Abstract
Lipopolysaccharide (LPS) fluorescently labeled with boron dipyrromethane (BODIPY) first binds to the plasma membrane of CD14-expressing cells and is subsequently internalized. Intracellular LPS appears in small vesicles near the cell surface and later in larger, punctate structures identified as the Golgi apparatus. To determine if membrane (m)CD14 directs the movement of LPS to the Golgi apparatus, an mCD14 chimera containing enhanced green fluorescent protein (mCD14–EGFP) was used to follow trafficking of mCD14 and BODIPY–LPS in stable transfectants. The chimera was expressed strongly on the cell surface and also in a Golgi complex–like structure. mCD14–EGFP was functional in mediating binding of and responses to LPS. BODIPY–LPS presented to the transfectants as complexes with soluble CD14 first colocalized with mCD14–EGFP on the cell surface. However, within 5–10 min, the BODIPY–LPS distributed to intracellular vesicles that did not contain mCD14–EGFP, indicating that mCD14 did not accompany LPS during endocytic movement. These results suggest that monomeric LPS is transferred out of mCD14 at the plasma membrane and traffics within the cell independently of mCD14. In contrast, aggregates of LPS were internalized in association with mCD14, suggesting that LPS clearance occurs via a pathway distinct from that which leads to signaling via monomeric LPS.
Keywords: enhanced green fluorescent protein, U373 cells, intracellular trafficking
Cells that express plasma membrane–bound (m)CD14,1 either naturally or through transfection, bind bacterial LPS. Monomeric LPS is removed from LPS aggregates and reaches mCD14 by a two-step process in serum 1. The serum protein LPS binding protein (LBP) binds to LPS aggregates and transfers monomers to a soluble (s) form of CD14, also present in serum. sCD14, in turn, may transfer LPS either to serum lipoprotein particles, causing LPS neutralization, or to cells, leading to production of cytokines and other responses. Transfer of LPS between CD14 molecules is a rapid process 2, suggesting that the movement of LPS from sCD14 to mCD14 on mCD14-bearing cells is likely to be more rapid than the movement of LPS from sCD14 to the surfaces of cells that do not express CD14.
The importance of mCD14 to the binding of LPS and the cellular responses that follow is underscored by a variety of observations. Blockade of LPS binding to mCD14 on leukocytes with mAbs 3 abrogates responses such as cytokine production by monocytes 4 and enhanced integrin-dependent adhesion by neutrophils 2 5. In addition, paroxysmal nocturnal hemoglobinuria, a disease characterized by a variable deficiency in the expression of mCD14 and other glycosylphosphatidyl inositol (GPI)-linked proteins on hemopoietic cells, renders monocytes less sensitive to LPS 6 7 8. Perhaps the strongest evidence for the primacy of mCD14 in mediating responses to LPS is the observation that monocytes from CD14-deficient mice show a strongly attenuated cytokine response to LPS 9. Moreover, transfection with mCD14 endows various cell types that do not otherwise express CD14 with the ability either to become responsive to LPS 10 or to become sensitive to much lower concentrations of LPS 11 12. Thus, mCD14 serves as the initial site of interaction between LPS and the surfaces of CD14-bearing cells.
Several lines of evidence suggest that after binding to mCD14, LPS must be internalized to initiate intracellular signaling leading to cellular responses. Recently, we and others have observed that fluorescently labeled LPS is rapidly endocytosed when it is presented to neutrophils or cultured human monocytes as monomeric LPS complexed with sCD14 13 14. In neutrophils, integrin-mediated adhesion in response to LPS exhibits a lag of 10–20 min and is completely blocked by phosphatidyl inositol 3 kinase inhibitors or by lowering the temperature to 19°C, two treatments that prevent vesicular transport 13. Thieblemont and Wright demonstrated that macrophages from mice with the LPS-hyporesponsive (Lpsd) trait are defective in vesicular transport of LPS 14. Furthermore, LPS antagonists block both the transport of LPS inside the cell 15 and cellular responses to LPS. Internalization of LPS may therefore be a key process for eliciting responses from cells expressing mCD14.
Although LPS internalization has been directly observed under a variety of circumstances, the precise role of mCD14 in this process is not known. The ability of CD14 to transfer LPS to high density lipoprotein particles 16 and phospholipid micelles 17 suggests that mCD14 might participate in the catalytic transfer of LPS to the lipid bilayer of the plasma membrane. LPS may then move within the cell independently of mCD14 or in a vesicle also containing mCD14. Alternatively, LPS may remain bound to mCD14 while being internalized, exhibiting the behavior of a classic ligand–receptor complex.
To determine whether mCD14 traffics along with LPS from the plasma membrane to intracellular compartments, we constructed a GPI-anchored fusion protein of CD14 and enhanced green fluorescent protein (EGFP) and used it to follow the location of mCD14 in the astrocytoma cell line U373. The fusion protein was expressed on the cell surface in a fully functional form. Expression of mCD14–EGFP endowed U373 cells with the ability to internalize detectable amounts of fluorescently labeled LPS (BODIPY–LPS). Using confocal microscopy, we observed that BODIPY–LPS presented in a monomeric form colocalized with mCD14 at the cell surface but distributed to intracellular locations without mCD14 as soon as internalization of LPS could be detected. Our observations suggest that monomeric LPS is rapidly transferred out of mCD14 at the plasma membrane and traffics within the cell independently of mCD14. Additional studies indicate that LPS aggregates are trafficked differently from monomers. LPS aggregates are known to be slowly internalized and directed to lysosomes 18, and we find that mCD14 remains associated with LPS aggregates during internalization.
Materials and Methods
Construction of the mCD14–EGFP and mEGFP Chimeras.
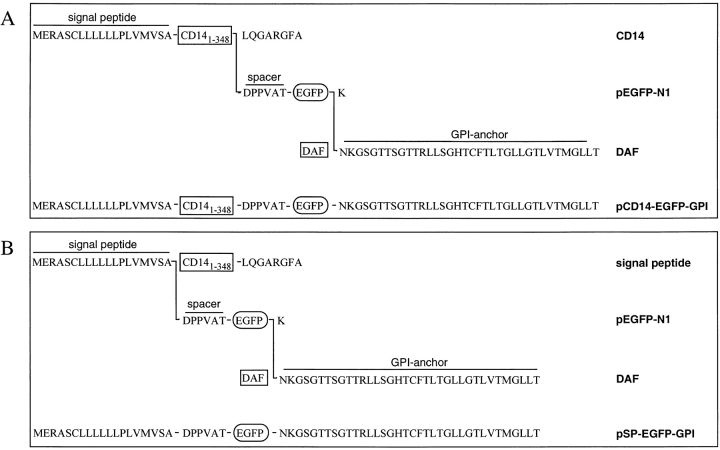
pEGFP-N1 (Clontech) encodes EGFP, a bright red–shifted variant of GFP containing the amino acid substitutions phenylaline-64→leucine and serine-65→threonine of GFPmut1 19. An mCD14–EGFP gene fusion was constructed in pEGFP-N1 in two steps. First, a 1.2-kb BamHI fragment coding for the entire CD14 protein minus the last eight COOH-terminal residues was inserted into BamHI-digested pEGFP-N1, yielding pCD14–EGFP. Second, a 116-bp BsrGI–NotI fragment, encoding the 36 COOH-terminal residues of decay accelerating factor (DAF) and a stop codon, was inserted into BsrGI- plus NotI-digested pCD14–EGFP, yielding pCD14–EGFP–GPI. The amino acid sequence of the fusion protein coded by this construct is shown (see Fig. 1 A).
Figure 1.
Structure of the mCD14–EGFP and mEGFP chimeras. (A) mCD14–EGFP. The gene fusion carried by pCD14–EGFP–GPI encoded a protein comprising the NH2-terminal signal peptide and the first 348 amino acids of CD14, a hexapeptide spacer DPPVAT, EGFP minus the last COOH-terminal amino acid (K), and a 36 COOH-terminal amino acid sequence from DAF signaling for attachment of a GPI anchor. (B) mEGFP. The gene fusion carried by pSP–EGFP–GPI encoded a protein comprising the NH2-terminal SP of CD14, a hexapeptide spacer DPPVAT, EGFP minus the last COOH-terminal amino acid (K), and a 36 COOH-terminal amino sequence from DAF for a GPI anchor.
Similarly, an mEGFP gene fusion was constructed by first inserting a 116-bp BsrGI–NotI fragment, encoding the 36 COOH-terminal residues of DAF and a stop codon, into BsrGI- plus NotI-digested pEGFP-N1, yielding pEGFP–GPI. Then, a 134-bp SalI–BamHI fragment coding for the first 19 NH2-terminal residues, the signal peptide (SP) of CD14, was inserted into SalI- plus BamHI-digested pEGFP–GPI, yielding pSP–EGFP–GPI. The sequence of the fusion protein coded by this construct is shown (see Fig. 1 B). These constructs placed the mCD14–EGFP and mEGFP chimeras under the control of the cytomegalovirus promoter/enhancer and permitted the selection of stable clones using geneticin.
