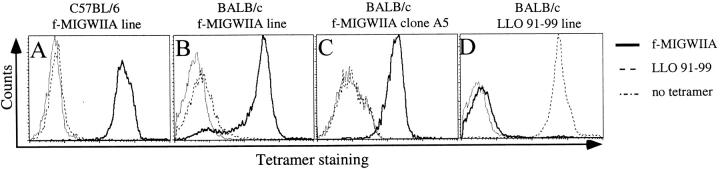
Tetrameric complexes refolded with the L. monocytogenes–derived f-MIGWIIA peptide were tested for specific staining of in vitro peptide-restimulated CTL lines and clones generated from L. monocytogenes–infected mice. All CTL lines and clones specifically killed target cells coated with the stimulating peptide when tested in chromium-release assays (data not shown). f-MIGWIIA–specific CTL lines derived from both C57BL/6 and BALB/c mouse strains demonstrated high intensity staining with H2-M3/f-MIGWIIA tetramers (Fig. 2A and Fig. B). Within some CTL lines, particularly those stimulated in vitro for a short time, a fraction of cells stained with low intensity or did not stain with the tetramers (Fig. 2 B, and data not shown). However, all three f-MIGWIIA–specific CTL clones stained with uniformly high intensity with H2-M3/f-MIGWIIA tetramers (Fig. 2 C, and data not shown). These data indicate that although H2-M3/f-MIGWIIA tetramers stain most f-MIGWIIA–specific CTLs, a small portion of T cells, perhaps those with lower affinity for the MHC/peptide complex, may not be detected. H2-M3/f-MIGWIIA tetramers did not stain CTLs specific for the H2-Kd–restricted peptide LLO91–99 (Fig. 2 D), demonstrating their specificity in staining, similar to the findings for H2-Kd/LLO91–99 tetramers (26; Fig. 2A–C). Some CTL lines specific for the other Listeria-derived, N-formylated peptides stained, to varying but generally small degrees, with H2-M3/f-MIGWIIA tetramers (data not shown). This staining correlated with cross-reactive killing in CTL assays and is not entirely surprising, given the cross-reactivity in recognition of N-formylated peptides demonstrated by Nataraj et al. 33. No staining of cell lines was detected with H2-M3 tetramers refolded with the COI self-peptide (data not shown). These studies show that H2-M3/f-MIGWIIA tetramers can be used to stain f-MIGWIIA–specific T cell populations and demonstrate that the staining is specific.
Figure 2.
H2-M3/f-MIGWIIA tetramers stain CTL lines and clones specific for the f-MIGWIIA peptide. f-MIGWIIA– and LLO91–99 specific CTL lines and clones were generated by in vitro peptide restimulation of splenocytes from L. monocytogenes–infected mice as described in Materials and Methods. Cells were stained with anti-CD8, anti-CD62L, PE-conjugated H2-M3/f-MIGWIIA or H2-Kd/LLO91–99 tetramers, and propidium iodide to identify dead cells. Histograms show tetramer staining gated on live CD8+ CTLs from f-MIGWIIA–specific C57BL/6 (A) and BALB/c (B) derived CTL lines, a BALB/c f-MIGWIIA–specific CTL clone (C), and a BALB/c LLO91–99 specific CTL line (D).
Comparison of H2-M3–restricted T Cell Populations in Different Mouse Strains.