The 1.2-kb BamHI fragment encoding most of CD14 was synthesized by PCR using pcDNAI-neo-CD14 as a template 20 and the primers 5′-GAG atg gat cca cca tgg agc gcg cgt cct gc-3′ and 5′-GAG ATG GAT CCA GCA CCA GGG TTC CCG A-3′. The 116-bp BsrGI–NotI fragment encoding part of DAF was synthesized by RT-PCR using total RNA from human monocytes as a template and the primers 5′-aat atg tac aat aaa gga agt gga acc ac-3′ and 5′-taa agc ggc cgc taa gtc agc aag ccc at-3′. The 134-bp SalI–BamHI fragment was synthesized by RT-PCR using total RNA from human neutrophils as a template and the primers 5′-ACG CGT CGA CGC CGC TGT GTA GGA AAG-3′ and 5′-CGC GGA TCC GCA GAG ACG TGC ACC Aat-3′. All syntheses were followed by digestion with the appropriate restriction enzymes and gel purification. Both RT-PCR amplifications were performed using the Gene Amp RNA PCR kit purchased from Perkin-Elmer Corp. The PCR insertions were sequenced to confirm the absence of PCR synthesis errors.
U373 Cell Lines.
U373 cells were grown as monolayers in RPMI (BioWhittaker, Inc.) supplemented with 10% heat-inactivated fetal bovine serum (FBS; BioWhittaker, Inc.), penicillin/streptomycin (100 U/ml and 100 μg/ml, respectively), and 2 mM glutamine. For making stable transfectants, 105 cells from a confluent culture of U373 cells were seeded on a 35-mm cell culture dish and grown to subconfluence for 24–48 h before transfection with either pCD14–EGFP–GPI or pSP–EGFP–GPI. For each dish, 1–2 μg of highly purified expression plasmid was used for transfection with 6 μl lipofectamine (GIBCO BRL) according to the manufacturer's instructions. The DNA–lipofectamine mixture remained on the cells for 6 h at 37°C and was then replaced by RPMI with 10% FBS and 2 mM glutamine without antibiotics. 72 h after transfection, the cells were trypsinized, plated at clonal density, and selected with 0.5 mg/ml geneticin (GIBCO BRL). After 3 wk, surviving cell colonies were visually screened for fluorescence. Several positive clones were identified, isolated using cloning rings, and expanded into cell lines for further analysis. U373–CD14 cells were obtained by selecting clones of U373 cells stably transfected with pcDNAI-neo-CD14 as described elsewhere 20.
sCD14 and LPS.
Recombinant human sCD14 was purified from conditioned medium of Schneider-2 insect cells transfected with cDNA encoding human CD14 as previously described 21. LPS from Salmonella minnesota R595 was purchased from List Biological Labs. The fluoroprobe BODIPY 558/568 (Molecular Probes, Inc.) was conjugated to unlabeled LPS micelles as previously described 22.
LPS–sCD14 and BODIPY–LPS–sCD14 complexes were formed by incubating LPS or BODIPY–LPS (20 μg/ml), respectively, with sCD14 (500 μg/ml) overnight at 37°C in Dulbecco's PBS (BioWhittaker, Inc.) with 0.5% pyrogen-free human serum albumin (Centeon, Armour, and Berring Pharmaceutical Co.). Previous work has shown that under these conditions all of the LPS forms stoichiometric complexes with monomeric sCD14 and that these complexes efficiently stimulate cells and deliver LPS to the plasma membrane 2 3.
BODIPY–LPS aggregates were prepared by incubating BODIPY–LPS at 1 μg/ml in FBS for 10 min at 37°C. The aggregation state of LPS was verified by monitoring its fluorescence emission at 568 nm before and after the addition of detergent, as described elsewhere 22. Adding 2% SDS to the BODIPY–LPS aggregates led to a 10-fold increase in fluorescence due to the loss of self-quenching as monomers were released from the aggregates. In addition, aggregates observed directly by fluorescence microscopy exhibited a pointillistic pattern of fluorescence, rather than the very diffuse fluorescence seen with BODIPY–LPS–sCD14 complexes.
Confocal Microscopy.
U373 transfectants were cultured for 24–48 h before experiments in RPMI without phenol red (BioWhittaker, Inc.) supplemented with 10% FBS, antibiotics, and 2 mM glutamine on glass chamber slides (Nunc, Inc.) precoated with 0.5% gelatin (Sigma Chemical Co.). The cells were washed twice in HAP buffer (Dulbecco's PBS, 0.05% human serum albumin, and 3 mM d-glucose, containing 0.5 U/ml of aprotinin) and incubated in HAP at 37°C with or without LPS–sCD14, BODIPY–LPS–sCD14, or BODIPY–LPS aggregates. Slides were washed twice with HAP and further incubated at 37°C. At the end of the incubation, the plastic chamber and silicone gasket were removed, and the slide was mounted in HAP for immediate microscopic observation. For removal of cell surface mCD14–EGFP with phosphatidyl inositol phospholipase C (PI-PLC), cells on slides were incubated for 1 h in 20 mM Hepes, pH 7.4, and 150 mM NaCl on ice with 5 U/ml of PI-PLC (Boehringer Mannheim).
When anti-CD14 mAb 26ic or 60b 23 was used, cells were incubated with the antibody at 10 μg/ml in HAP buffer at 4°C for 30 min and washed twice with ice-cold HAP and once with HAP at 37°C just before adding the BODIPY–LPS–sCD14 complexes.
Confocal scanning laser microscopy was performed using a Nikon Optiphot-2 microscope with a ×60 objective (NA 1.4) and Bio-Rad MRC 1024 instrumentation with a krypton/argon laser. Unless otherwise noted, each image represents a single Kalman averaged (6–10 scans) optical section collected with a 2–3-mm-diameter iris aperture. Optical sections were collected digitally and analyzed using LaserSharp software (Bio-Rad Labs.). For two-color images, each color was acquired sequentially. This was necessary because EGFP has a broad peak of fluorescence, and some signal bleedthrough was observed in the BODIPY–LPS channel when simultaneous collection was attempted.
Intracellular Fluorescence.
Quenching of cell surface fluorescence by trypan blue was employed to both quantitate and observe the distribution of intracellular mCD14–EGFP and mEGFP. U373 transfectants were grown to confluence in a 96-well culture plate and, after the experiment, the total fluorescence associated with the cells was measured using a Cytofluor 4000 (PE Biosystems) (excitation 485 nm, emission 530 nm). Trypan blue (200 μg/ml, ambient temperature) was added to the wells to quench fluorescence from cell surface EGFP, and the remaining fluorescence from intracellular mCD14–EGFP or mEGFP was immediately measured. Intracellular fluorescence is expressed as a percent of total fluorescence from triplicate samples. Trypan blue was also used to quench cell surface EGFP on U373 transfectants before observing the cells by confocal microscopy. After the experimental manipulation, U373 transfectants cultured on glass chamber slides were washed once and mounted in HAP containing 200 μg/ml trypan blue before immediate observation.
IL-6 Production by U373 Cells.
U373 transfectants grown in 96-well cell culture plates were washed extensively with AIM-V serum–free medium (GIBCO BRL) and incubated as indicated in AIM-V medium containing 0.5 mg/ml human serum albumin. After 16 h at 37°C, the overlying medium was collected from each well and assayed for IL-6 by ELISA as described 24.
SDS-PAGE and Western Blotting.
For each condition, 106 cells were lysed by incubation on ice for 20 min in 300 μl of 100 mM Tris/HCl, pH 8.0, 100 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.3 U/ml aprotinin, 2 mM PMSF, 3 mM diisopropyl fluorophosphate, 50 μg/ml benzamidine, and 5 μg/ml each of antipain, leupeptin, chymostatin, and pepstatin A. Lysates were centrifuged for 15 min at 12,000 g, and the supernatants were prepared for SDS-PAGE under reducing conditions. SDS-PAGE was run on a 4–20% gradient Tris–glycine gel (Novex). Proteins were electrotransferred to nitrocellulose membranes and detected with either an anti-EGFP rabbit pAb (Clontech) or an anti-CD14 pAb 25. Horseradish peroxidase–conjugated goat anti–rabbit IgG was used as the secondary antibody, and the enzymatic reaction was detected with an ECL kit (Amersham Corp.). Purified rEGFP was obtained from Clontech.
Results
EGFP-tagged mCD14 Is Expressed on the Surfaces of U373 Cells.
To generate a fluorescently tagged CD14 that was attached to the membrane via a GPI anchor, we fused a sequence coding for CD14 to one coding for EGFP and added the sequence for the 36 COOH-terminal residues of DAF. The chimeric protein resulting from the gene fusion pCD14–EGFP–GPI, shown in Fig. 1, contains the NH2-terminal signal peptide of CD14 and the COOH-terminal signal peptide of DAF. Thus, when it was expressed and processed, it would translocate normally into the endoplasmic reticulum and have a GPI anchor attached. For a control, we also engineered a chimeric EGFP with a GPI anchor that contained the NH2-terminal signal peptide (SP) of CD14 and the COOH-terminal signal peptide of DAF. This second chimera was used to compare the distribution of EGFP-tagged CD14 (mCD14–EGFP) with a generic GPI-anchored protein (mEGFP).
The two gene fusion products were transfected into the U373 astrocytoma cell line, which does not express mCD14. Stable transfectants for mCD14–EGFP and mEGFP were designated U373–CD14–EGFP and U373–EGFP, respectively. One clone of U373–CD14–EGFP was used for the results presented, but a second clone gave identical results.
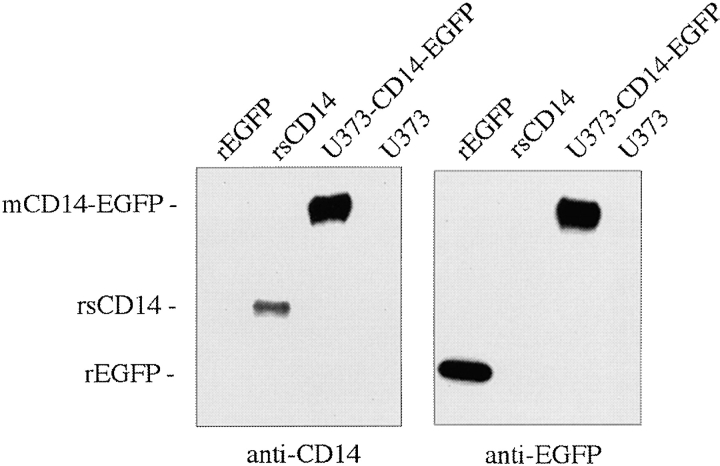
Expression of mCD14–EGFP as an intact polypeptide in U373–CD14–EGFP was tested in Western blots of cell lysates run in parallel with rsCD14 and EGFP. Antibodies against either CD14 or EGFP recognized the appropriate control protein and the same single band at 80 kD in U373–CD14–EGFP (Fig. 2). The band corresponded to a protein of the expected mass for mCD14–EGFP and was not present in cell lysates of untransfected U373. Thus, mCD14–EGFP was expressed in U373 cells as an intact polypeptide, and we can be confident that by observing EGFP fluorescence we are also observing the location of mCD14.
Figure 2.
mCD14–EGFP is expressed as a single polypeptide in U373 cells. Lysates from U373–CD14–EGFP and U373 cells (10 μl from each) were run on SDS-PAGE along with 50 ng each of recombinant purified EGFP and sCD14 in the indicated lanes. Western blots were probed with either anti-CD14 or anti-EGFP pAb.
We further confirmed by confocal microscopy that both mCD14–EGFP and mEGFP were expressed on the surfaces of U373 cells. Optical sections of live U373–CD14–EGFP revealed fluorescence associated with the plasma membrane that was relatively uniform in distribution (Fig. 3). Fluorescence was also observed on fine, filamentous projections from the cell surface. Intracellular mCD14–EGFP was observed in a juxtanuclear reticulum, a structure characteristic of the Golgi apparatus. A similar labeling pattern was observed with U373–EGFP (data not shown), suggesting that this is a normal distribution of GPI-anchored proteins in U373 cells.
Figure 3.
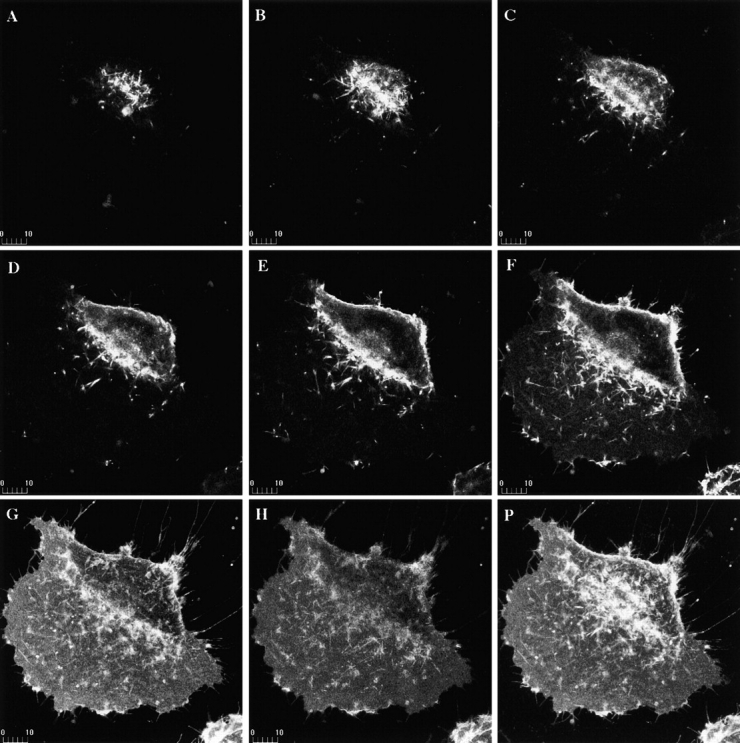
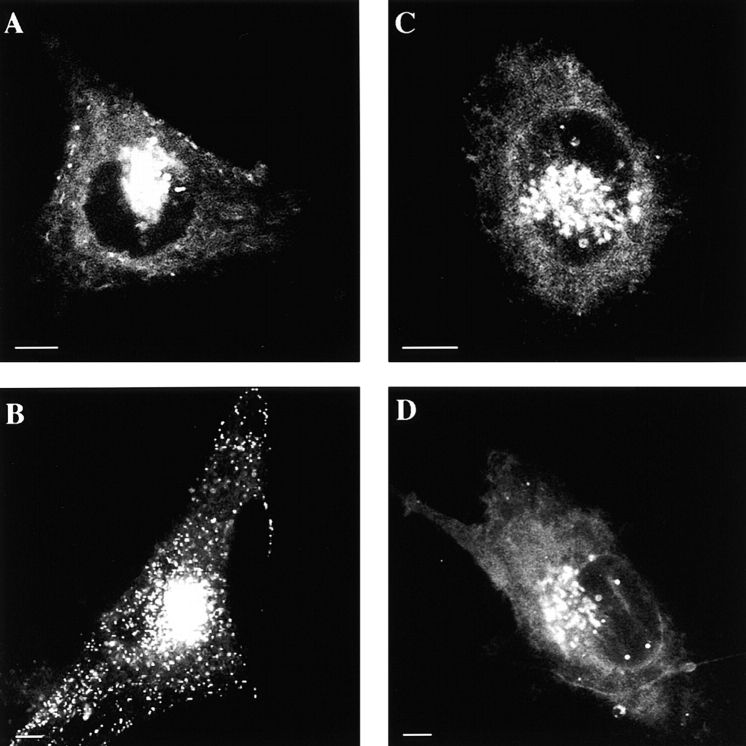
mCD14–EGFP is expressed on the surfaces of U373 cells. Confocal microscopy was used to collect serial optical sections of a U373–CD14–EGFP cell at 1-μm intervals, beginning at the top of the cell (A–H). A projection of all the sections is shown (P). mCD14–EGFP was localized to the cell surface and Golgi apparatus, consistent with its being expressed and transported to the plasma membrane.
To confirm that the mCD14–EGFP was attached to the plasma membrane via a GPI anchor, we digested U373–CD14–EGFP cells with PI-PLC, which cleaves GPI-anchored proteins from their GPI anchors, before observing them by confocal microscopy. After 1-h treatment at 4°C with PI-PLC, cell surface fluorescence on U373–CD14–EGFP was barely detectable (not shown), suggesting that most of the cell surface associated mCD14–EGFP was removed.
EGFP-tagged mCD14 Enhances Responses of U373 Cells to LPS.
U373 astrocytoma cells do not respond to LPS alone, but when LPS is added in the presence of sCD14, they produce IL-6 26. Transfection of U373 cells with mCD14 allows responses to LPS in the absence of sCD14. More importantly, it greatly increases the sensitivity of these cells to LPS–sCD14 complexes and allows more rapid responses to LPS–sCD14. This is consistent with observations that mCD14 on neutrophils and monocytes is necessary for responses to LPS in the absence of sCD14 and greatly increases the sensitivity of their responses to LPS–sCD14 complexes 2 9.