Most common laboratory mouse strains share the same allele of H2-M3 12. To determine whether this translates into similar H2-M3–restricted T cell responses to bacterial infection, H2-M3/f-MIGWIIA and H2-M3/COI tetramers were used to stain splenocytes from L. monocytogenes–infected mice. Interestingly, direct ex vivo staining for H2-M3–restricted T cells revealed relatively high numbers of activated (CD62Llow) f-MIGWIIA–specific T cells compared with previously reported findings for the immunodominant LLO91–99 specific response 26, although the magnitude of H2-M3–restricted responses differed among mouse strains. Throughout the peak phase of the primary response (days 5–9 after infection), C57BL/6 and C3H/HeJ mice had significantly higher numbers of H2-M3/f-MIGWIIA tetramer–staining T cells than BALB/c mice (Fig. 3). In this set of experiments, the differences were most evident 7 d after infection, when C57BL/6 and C3H/HeJ mice had large, distinct populations of activated T cells staining with H2-M3/f-MIGWIIA tetramers (Fig. 3 A). At the same time point, BALB/c mice had smaller populations of f-MIGWIIA–specific cells that stained with lower intensity (Fig. 3 A). A diffuse staining of cells within the CD8+CD62Lhi (unactivated) population with H2-M3/f-MIGWIIA tetramers was also detected, although at very similar levels among the mouse strains investigated (Fig. 3 A, and data not shown). A comparable degree of staining of CD8+CD62Lhi cells was also found with H2-Kd tetramers (26; Fig. 4 and Fig. 5). Further experiments will be needed to determine the origin and specificity of this “background” staining. Nevertheless, this experiment demonstrates that L. monocytogenes–infected mice of different strains generate H2-M3–restricted T cell responses of different sizes that can be detected directly ex vivo with H2-M3/f-MIGWIIA tetramers. In addition, despite the large differences in magnitude, H2-M3–restricted, f-MIGWIIA–specific T cell responses showed similar kinetics among the mouse strains (Fig. 3 B).
Figure 3.

The magnitude of f-MIGWIIA–specific CTL responses to L. monocytogenes infection is different among mouse strains. Female age-matched C57BL/6, C3H/HeJ, and BALB/c mice were infected with 2,000 L. monocytogenes, and splenocytes were obtained on the indicated day after primary infection, enriched for CD8+ T cells, and stained with antibodies to CD8 and CD62L as well as one of the PE-conjugated tetramers. Two mice per strain were analyzed each day. (A) The dot plots show H2-M3/f-MIGWIIA (left panels) and H2-M3/COI self (right panels) tetramer stainings for one mouse per strain on day 7 after infection. Staining for CD62L (l-selectin) is shown on the x-axis, and tetramer staining on the y-axis. Dot plots are gated on CD8+ lymphocytes. Percentages in the upper left quadrants represent the percentage of CD8+ CD62Llow (activated) cells that are tetramer positive. (B) The average number of H2-M3/f-MIGWIIA and H2-M3/COI (self) tetramer–positive cells per spleen is shown for two mice of each strain on days 5, 7, and 9 after infection. SD are indicated.
Figure 4.

H2-M3–restricted T cell responses peak earlier than H2-Kd–restricted responses during primary L. monocytogenes infection. Female, age-matched CB6/F1 mice were infected with 2,000 L. monocytogenes. Splenocytes were taken from uninfected mice (day 0) and infected mice on the indicated day after infection. CD8+-enriched splenocytes were stained with anti-CD8, anti-CD62L, and one of three PE-conjugated tetramers: H2-M3/f-MIGWIIA, H2-M3/COI self, or H2-Kd/LLO91–99. Dot plots are gated on CD8+ lymphocytes, with CD62L staining on the x-axis and tetramer staining on the y-axis. The percentage of CD62Llow (activated), tetramer-staining cells is indicated in the upper left quadrant. Dot plots for each day are from one representative mouse. Four log phases are shown.
Figure 5.

H2-M3–restricted T cells undergo little expansion during recall L. monocytogenes infection compared with H2-Kd–restricted T cells. Female, age-matched CB6/F1 mice were infected with 2,000 L. monocytogenes. After 48 d, the mice were reinfected with 100,000 bacteria. Day 0 represents a mouse not reinfected with L. monocytogenes, and is the same as day 48 from Fig. 4. Splenocytes were taken on the indicated days and were stained and analyzed as described in the legend to Fig. 4.
H2-M3–restricted T Cell Responses to Primary Infection with L. monocytogenes.