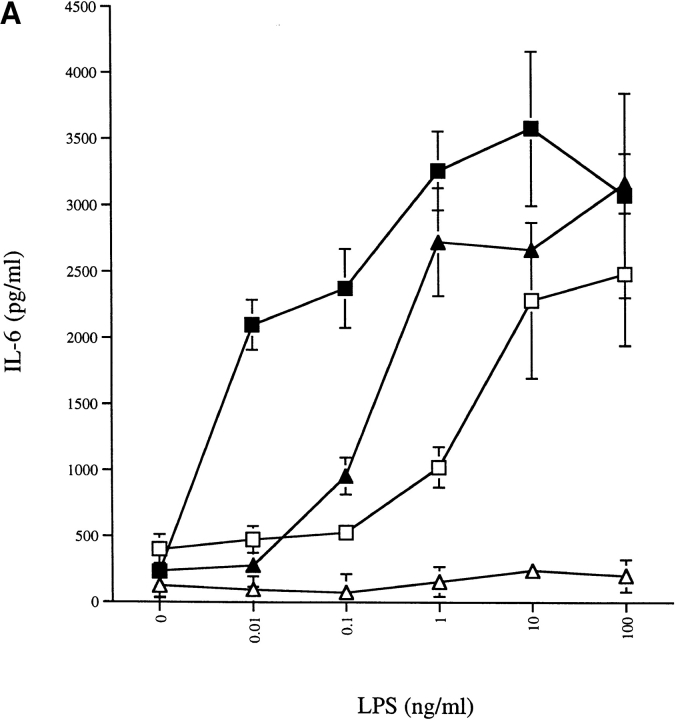
To show that the mCD14–EGFP chimera was functional, we measured secretion of IL-6 by U373–CD14–EGFP and U373–EGFP cells in response to either LPS or LPS–sCD14 complexes. The U373–EGFP cells did not respond to LPS alone up to 100 ng/ml (Fig. 4 A). In contrast, expression of mCD14–EGFP in U373 cells led to IL-6 production in response to concentrations of LPS of 1 ng/ml or higher. Similarly, U373–CD14–EGFP cells responded to concentrations of LPS–sCD14 ∼100-fold lower than those required to elicit the same response in U373–EGFP cells (Fig. 4 A). The enhancement of sensitivity by mCD14–EGFP was quantitatively similar to that previously observed with mCD14. Thus, the presence of EGFP does not affect the function of mCD14–EGFP on cells.
Figure 4.
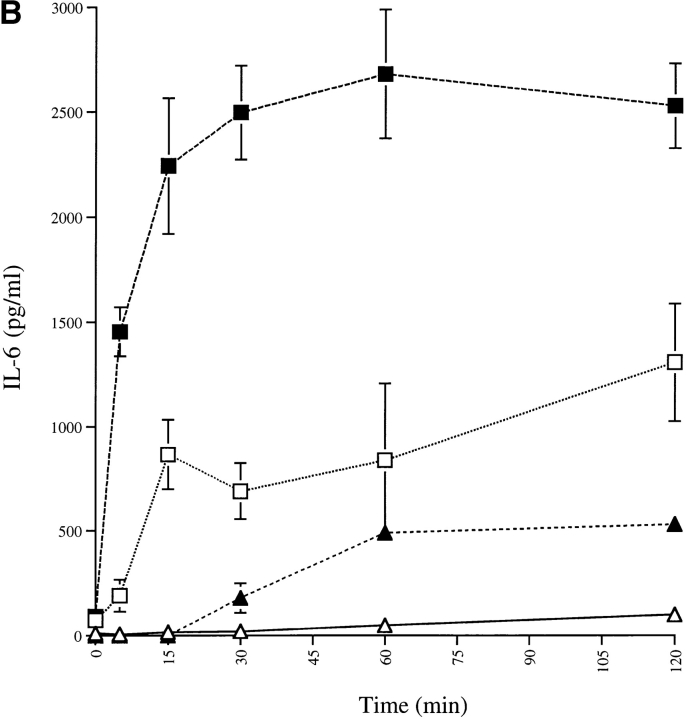
Expression of mCD14–EGFP in U373 cells enhances the sensitivity and speed of responses to LPS. (A) U373–CD14–EGFP respond to lower concentrations of LPS than U373–EGFP. U373–CD14–EGFP (□, ▪) and U373–EGFP (▵, ▴) cells were incubated with the indicated concentrations of LPS (□, ▵) or LPS–sCD14 complexes (▪, ▴) for 16 h at 37°C, and IL-6 secretion was measured by ELISA. (B) U373–CD14–EGFP respond to LPS more rapidly than U373–EGFP. U373–CD14–EGFP (□, ▪) and U373–EGFP (▵, ▴) cells were incubated with LPS (□, ▵; 40 ng/ml) or LPS–sCD14 complexes (▪, ▴; 40 ng/ml LPS, 1 μg/ml sCD14) for the indicated times at 37°C. Cells were then washed extensively and further incubated for 16 h at 37°C. IL-6 secretion was measured by ELISA. Results in A and B are expressed as the means of triplicate wells ± SEM and are from an experiment performed three times with the same result.
To determine the time of exposure required to elicit a response to LPS, U373–CD14–EGFP and U373–EGFP cells were given a fixed concentration of LPS (40 ng/ml), either alone or in sCD14 complexes, for increasing intervals of time before washing and incubation to allow synthesis of IL-6. U373–EGFP cells were essentially unresponsive to LPS alone after up to 2 h of exposure (Fig. 4 B). As previously seen with mCD14, the presence of mCD14–EGFP dramatically enhanced the magnitude and speed of cellular responses to LPS. In particular, after as little as 5 min of exposure to LPS, U373–CD14–EGFP cells responded with IL-6 production to both LPS–sCD14 complexes (Fig. 4 B) and LPS aggregates (data not shown).
Taken together, these results suggest that mCD14–EGFP was able to bind LPS and mediate cellular responses to it. All of the properties exhibited by U373–CD14–EGFP are the same as those of mCD14 expressed in U373, indicating that mCD14 expressed as a chimera with EGFP was still a functional entity. These results further suggest that the distribution and trafficking pattern of mCD14–EGFP in U373 would reflect that of a fully functional protein.
LPS Binds to U373–CD14–EGFP and Colocalizes with mCD14 on the Cell Surface.
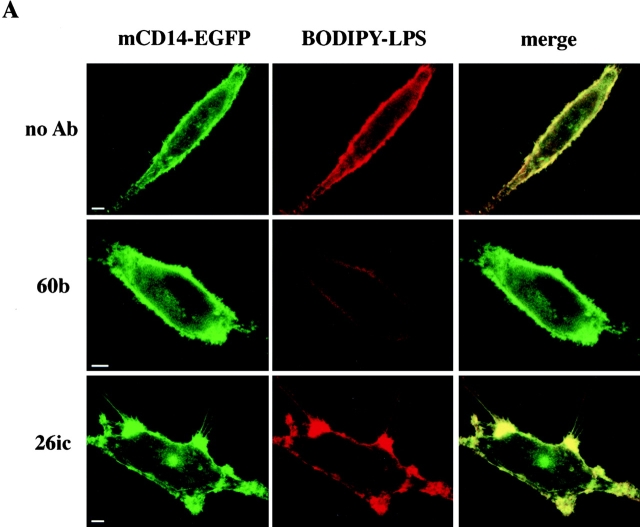
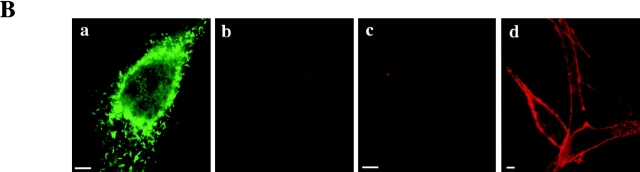
The association of LPS with mCD14–EGFP was confirmed by confocal microscopy. BODIPY–LPS in the form of monomeric complexes with sCD14 was incubated with U373–CD14–EGFP for 2–3 min at 37°C before washing and observation. Fluorescence from BODIPY–LPS was seen associated with the plasma membrane of U373–CD14–EGFP (Fig. 5 A, no Ab, red panel), indicating that it was successfully transferred from sCD14. There was colocalization of BODIPY–LPS with mCD14–EGFP on the cell surface, as demonstrated by the overlap in fluorescence signals (Fig. 5 A, no Ab, merge panel). The same cell surface distribution of BODIPY–LPS was observed with U373 transfectants expressing mCD14 without EGFP attached (Fig. 5 B, panel d). In contrast, U373–EGFP cells incubated briefly with BODIPY–LPS–sCD14 complexes did not have detectable BODIPY–LPS fluorescence associated with their cell surfaces (Fig. 5 B, panels a and b).
Figure 5.
LPS and mCD14–EGFP colocalize on the cell surface. (A) U373–CD14–EGFP cells were incubated in HAP buffer with BODIPY–LPS–sCD14 (40 ng/ml LPS) complexes for 3 min at 37°C in the presence or absence of anti-CD14 and either the blocking mAb 60b or the nonblocking mAb 26ic. Confocal optical sections for mCD14–EGFP and BODIPY–LPS fluorescence are shown for representative cells in the left and center panels, respectively, and the merged images are shown in the right panels. Yellow indicates regions where the signals overlap. In the absence of antibody or presence of 26ic, there was colocalization of mCD14–EGFP and BODIPY–LPS on the cell surface. In the presence of 60b, binding of LPS was inhibited, and there was no colocalization of label. (B) U373–EGFP (a and b), U373 (c), and U373-CD14 (d) were incubated in HAP buffer with BODIPY–LPS–sCD14 (40 ng/ml LPS) complexes for 3 min at 37°C. Confocal optical sections for mEGFP (a) and BODIPY–LPS (b–d) are shown for representative cells. Bars, 10 μm.
To confirm that the transfer of BODIPY–LPS from sCD14 was mCD14 dependent, we pretreated U373–CD14–EGFP cells with a blocking anti-CD14 mAb, 60b 3. The antibody prevented binding of BODIPY–LPS, and no colocalization with mCD14–EGFP was observed (Fig. 5 A). Colocalization of BODIPY–LPS and mCD14–EGFP was still observed when the cells were pretreated with 26ic, a nonblocking anti-CD14 antibody 3 (Fig. 5 A). Thus, colocalization of BODIPY–LPS and mCD14 required binding of LPS to mCD14.
When Presented in a Monomeric Form, LPS Is Internalized Separately from mCD14.
To determine whether LPS remained bound to mCD14 after internalization, we observed the trafficking of mCD14–EGFP and BODIPY–LPS simultaneously in live cells. U373–CD14–EGFP cells were incubated with BODIPY–LPS–sCD14 complexes for 2–3 min at 37°C, the complexes were removed by washing, and the cells were incubated further at 37°C before observation. Particular attention was devoted to observations made 5–10 min after the incubation began. At the earliest time points, BODIPY–LPS was clearly visible in small intracellular vesicles (Fig. 6, 10 min, red panel). These vesicles were visible at times as early as 5 min (data not shown). However, the vesicles containing BODIPY–LPS did not contain mCD14–EGFP, as evidenced by the lack of overlap in fluorescent signals (Fig. 6, 10 min, merge panel). This indicates that in the brief time required for LPS to reach a vesicular location, LPS had separated from mCD14.
Figure 6.
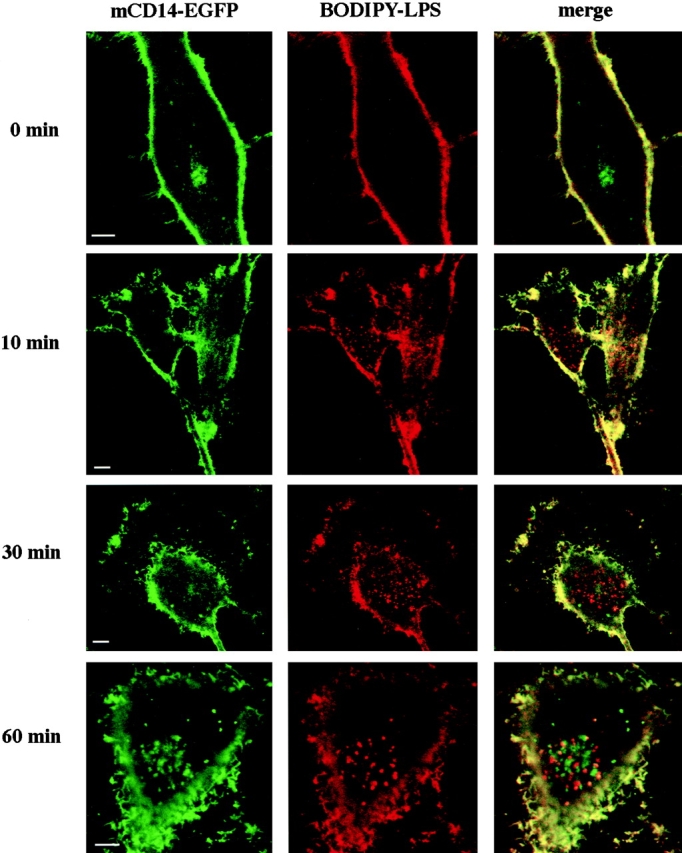
LPS moves into the cell without mCD14–EGFP. U373–CD14–EGFP cells were incubated in HAP buffer with BODIPY–LPS–sCD14 (40 ng/ml LPS) complexes for 3 min at 37°C, washed, and incubated further for 0, 10, 30, or 60 min at 37°C. Confocal optical sections for mCD14–EGFP and BODIPY–LPS fluorescence are shown for representative cells in the left and center panels, respectively, and the merged images are shown in the right panels. Yellow indicates the regions where the signals overlap. Although there was colocalization of mCD14–EGFP and BODIPY–LPS on the cell surface, internalized BODIPY–LPS did not colocalize with mCD14–EGFP. Bars, 10 μm.
At later times, vesicles containing BODIPY–LPS accumulated predominantly in the perinuclear area (Fig. 6 and 60 min, red panels). Although by 60 min a punctate localization of mCD14–EGFP in the same vicinity was observed (Fig. 6, 60 min, green panel), repeated observations with LPS concentrations between 10 and 200 ng/ml demonstrated that colocalization of BODIPY–LPS and mCD14–EGFP in this area did not occur (not shown). LPS concentration therefore had no effect on its localization. Thus, not only did LPS leave mCD14 upon entering the cells, but it also did not reassociate with mCD14 in any intracellular compartment.
The kinetics of BODIPY–LPS internalization in U373–CD14–EGFP were identical to those in U373 expressing mCD14 without EGFP (data not shown). Furthermore, the distribution of intracellular BODIPY–LPS in vesicles and their subcellular localization was the same in both cell types (data not shown). This indicates that the presence of EGFP on mCD14 did not disturb the normal trafficking pattern of LPS in U373 cells.
Intracellular CD14 Is Exocytosed in Response to LPS.
Although there was no apparent change in the distribution of mCD14–EGFP in response to LPS in the colocalization studies, the bright cell surface fluorescence prevented observation of any changes in the distribution of intracellular mCD14–EGFP. To better observe the distribution of intracellular mCD14–EGFP, we quenched the cell surface fluorescence on U373–CD14–EGFP cells with trypan blue (Fig. 7A and Fig. B). In addition to the bright fluorescence emanating from the Golgi apparatus area, numerous fluorescent vesicles of ∼50–100-nm average diameter were distributed throughout the cytoplasm. The vesicles were particularly evident near the basal aspects of the cells (Fig. 7 B). A similar intracellular localization of mEGFP in the Golgi complex and cytoplasmic vesicles was also observed (data not shown). Thus, the vesicular compartment may represent either a component of the secretory pathway en route to the cell surface or a recycling compartment for GPI-anchored proteins.
Figure 7.
CD14 is present in intracellular vesicles that are exocytosed in response to LPS. U373–CD14–EGFP cells were incubated for 30 min at 37°C in HAP buffer in the absence (A and B) or presence (C and D) of preformed LPS–sCD14 complexes (40 ng/ml LPS). Trypan blue (200 μg/ml) was added to quench mCD14–EGFP on the cell surface, and the cells were examined by confocal microscopy. Most of the intracellular mCD14–EGFP was localized to a juxtanuclear reticulum (A and C), presumably the Golgi apparatus. In untreated cells, a pool of mCD14–EGFP was also observed in vesicles located throughout the cell but especially prevalent near the basal aspect (B). Most of the vesicular pool of intracellular mCD14–EGFP was no longer visible after stimulation of the cells with preformed LPS–sCD14 complexes (D). Bars, 10 μm.