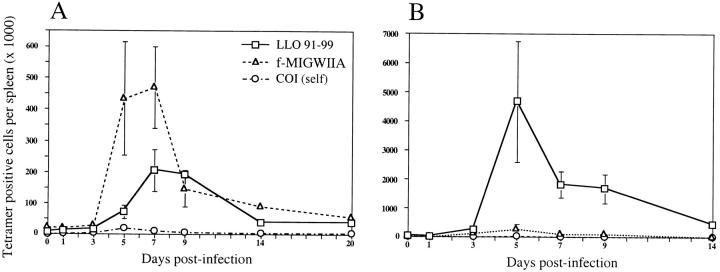
CD8+ T cell populations specific for different peptides bound to the MHC class Ia molecule H2-Kd expand and contract in a coordinated fashion in response to infection with L. monocytogenes, despite very different magnitudes of the T cell responses 26. To investigate whether T cells restricted by H2-M3 follow the same kinetics, we studied the expansion and contraction of f-MIGWIIA–specific T cells in CB6/F1 mice (H2b×d). This F1 hybrid of C57BL/6 and BALB/c strains generates H2-M3–restricted, f-MIGWIIA–specific T cell responses of intermediate size compared with the parental strains (Fig. 3 and Fig. 4) and also develops H2-Kd–restricted, LLO91–99 specific T cell responses. As described previously in BALB/c mice 26, a distinct LLO91–99 specific T cell population was first detectable on day 5 after infection with L. monocytogenes (Fig. 4 and Fig. 6 A). Further expansion resulted in a peak of the LLO91–99 specific T cell response 7–9 d after infection; the percentage of tetramer-staining T cells continued to increase until day 9, but the absolute number reached a plateau due to a decreasing number of total splenocytes after day 7 (Fig. 6 A). H2-M3–restricted T cells became visible 3 d after primary infection and expanded dramatically between days 3 and 5 after infection. Strikingly, these f-MIGWIIA–specific T cells were already at or near their peak magnitude at this early time point; the absolute number of specific T cells was highest between days 5 and 7 after infection (Fig. 6 A). No significant staining with H2-M3 tetramers complexed with the COI self-peptide was detected at any point. T cell responses to two other H2-M3–restricted, Listeria-derived peptides, though smaller than f-MIGWIIA–specific responses, followed the same kinetics as determined by tetramer staining (data not shown). For each peptide-specific T cell population, the peak response was followed by a contraction phase, and by 7 wk after infection, the low numbers of remaining f-MIGWIIA– and LLO91–99 specific T cells were near the limits of detection. Although a certain degree of variability is inherent in T cell responses to infection, the relative kinetics of H2-Kd– and H2-M3–restricted T cell populations are highly reproducible, with the climax of H2-M3–restricted T cells preceding that of H2-Kd–restricted T cells by ∼2 d in several studies (Fig. 3 and Fig. 4, and data not shown). Indeed, in the strain comparison reported here (Fig. 3), f-MIGWIIA–specific cells from three strains synchronously peaked on day 7 after infection (Fig. 3 B), whereas LLO91–99 specific cells in H2d BALB/c mice reached their highest magnitude around day 9 after infection (data not shown). These experiments indicate that although magnitude and exact kinetics of H2-M3–restricted responses vary somewhat among mice investigated, f-MIGWIIA–specific CD8+ T cell populations respond earlier than H2-Kd–restricted T cell populations upon infection with L. monocytogenes.
Figure 6.
H2-M3–restricted T cell responses differ from H2-Kd–restricted T cells during both primary infection and reinfection with L. monocytogenes. The total number of CD8+ CD62Llow (activated), tetramer-positive cells (for H2-M3/f-MIGWIIA, H2-M3/COI self, and H2-Kd/LLO91–99 tetramers) was quantified from mice at each time point during primary and recall studies. Absolute numbers (y-axis) are shown for primary infection (A) and recall infection (B) with days after infection marked on the x-axis. Averages and SD are shown for three mice per day except primary days 0 (uninfected), 20, and 48 (recall day 0), at which times there were two mice. Note that y-axes have different scales for primary and recall infections.
Memory T Cell Responses upon Reinfection with L. monocytogenes.