The effect of LPS on the intracellular distribution of mCD14–EGFP was examined by adding LPS–sCD14 complexes to U373–CD14–EGFP cells and observing mCD14–EGFP localization by confocal microscopy after trypan blue quenching. The localization of mCD14–EGFP in the Golgi complex was unaffected by LPS stimulation (Fig. 7 C). However, after addition of LPS, we observed a steady decrease in the number of mCD14–EGFP-containing vesicles from the earliest times observed (2–5 min), with almost no mCD14–EGFP-containing vesicles remaining after 45–60 min (Fig. 7 D), suggesting that the compartments containing mCD14–EGFP were exocytosed upon exposure of the cells to LPS. Although the resolution of these experiments does not allow us to rule out the possibility that a small fraction of mCD14–EGFP was internalized, the decline in the number of intracellular vesicles suggests that exocytosis was the preferred route. A similar loss of fluorescent vesicles was observed in U373–EGFP cells exposed to LPS–sCD14 complexes for 45–60 min (data not shown), indicating that the vesicle pool in question is not defined by the presence of CD14 per se. These results suggest that LPS either induces the release of a compartment in U373 cells that contains GPI-anchored proteins or that it prevents the reinternalization of a recycled pool of membrane containing GPI-anchored proteins. Thus, rather than being internalized with LPS from the cell surface, mCD14 apparently moved to the cell surface from an intracellular store in response to LPS.
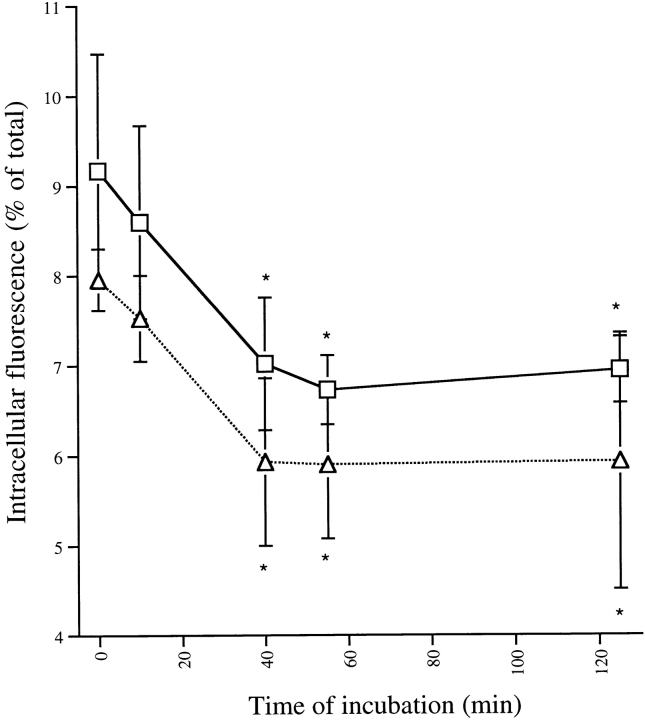
Exocytosis of the mCD14-containing vesicles was confirmed by quantitative measurements of the time course. U373–CD14–EGFP cells were grown in 96-well tissue culture plates, and both total and intracellular fluorescence was measured before and at various times after exposure to LPS–sCD14 complexes (see Materials and Methods). Stimulation with LPS did not induce any change in total fluorescence associated with the cells (data not shown) but did induce a rapid decrease in intracellular mCD14–EGFP (Fig. 8). A decrease of ∼20% in the intracellular fluorescence was observed after a 15-min incubation of U373–CD14–EGFP cells with LPS–sCD14 complexes. A similar decrease in intracellular fluorescence associated with U373–EGFP cells was observed in response to LPS–sCD14 complexes (Fig. 8), indicating that the compartments released contained GPI-anchored proteins in addition to mCD14–EGFP.
Figure 8.
Intracellular mCD14–EGFP is rapidly exocytosed. Adherent U373–CD14–EGFP (□) and U373–EGFP (▵) cells cultured in 96-well plates were incubated with LPS–sCD14 complexes (40 ng/ml LPS) at 37°C for the times indicated. At 0 min, no LPS was added. At the end of the incubation, total fluorescence associated with the cells was measured in a fluorescent plate reader. Trypan blue (200 μg/ml) was added to quench extracellular fluorescence, and the remaining intracellular fluorescence was measured. Intracellular fluorescence was calculated as a percentage of total fluorescence in each well. Results are expressed as the means of triplicate wells ± SEM and are from an experiment performed three times with the same result. *Direct comparison using Student's t test shows that these values are significantly different from the value of cells incubated 0 min with LPS (P < 0.003).
LPS Aggregates and mCD14 Colocalize on the Cell Surface and Are Cointernalized.
LPS aggregates are formed when LPS micelles and LBP are incubated with little or no sCD14. These LPS–LBP aggregates can bind to mCD14 on the surfaces of cells, and they are subsequently internalized 27 28 29. However, binding of LPS–LBP aggregates by mCD14 differs from binding of monomeric LPS presented as a complex with sCD14. Aggregates bind at 4°C, but monomeric LPS cannot be transferred from sCD14 to mCD14 at this temperature. In addition, although internalization of monomeric LPS correlates with intracellular signaling 13 14, internalization of LPS aggregates can be dissociated from the generation of signals 27. These results suggest that LPS aggregates may have a pathway for internalization that is distinct from that of monomeric LPS.
To determine whether mCD14 traffics with LPS aggregates during internalization, we incubated U373–CD14–EGFP with BODIPY–LPS aggregates at concentrations between 40 and 100 ng/ml and followed both BODIPY–LPS and mCD14–EGFP fluorescence by confocal microscopy. After incubation for 5–10 min at 37°C with BODIPY–LPS aggregates, the cells were washed and incubated further at 37°C. The aggregates bound to the cell surface and colocalized with mCD14–EGFP (Fig. 9). At least 15 min at 37°C was required to detect BODIPY–LPS in intracellular vesicles, suggesting that internalization of LPS aggregates was somewhat slower than internalization of LPS monomers in U373–CD14–EGFP. The mCD14–EGFP colocalized with BODIPY–LPS in intracellular vesicles detected at the earliest times, although not all of the vesicles that contained BODIPY–LPS also contained mCD14–EGFP. These results suggest that, in contrast with LPS monomers, LPS aggregates can remain bound to mCD14 during internalization. This supports the idea that there is more than one pathway for internalization of LPS.
Figure 9.
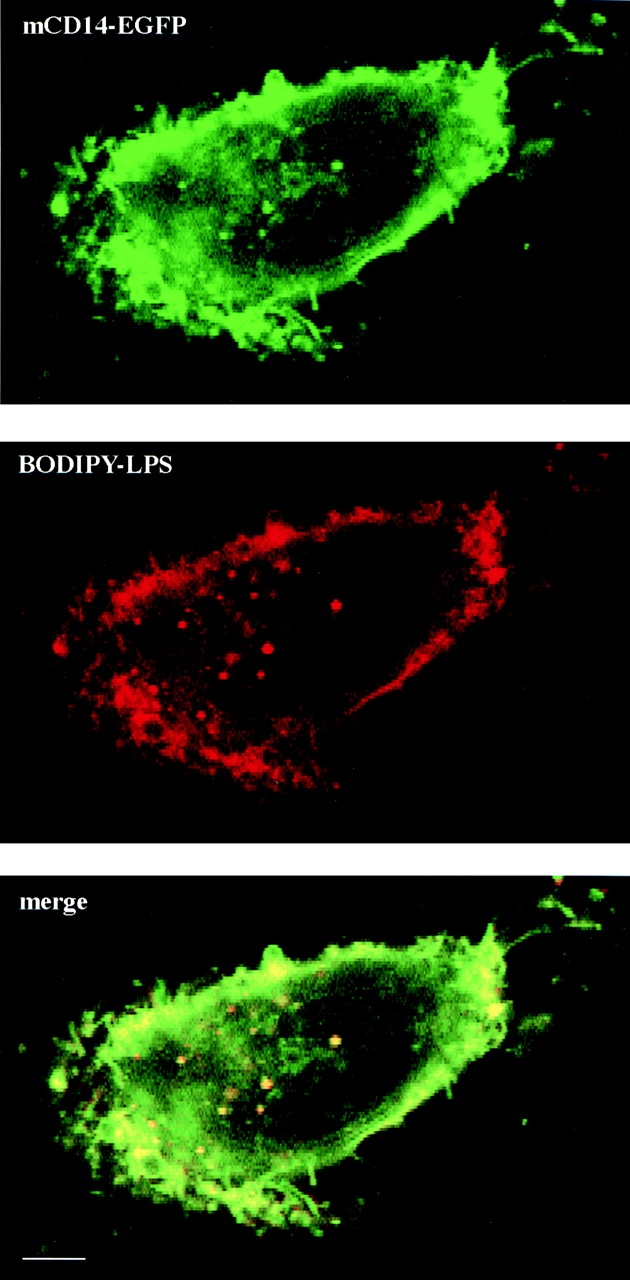
LPS aggregates formed in serum are internalized with mCD14–EGFP. U373–CD14–EGFP cells were incubated in HAP buffer with aggregated BODIPY–LPS (100 ng/ml) for 10 min at 37°C, washed, and incubated for an additional 30 min at 37°C before examination by confocal microscopy. The images for mCD14–EGFP fluorescence and BODIPY–LPS are shown in the top and center panels, respectively, and the merged image is in the bottom panel. There was colocalization of BODIPY–LPS and mCD14–EGFP on the cell surface and within many intracellular compartments.
Discussion
Here we have used a chimeric construct of mCD14 with EGFP transfected into the astrocytoma cell line U373 as a probe for observing the distribution of mCD14 in living cells. Labeling mCD14 directly with a fluorescent tag avoided any perturbation of the distribution of mCD14, a GPI-anchored protein, that might be caused by antibody cross-linking 30. A variety of functional studies confirmed that the mCD14–EGFP chimera was an intact and functional protein that was attached to the cell surface via a GPI anchor.
Using this method, we observed that mCD14 was not only expressed on the cell surface but was also present intracellularly both in the perinuclear area and in small vesicles near the basal aspect of the cells. These vesicles might represent compartments in which mCD14, and perhaps other GPI-anchored proteins, traffic from the Golgi apparatus to the plasma membrane. They may also represent a recycling compartment for GPI-linked proteins that are internalized from the plasma membrane and then returned to it 31. Most of these vesicles were exocytosed when U373–CD14–EGFP cells were exposed to LPS–sCD14 complexes, indicating that they are capable of fusion with the plasma membrane.
The rapid exocytosis of the vesicles containing mCD14–EGFP upon stimulation of U373–CD14–EGFP cells with LPS was reminiscent of the exocytosis of secretory vesicles containing mCD14 and other GPI-anchored proteins upon stimulation of neutrophils with an agonist 25. Secretory vesicles are thought to be an endocytic compartment, as they also contain the serum proteins albumin and tetranectin 32 33. In response to formyl peptide, secretory vesicles are brought rapidly to the neutrophil surface, augmenting the expression of GPI-anchored proteins on the plasma membrane 25. Additional studies will be required to determine whether any of the mCD14-containing vesicles in U373–CD14–EGFP cells represent a recycling compartment for GPI-anchored proteins similar to the secretory vesicles of neutrophils.
Expression of mCD14–EGFP on the plasma membrane enabled uptake of BODIPY–LPS (Fig. 5 and Fig. 6) and cellular responses to LPS (Fig. 4 A). CD14 binds LPS 34 and, not surprisingly, added LPS was found to colocalize with plasma membrane mCD14–EGFP (Fig. 5 and Fig. 6). After association with the membrane, monomeric LPS is known to be rapidly internalized 13 14. Here we show that when LPS was internalized, it moved into intracellular vesicles that did not contain mCD14. Thus, we have demonstrated in living cells that LPS, once it is bound by mCD14 on the cell surface, is internalized without being accompanied by its receptor. Together with our previous observations that each mCD14 on the surfaces of human monocytes enables the uptake of 15 LPS molecules in 30 min 3 and that this uptake depends on an additional cell surface protein 3, these observations support a model for LPS trafficking that involves transfer of LPS from mCD14 to another cell surface protein or to the lipid bilayer of the plasma membrane.
In addition to our studies with monomeric LPS, we have also observed the trafficking of LPS aggregates. Aggregated LPS may engage multiple copies of mCD14 on the cell surface at the same time, and it may be more difficult for mCD14 to transfer LPS from aggregates to the plasma membrane. Using the mCD14–EGFP chimera, we observed that mCD14 was internalized with aggregated LPS in U373 cells. This behavior is opposite to that of LPS monomers. It is, however, consistent with a variety of other studies documenting different fates of monomeric and aggregated LPS. After internalization, primarily through noncoated structures 35, LPS aggregates move over the course of several hours into a compartment that is likely to be lysosomal in nature. There, acyloxyacyl hydrolase deacylates and thus detoxifies LPS 18. Internalization of aggregates can be disassociated from signaling 27 29 and thus appears more relevant to the detoxification and clearance of LPS rather than signaling.
Several observations indicate a close correlation between signaling and LPS transit to the Golgi complex. For example, inactive structural analogues of LPS are not transported to the Golgi complex 15, and cells from Lpsd mice, which exhibit a defect in LPS signaling, fail to transport LPS to the Golgi complex 14. Recent work has shown that Lpsd mice are defective in Toll-like receptor (TLR)4, a member of the IL-1 receptor family 36. In this regard, it is interesting to note that the ligated IL-1 receptor type I (IL-1RI) may require trafficking to an intracellular compartment to generate signals 37. A thymocyte cell line has been identified that is defective in its responses to IL-1 and also does not internalize IL-1RI. The defect can be overcome by intracellular delivery of IL-1 38 or by transfection with IL-1R accessory protein (IL-1RAcP), which restores both IL-1RI internalization 39 and IL-1 responses 39. Interaction of IL-1 with IL-1RI and IL-1RAcP triggers a cascade of signaling events, including activation of the stress-activated, mitogen-activated protein (MAP) kinase pathways and transcription factor NF-κB. Of interest in this regard is that TLR4 shares homology in its cytoplasmic domain with the IL-1R family 40, suggesting the possibility that it may also share similar directions of intracellular trafficking. Whether TLR4 colocalizes with LPS before or after transport to the Golgi complex, however, will have to await further studies.
Acknowledgments
We thank Drs. Samuel Wright, Nathalie Thieblemont, and Norbert Lamping for critical reading of this manuscript.
Footnotes
1used in this paper: BODIPY, boron dipyrromethane; DAF, decay accelerating factor; EGFP, enhanced green fluorescent protein; FBS, fetal bovine serum; GPI, glycosylphosphatidyl inositol; LBP, LPS binding protein; m, membrane-bound; PI-PLC, phosphatidyl inositol phospholipase C; s, soluble; SP, signal peptide; TLR, Toll-like receptor
A preliminary version of this work was presented at the Fifth Conference of the International Endotoxin Society, Santa Fe, NM, September 12–15, 1998.
Le Grand, C.B., N. Lamping, T. Sugiyama, S.D. Wright, and R. Thieringer, manuscript submitted for publication.