Exposure to pathogens can result in immunological memory, which leads to faster and more effective immune responses when the same pathogen is encountered at a later time 34. We have previously shown that in BALB/c mice reinfected with L. monocytogenes, H2-Kd–restricted, Listeria-specific T cells respond more quickly and reach much higher frequencies than seen during primary infection 26 27 35. The rapid expansion of LLO91–99 specific T cells during recall infection is also seen in CB6/F1 mice; by day 3 after reinfection, the percentage of LLO91–99 specific T cells was as high as the peak primary response (Fig. 4 and Fig. 5). Between days 3 and 5, there was a dramatic expansion of LLO91–99 specific T cells, and at the peak of the recall response, 5 d after reinfection, the population of LLO91–99 specific T cells constituted 10–20% of the entire CD8+ T cell compartment (Fig. 5 and Fig. 6 B, and data not shown). H2-M3–restricted, f–MIGWIIA peptide–specific T cell responses were remarkably different from LLO91–99 specific responses in the same mouse. The two epitope-specific T cell populations started out at approximately the same frequency before reinfection (day 0), and numbers decreased slightly in the spleen on day 1 after infection, as reported previously 26. By the third day after reinfection, a population of H2-M3/f-MIGWIIA tetramer–staining cells was clearly detected; this contrasts with primary infection, during which no clear population of H2-M3/f-MIGWIIA–staining cells was seen 3 d after infection. Although the peak of both primary and recall H2-M3–restricted T cell responses occurred ∼5 d after infection, the magnitude of the f-MIGWIIA–specific T cell population after reinfection was lower than the peak primary response specific for the peptide and >10-fold lower than the recall LLO91–99 specific T cell response in the same mouse (Fig. 5 and Fig. 6B). Similar results were obtained in the B10.D2 mouse strain (data not shown). These experiments show that H2-M3– and H2-Kd–restricted T cell populations differ greatly in their extent of expansion after reinfection with L. monocytogenes. In addition, unlike the primary response, during which f-MIGWIIA and the LLO91–99 specific T cell responses have remarkably different timing in expansion and contraction, after reinfection with L. monocytogenes, the two populations have similar kinetics.
Functional Qualities of H2-M3–restricted T Cells.
The previous experiments described in this report demonstrate that Listeria-specific H2-M3–restricted T cells are detectable and peak earlier than H2-Kd–restricted responses after primary infection, but they do not indicate whether this early expansion might have implications for bacterial clearance. To determine whether H2-M3–restricted L. monocytogenes–specific T cells present 5 d after primary infection have characteristics of functional CD8+ T cells, we placed immune splenocytes directly into a chromium-release assay to assess cytolytic ability (Fig. 7 A) and an ELISPOT assay to detect IFN-γ production (Fig. 7 B). FACS®-sorted CD62Llow (activated), f-MIGWIIA–specific CD8+ T cells demonstrated significant specific killing activity against f-MIGWIIA–coated target cells (Fig. 7 A, left panel). Splenocytes depleted of H2-M3/f-MIGWIIA tetramer–staining cells (Fig. 7 A, right panel) had little activity against f-MIGWIIA peptide, indicating that essentially all f-MIGWIIA–specific T cells were detected with the tetramers. No LLO91–99 specific killing activity was detected at this early time point after infection. Splenocytes from another mouse were stained with H2-Kd/LLO91–99 tetramers at the same time and subjected to an identical sorting process for tetramer-positive and tetramer-negative cells within the CD8+CD62Llow population. Too few H2-Kd/LLO91–99 tetramer–positive cells were detected to sort effectively. However, within the tetramer-negative population, significant f-MIGWIIA–specific killing was found, with >15% specific lysis of target cells (data not shown). H2-M3–restricted T cells also secreted IFN-γ 5 d after primary infection (Fig. 7 B). Approximately four times more IFN-γ–secreting f-MIGWIIA peptide–specific cells than LLO91–99 specific cells were detected. These experiments demonstrate that H2-M3–restricted T cells detected early (day 5) after primary infection are functional CD8+ T cells, with both cytolytic and IFN-γ–secreting abilities.
Figure 7.
H2-M3–restricted T cells detected 5 d after primary infection are effective killers and secrete IFN-γ. (A) Splenocytes from a CB6/F1 mouse infected with 2,000 L. monocytogenes 5 d previously were enriched for CD8+ T cells followed by staining with anti-CD8, anti-CD62L, and PE-conjugated H2-M3/f-MIGWIIA tetramers. Tetramer-positive (left panel) and tetramer-negative (right panel) cells were sorted separately from the CD8+CD62Llow staining population and placed in a CTL assay with peptide-coated p815 target cells. The peptide used and percent specific lysis are shown on the x-axis and y-axis, respectively. The E/T ratios were ∼1:1 for the tetramer-positive cells and 10:1 for the tetramer-negative population. (B) Unsorted splenocytes from both immunized mice were placed in an ELISPOT assay for detection of IFN-γ–secreting cells. The number of IFN-γ–secreting cells detected is shown as the average of two mice (y-axis), with SD indicated. Four replicate wells were counted for each mouse. The stimulating peptide is indicated on the x-axis.