References
- Wright S.D. CD14 and innate recognition of bacteria. J. Immunol. 1995;155:6–8. [PubMed] [Google Scholar]
- Hailman E., Vasselon T., Kelley M., Busse L.A., Hu M.C.-T., Lichenstein H.S., Detmers P.A., Wright S.D. Stimulation of macrophages and neutrophils by complexes of lipopolysaccharide and soluble CD14. J. Immunol. 1996;156:4384–4390. [PubMed] [Google Scholar]
- Vasselon T., Pironkova R., Detmers P.A. Sensitive responses of leukocytes to lipopolysaccharide require a protein distinct from CD14 at the cell surface. J. Immunol. 1997;159:4498–4505. [PubMed] [Google Scholar]
- Wright S.D., Ramos R.A., Tobias P.S., Ulevitch R.J., Mathison J.C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- Wright S.D., Ramos R.A., Hermanowski-Vosatka A., Rockwell P., Detmers P.A. Activation of the adhesive capacity of CR3 on neutrophils by endotoxindependence on lipopolysaccharide binding protein and CD14. J. Exp. Med. 1991;173:1281–1286. doi: 10.1084/jem.173.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundan A., Gullstein-Jahr T., Otterlei M., Ryan L., Bazil V., Wright S.D., Espevik T. Soluble CD14 from urine copurifies with a potent inducer of cytokines. Eur. J. Immunol. 1994;24:1779–1784. doi: 10.1002/eji.1830240809. [DOI] [PubMed] [Google Scholar]
- Golenbock D.T., Bach R.R., Lichenstein H., Juan T.S.-C., Tadavarthy A., Moldow C.F. Soluble CD14 promotes LPS activation of CD14-deficient PNH monocytes and endothelial cells. J. Lab. Clin. Med. 1995;125:662–671. [PubMed] [Google Scholar]
- Duchow J., Marchant A., Crusiaux A., Husson C., Alonso-Vega C., De Groote D., Neve P., Goldman M. Impaired phagocyte responses to lipopolysaccharide in paroxysmal nocturnal hemoglobinuria. Infect. Immun. 1993;61:4280–4285. doi: 10.1128/iai.61.10.4280-4285.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haziot A., Ferrero E., Kontgen F., Hijiya N., Yamamoto S., Silver J., Stewart C.L., Goyert S.M. Resistance to endotoxin shock and reduced dissemination of gram-negative bacteria in CD14-deficient mice. Immunity. 1996;4:407–414. doi: 10.1016/s1074-7613(00)80254-x. [DOI] [PubMed] [Google Scholar]
- Golenbock D.T., Liu Y., Millham F.H., Freeman M.W., Zoeller R.A. Surface expression of human CD14 in Chinese hamster ovary fibroblasts imparts macrophage-like responsiveness to bacterial endotoxin. J. Biol. Chem. 1993;268:22055–22059. [PubMed] [Google Scholar]
- Lee J.-D., Kato K., Tobias P.S., Kirkland T.N., Ulevitch R.J. Transfection of CD14 into 70Z/3 cells dramatically enhances the sensitivity to complexes of lipopolysaccharide (LPS) and LPS binding protein. J. Exp. Med. 1992;175:1697–1703. doi: 10.1084/jem.175.6.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.Y., Kitchens R.L., Munford R.S. Phosphatidylinositides bind to plasma membrane CD14 and can prevent monocyte activation by bacterial lipopolysaccharide. J. Biol. Chem. 1998;273:24309–24313. doi: 10.1074/jbc.273.38.24309. [DOI] [PubMed] [Google Scholar]
- Detmers P.A., Thieblemont N., Vasselon T., Pironkova R., Miller D.S., Wright S.D. Potential role of membrane internalization and vesicle fusion in adhesion of neutrophils in response to lipopolysaccharide and TNF. J. Immunol. 1996;157:5589–5596. [PubMed] [Google Scholar]
- Thieblemont N., Wright S.D. Mice genetically hyporesponsive to lipopolysaccharide (LPS) exhibit a defect in endocytic uptake of LPS and ceramide. J. Exp. Med. 1997;185:2095–2100. doi: 10.1084/jem.185.12.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieblemont N., Thieringer R., Wright S.D. Innate immune recognition of bacterial lipopolysaccharidedependence on interactions with membrane lipids and endocytic movement. Immunity. 1998;8:771–777. doi: 10.1016/s1074-7613(00)80582-8. [DOI] [PubMed] [Google Scholar]
- Wurfel M.M., Hailman E., Wright S.D. Soluble CD14 acts as a shuttle in the neutralization of LPS by LPS-binding protein and reconstituted high-density lipoprotein. J. Exp. Med. 1995;181:1743–1754. doi: 10.1084/jem.181.5.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurfel M.M., Wright S.D. Lipopolysaccharide-binding protein and soluble CD14 transfer lipopolysaccharide to phospholipid bilayerspreferential interaction with particular classes of lipid. J. Immunol. 1997;158:3925–3934. [PubMed] [Google Scholar]
- Luchi M., Munford R.S. Binding, internalization, and deacylation of bacterial lipopolysaccharide by human neutrophils. J. Immunol. 1993;151:959–969. [PubMed] [Google Scholar]
- Cormack B.P., Valdivia R.H., Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- LeGrand C.B., Thieringer R. CD14-dependent induction of protein tyrosine phosphorylation by lipopolysaccharide in murine B-lymphoma cells. Biochim. Biophys. Acta. 1994;1223:36–46. doi: 10.1016/0167-4889(94)90071-x. [DOI] [PubMed] [Google Scholar]
- Detmers P.A., Zhou D., Polizzi E., Thieringer R., Hanlon W.A., Vaidya S., Bansal V. Role of stress-activated mitogen-activated protein kinase (p38) in beta 2-integrin-dependent neutrophil adhesion and the adhesion-dependent oxidative burst. J. Immunol. 1998;161:1921–1929. [PubMed] [Google Scholar]
- Yu B., Wright S.D. Catalytic properties of lipopolysaccharide (LPS) binding protein (LBP). Transfer of LPS to soluble CD14. J. Biol. Chem. 1996;271:4100–4105. doi: 10.1074/jbc.271.8.4100. [DOI] [PubMed] [Google Scholar]
- Todd R.F., Van A.A., Schlossman S.F., Terhorst C. Structural analysis of differentiation antigens Mo1 and Mo2 on human monocytes. Hybridoma. 1982;1:329–337. doi: 10.1089/hyb.1.1982.1.329. [DOI] [PubMed] [Google Scholar]
- Kusunoki T., Hailman E., Juan T.S.-C., Lichenstein H.S., Wright S.D. Molecules from Staphylococcus aureus that bind CD14 and stimulate innate immune responses. J. Exp. Med. 1995;182:1673–1682. doi: 10.1084/jem.182.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmers P.A., Zhou D., Powell D., Lichenstein H., Kelley M., Pironkova R. Endotoxin receptors (CD14) are found with CD16 (FcγRIII) in an intracellular compartment of neutrophils that contains alkaline phosphatase. J. Immunol. 1995;155:2085–2095. [PubMed] [Google Scholar]
- Frey E.A., Miller D.S., Jahr T.G., Sundan A., Bazil V., Espevik T., Finlay B.B., Wright S.D. Soluble CD14 participates in the response of cells to lipopolysaccharide. J. Exp. Med. 1992;176:1665–1671. doi: 10.1084/jem.176.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegner J.A., Ulevitch R.J., Tobias P.S. Lipopolysaccharide (LPS) signal transduction and clearance. Dual roles for LPS binding protein and membrane CD14. J. Biol. Chem. 1995;270:5320–5325. doi: 10.1074/jbc.270.10.5320. [DOI] [PubMed] [Google Scholar]
- Poussin C., Foti M., Carpentier J.L., Pugin J. CD14-dependent endotoxin internalization via a macropinocytic pathway. J. Biol. Chem. 1998;273:20285–20291. doi: 10.1074/jbc.273.32.20285. [DOI] [PubMed] [Google Scholar]
- Kitchens R.L., Munford R.S. CD14-dependent internalization of bacterial lipopolysaccharide (LPS) is strongly influenced by LPS aggregation but not by cellular responses to LPS. J. Immunol. 1998;160:1920–1928. [PubMed] [Google Scholar]
- Mayor S., Rothberg K.G., Maxfield F.R. Sequestration of GPI-anchored proteins in caveolae triggered by cross-linking. Science. 1994;264:1948–1951. doi: 10.1126/science.7516582. [DOI] [PubMed] [Google Scholar]
- Maxfield F.R., Mayor S. Cell surface dynamics of GPI-anchored proteins. Adv. Exp. Med. Biol. 1997;419:355–364. doi: 10.1007/978-1-4419-8632-0_47. [DOI] [PubMed] [Google Scholar]
- Borregaard N., Christensen L., Bejerrum O.W., Birgens H.S., Clemmensen I. Identification of a highly mobilizable subset of human neutrophil intracellular vesicles that contains tetranectin and latent alkaline phosphatase. J. Clin. Invest. 1990;85:408–416. doi: 10.1172/JCI114453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borregaard N., Kjeldsen L., Rygaard K., Bastholm L., Nielsen M.H., Sengelov H., Bjerrum O.W., Johnsen A.H. Stimulus-dependent secretion of plasma proteins from human neutrophils. J. Clin. Invest. 1992;90:86–96. doi: 10.1172/JCI115860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailman E., Lichenstein H.S., Wurfel M.M., Miller D.S., Johnson D.A., Kelley M., Busse L.A., Zukowski M.M., Wright S.D. Lipopolysaccharide (LPS)-binding protein accelerates the binding of LPS to CD14. J. Exp. Med. 1994;179:269–277. doi: 10.1084/jem.179.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchens R.L., Wang P., Munford R.S. Bacterial lipopolysaccharide can enter monocytes via two CD14-dependent pathways. J. Immunol. 1998;161:5534–5545. [PubMed] [Google Scholar]
- Poltorak A., He X., Smirnova I., Liu M.Y., Huffel C.V., Du X., Birdwell D., Alejos E., Silva M., Galanos C. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr micemutations in tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Hofmeister R., Wiegmann K., Korherr C., Bernardo K., Kronke M., Falk W. Activation of acid sphingomyelinase by interleukin-1 (IL-1) requires the IL-1 receptor accessory protein. J. Biol. Chem. 1997;272:27730–27736. doi: 10.1074/jbc.272.44.27730. [DOI] [PubMed] [Google Scholar]
- Hofmeister R., Mannel D.N., Falk W. Evidence for an intracellular activation loop in the IL-1 system. J. Inflamm. 1995;47:151–163. [PubMed] [Google Scholar]
- Wesche H., Korherr C., Kracht M., Falk W., Resch K., Martin M.U. The interleukin-1 receptor accessory protein (IL-1RAcP) is essential for IL-1-induced activation of interleukin-1 receptor-associated kinase (IRAK) and stress-activated protein kinases (SAP kinases) J. Biol. Chem. 1997;272:7727–7731. doi: 10.1074/jbc.272.12.7727. [DOI] [PubMed] [Google Scholar]
- Rock F.L., Hardiman G., Timans J.C., Kastelein R.A., Bazan J.F. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. USA. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]