Discussion
In this report, we characterize H2-M3–restricted CD8+ T cell responses after infection with the intracellular bacterium L. monocytogenes. Our studies demonstrate that these MHC class Ib–restricted T cell responses are distinctive, differing in several ways from MHC class Ia–restricted responses. More specifically, we show that (a) functional H2-M3–restricted T cell populations are detectable and achieve their maximal size earlier than H2-Kd–restricted T cell populations after primary infection; (b) H2-M3–restricted T cell responses after recall bacterial infection do not exhibit the typical memory features of faster expansion and greater magnitude; and (c) mouse strains that share the same H2-M3 allele generate H2-M3–restricted T cell responses with similar kinetics but different magnitudes. Our results suggest that CD8+ T cells restricted by the class Ib molecule H2-M3 have an early and perhaps unique role in primary defense against infection with intracellular bacteria but may play only a minor role in memory responses.
Why do H2-M3–restricted T cells expand more rapidly during a primary immune response to L. monocytogenes than other T cells? One possibility is that N-formylated peptides are presented earlier during infection than other epitopes, giving H2-M3–restricted T cells a temporal advantage over other T cells. The efficiency of L. monocytogenes N-formyl peptide presentation by H2-M3 is not known. However, bacterially secreted N-formyl peptides are likely to require much less processing to be presented by class I molecules than epitopes occupying internal positions in proteins. In addition, it is known that H2-M3 is poorly represented on the cell surface due to the paucity of mitochondrial self-peptides 36. Therefore, it is possible that bacterial infection induces the surface expression of H2-M3 due to a sudden abundance of N-formylated peptides. In this setting, bacterially derived N-formylated epitopes could be presented quickly, whereas presentation of bacterial peptides bound by class Ia molecules might be delayed as they compete for presentation with a plethora of self-peptides. Although these mechanisms might provide H2-M3–restricted T cells with a relative time advantage, it seems unlikely that they could account for the 2-d lead over the H2-Kd–restricted T cell responses.
An alternative explanation for the early response of H2-M3–restricted T cells is that they may have been primed in vivo before L. monocytogenes infection. Indeed, in its relatively rapid expansion after L. monocytogenes infection, the H2-M3–restricted T cell response more closely resembles a memory response than a primary response to antigen. The notion that commensal bacteria may prime H2-M3–restricted T cells was proposed by Lenz and Bevan when they found that mice housed in non-specific pathogen–free (SPF) facilities had established immunity to the Listeria-derived f-MIGWII and f-MIVIL peptides in the absence of previous infection with L. monocytogenes 37. We have also noted that older mice often generate larger H2-M3–restricted T cell responses than younger mice (Kerksiek, K., and E.G. Pamer, unpublished results), suggesting that exposure to environmental bacteria, such as the normal flora of the gut, may modulate the repertoire of T cells specific for foreign bacteria. This hypothesis requires the assumption that environmental bacteria either express the same formyl peptides expressed by L. monocytogenes and other foreign bacteria, or that formyl peptides are promiscuously recognized by H2-M3–restricted T cells, perhaps due to their relative shortness or hydrophobicity. Recent studies of f-MIGWII–specific T cell clones demonstrated a remarkable degree of peptide cross-recognition, supporting the latter of these two possibilities 33. Although T cells responding to f-MIGWIIA display the characteristic memory phenotype of rapid in vivo expansion, further experiments will have to be performed to dissect the role of bacterial exposure in the generation of H2-M3–restricted T cell responses.
Although a relatively rapid H2-M3–restricted T cell response is generated during primary infection with L. monocytogenes, these cells do not expand dramatically upon reinfection with the bacterium. It is possible that H2-M3–restricted memory T cells are not generated during the primary immune response or are not maintained after the resolution of infection. However, our tetramer stainings, while near the limits of detection, suggest that f-MIGWIIA– and LLO91–99 specific T cells are present at comparable levels 7 wk after primary infection. We favor the possibility that expansion of H2-M3–restricted memory T cells requires a greater duration of antigen exposure than required by LLO91–99 specific T cells, which undergo dramatic in vivo expansion after reinfection. Bacterial infection during the recall response is limited to 1 or 2 d (26; and data not shown), which might be insufficient for f-MIGWIIA–specific T cell expansion. Thus, while a “memory response” during primary infection and the lack of such during recall infection may appear paradoxical, the vastly different kinetics of bacterial infection in these two circumstances may provide an explanation.
Common laboratory mouse strains share the same H2-M3 allele and should therefore be capable of generating functional H2-M3–restricted T cells in response to bacterial infection. Indeed, all of the mouse strains investigated here show such a response. However, the magnitudes of these responses differed quite dramatically; in particular, BALB/c mice generated significantly fewer H2-M3–restricted, L. monocytogenes–specific T cells than did C57BL/6 and C3H/HeJ mice. There are several reasons why these differences might occur. One possibility is that thymic or peripheral selection of the T cells occurs differently among these mouse strains; it is unclear how the repertoire of H2-M3–restricted T cells is selected 12. If selection of H2-M3–restricted T cells occurs on H2-M3/self-peptide complexes expressed in the thymus, the different mouse strains should possess similar repertoires, as the same mitochondrial proteins are present in these strains. However, if the repertoire of H2-M3–restricted cells is influenced by resident bacterial flora such as those found in the gut, then distinct subsets of H2-M3–restricted T cells might be selected in different mouse strains. Another factor that might account for the varied H2-M3–restricted T cell responses in different mouse strains is the genetic background of the host. Indeed, mouse strains are known to vary dramatically in susceptibility to L. monocytogenes infection 38, and the basis for this remains largely unexplored. It is possible that complex genetic factors that influence infectious disease susceptibility can affect the magnitude of T cell responses.
Primary L. monocytogenes infection of immunocompetent mice generally resolves within 6–7 d. Although CD8+ T cells are known to be important in the clearance of infection, maximal T cell responses restricted by H2-Kd occur after the infection has been cured. However, H2-M3–restricted CD8+ T cells respond to primary L. monocytogenes infection ∼2 d earlier than their MHC class Ia–restricted cousins, resulting in significant numbers of cytolytic, IFN-γ–producing H2-M3–restricted T cells on the fifth day after infection (Fig. 7). This finding suggests that H2-M3–restricted T cells play an important role in the early control of primary infection with L. monocytogenes. Remarkably, although the H2-M3–restricted T cell population appears very vigorous during primary infection, MHC class Ia–restricted T cells seem to be more important for providing T cell memory. This is the first demonstration, to our knowledge, of temporal diversity and differential memory capacity within CD8+ T cell populations responding to bacterial infection.
Acknowledgments
We thank David Garboczi for his advice regarding the expression of H2-M3, and Peter Cresswell for valuable discussion.
Footnotes
1used in this paper: β2m, β2-microglobulin; COI, cytochrome c oxidase subunit I; ELISPOT, enzyme-linked immunospot; IPTG, isopropyl-β-d-thiogalactopyranoside; LLO, listeriolysin O
K.M. Kerksiek is supported by National Institutes of Health Training Grant 5T32-AI07019. D.H. Busch is a recipient of a Howard Hughes Fellowship for Physicians. This work was supported by Public Health Service Grant 1RO1 AI-42135.
References
- Medzhitov R., Janeway C.A., Jr. Innate immunityimpact on the adaptive immune response. Curr. Opin. Immunol. 1997;9:4–9. doi: 10.1016/s0952-7915(97)80152-5. [DOI] [PubMed] [Google Scholar]
- Schumann R.R., Leong S.R., Flaggs G.W., Gray P.W., Wright S.D., Mathison J.C., Tobias P.S., Ulevitch R.J. Structure and function of lipopolysaccharide binding protein. Science. 1990;249:1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- Wright S.D., Ramos R.A., Tobias P.S., Ulevitch R.J., Mathison J.C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- Ingalls R.R., Golenbock D.T. CD11c/CD18, a transmembrane signaling receptor for lipopolysaccharide. J. Exp. Med. 1995;181:1473–1479. doi: 10.1084/jem.181.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne D.W., Resnick D., Greenberg J., Krieger M., Joiner K.A. The type I macrophage scavenger receptor binds to gram-positive bacteria and recognizes lipoteichoic acid. Proc. Natl. Acad. Sci. USA. 1994;91:1863–1867. doi: 10.1073/pnas.91.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger M., Herz J. Structures and functions of multiligand lipoprotein receptorsmacrophage scavenger receptors and LDL receptor-related protein (LRP) Annu. Rev. Biochem. 1994;63:601–637. doi: 10.1146/annurev.bi.63.070194.003125. [DOI] [PubMed] [Google Scholar]
- Murphy P.M. The molecular biology of leukocyte chemoattractant receptors. Annu. Rev. Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- Janeway C.A., Jr., Travers P. ImmunobiologyThe Immune System in Health and Disease. 3rd ed. Garland Publishing Inc; New York: 1997. [Google Scholar]
- Germain R.N. MHC-dependent antigen processing and peptide presentationproviding ligands for T lymphocyte activation. Cell. 1994;76:287–299. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- Shawar S.M., Vyas J.M., Rodgers J.R., Rich R.R. Antigen presentation by major histocompatibility complex class I-B molecules. Annu. Rev. Immunol. 1994;12:839–880. doi: 10.1146/annurev.iy.12.040194.004203. [DOI] [PubMed] [Google Scholar]
- Stroynowski I. Molecules related to class-I major histocompatibility complex antigens. Annu. Rev. Immunol. 1990;8:501–530. doi: 10.1146/annurev.iy.08.040190.002441. [DOI] [PubMed] [Google Scholar]
- Lindahl K.F., Byers D.E., Dabhi V., Hovik R., Jones E.P., Smith G.P., Wang C.-R., Xiao H., Yoshino M. H2-M3, a full service class Ib histocompatibility antigen. Annu. Rev. Immunol. 1997;15:851–879. doi: 10.1146/annurev.immunol.15.1.851. [DOI] [PubMed] [Google Scholar]
- Porcelli S.A. The CD1 familya third lineage of antigen-presenting molecules. Adv. Immunol. 1995;59:1–98. doi: 10.1016/s0065-2776(08)60629-x. [DOI] [PubMed] [Google Scholar]
- Zeng Z.-H., Castano A.R., Segelke B.W., Stura E.A., Peterson P.A., Wilson I.A. Crystal structure of mouse CD1an MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- Beckman E.M., Porcelli S.A., Morita C.T., Behar S.M., Furlong S.T., Brenner M.B. Recognition of a lipid antigen by CD1-restricted alpha/beta T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- Sieling P.A., Chatterjee D., Porcelli S.A., Prigozy T.I., Mazzaccaro R.J., Soriano T., Bloom B.R., Brenner M.B., Kronenberg M., Brennan P.J., Modlin R.L. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227–230. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- Wang C.R., Castano A.R., Peterson P.A., Slaughter C., Lindahl K.F., Deisenhofer J. Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3. Cell. 1995;82:655–664. doi: 10.1016/0092-8674(95)90037-3. [DOI] [PubMed] [Google Scholar]
- Kaufmann S.H.E., Hug E., DeLibero G. Listeria monocytogenes reactive T lymphocyte clones with cytolytic activity against infected target cells. J. Exp. Med. 1986;164:363–368. doi: 10.1084/jem.164.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamer E.G., Sijts A.J.A.M., Villanueva M.S., Busch D.H., Vijh S. MHC class I antigen processing of Listeria monocytogenes proteinsimplications for dominant and subdominant CTL responses. Immunol. Rev. 1997;158:129–136. doi: 10.1111/j.1600-065x.1997.tb00999.x. [DOI] [PubMed] [Google Scholar]
- Pamer E.G., Wang C.R., Flaherty L., Lindahl K.F., Bevan M.J. H2-M3 presents a Listeria monocytogenes peptide to cytotoxic T lymphocytes. Cell. 1992;70:215–223. doi: 10.1016/0092-8674(92)90097-v. [DOI] [PubMed] [Google Scholar]
- Kurlander R.J., Shawar S.M., Brown M.L., Rich R.R. Specialized role for a murine class I-b MHC molecule in prokaryotic host defenses. Science. 1992;257:678–679. doi: 10.1126/science.1496381. [DOI] [PubMed] [Google Scholar]
- Kaufmann S.H.E., Rodewald H.R., Hug E., Libero G.D. Cloned Listeria monocytogenes specific non-MHC-restricted Lyt-2+ T cells with cytolytic and protective activity. J. Immunol. 1988;140:3173–3179. [PubMed] [Google Scholar]
- Lenz L.L., Dere B., Bevan M.J. Identification of an H2-M3-restricted Listeria epitopeimplications for antigen presentation by M3. Immunity. 1996;5:63–72. doi: 10.1016/s1074-7613(00)80310-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulden P.H., Fischer P., Sherman N.E., Wang W., Engelhard V.H., Shabanowitz J., Hunt D.H., Pamer E.G. A Listeria monocytogenes pentapeptide is presented to cytolytic T lymphocytes by the H2-M3 MHC class Ib molecule. Immunity. 1996;5:73–79. doi: 10.1016/s1074-7613(00)80311-8. [DOI] [PubMed] [Google Scholar]
- Princiotta M.F., Lenz L.L., Bevan M.J., Staerz U.D. H2-M3 restricted presentation of a Listeria-derived leader peptide. J. Exp. Med. 1998;187:1711–1720. doi: 10.1084/jem.187.10.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch D.H., Pilip I.M., Vijh S., Pamer E.G. Coordinate regulation of complex T cell populations responding to bacterial infection. Immunity. 1998;8:353–362. doi: 10.1016/s1074-7613(00)80540-3. [DOI] [PubMed] [Google Scholar]
- Busch D.H., Pilip I., Pamer E.G. Evolution of a complex T cell receptor repertoire during primary and recall bacterial infection. J. Exp. Med. 1998;188:61–70. doi: 10.1084/jem.188.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garboczi D.N., Utz U., Ghosh P., Seth A., Kim J., VanTienhoven E.A.E., Biddison W.E., Wiley D.C. Assembly, specific binding, and crystallization of a human TCR-alphabeta with an antigenic Tax peptide from human T lymphotropic virus type 1 and the class I MHC molecule HLA-A2. J. Immunol. 1996;157:5403–5410. [PubMed] [Google Scholar]
- Vijh S., Pamer E.G. Immunodominant and subdominant CTL responses to Listeria monocytogenes infection. J. Immunol. 1997;158:3366–3371. [PubMed] [Google Scholar]
- Morse M.-C., Bleau G., Dabhi V.M., Hetu F., Drobetsky E.A., Lindahl K.F., Perreault C. The COI mitochondrial gene encodes a minor histocompatibility antigen presented by H2-M3. J. Immunol. 1996;156:3301–3307. [PubMed] [Google Scholar]
- Schatz P.J. Use of peptide libraries to map the substrate specificity of a peptide modifying enzymea 13 residue consensus peptide specifies biotinylation in Escherichia coli . Biotechnology. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Altman J.D., Moss P.A.H., Goulder P.J.R., Barouch D.H., McHeyzer-Williams M.G., Bell J.I., McMichael A.J., Davis M.M. Phenotypic analysis of antigen specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- Nataraj C., Huffman G.R., Kurlander R.J. H2M3wt-restricted, Listeria monocytogenes-immune CD8 T cells respond to multiple formylated peptides and to a variety of gram-positive and gram-negative bacteria. Int. Immunol. 1998;10:7–15. doi: 10.1093/intimm/10.1.7. [DOI] [PubMed] [Google Scholar]
- Ahmed R., Gray D. Immunological memory and protective immunityunderstanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- Busch D.H., Pamer E.G. T cell affinity maturation by selective expansion during infection. J. Exp. Med. 1999;189:701–709. doi: 10.1084/jem.189.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas J.M., Rich R.R., Howell D.D., Shawar S.M., Rodgers J.R. Availability of endogenous peptides limits expression of an M3a-Ld major histocompatibility complex class I chimera. J. Exp. Med. 1994;179:155–165. doi: 10.1084/jem.179.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz L.L., Bevan M.J. CTL responses to H2-M3-restricted Listeria epitopes. Immunol. Rev. 1997;158:115–121. doi: 10.1111/j.1600-065x.1997.tb00997.x. [DOI] [PubMed] [Google Scholar]
- Cheers C., McKenzie L., Pavlov H., Waid C., York J. Resistance and susceptibility of mice to bacterial infectioncourse of Listeriosis in resistant or susceptible mice. Infect. Immun. 1978;19:763–770. doi: 10.1128/iai.19.3.763-770.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]