Abstract
In this study, by the generation of a specific monoclonal antibody, we identified p75/AIRM1 (for adhesion inhibitory receptor molecule 1), a novel inhibitory receptor that is mostly confined to human natural killer cells. p75/AIRM1 is a 75-kD glycoprotein that, upon sodium pervanadate treatment, becomes tyrosine phosphorylated and associates to src homology 2 domain–bearing protein tyrosine phosphatase (SHP)-1. The p75/AIRM1 gene is located on human chromosome 19 and encodes a novel member of the sialoadhesin family characterized by three immunoglobulin-like extracellular domains (one NH2-terminal V-type and two C2-type) and a classical immunoreceptor tyrosine–based inhibitory motif (ITIM) in the cytoplasmic portion. The highest amino acid sequence similarity has been found with the myeloid-specific CD33 molecule and the placental CD33L1 protein. Similar to other sialoadhesin molecules, p75/AIRM1 appears to mediate sialic acid–dependent ligand recognition.
Keywords: molecular cloning, natural killer cells, sialoadhesin, inhibitory receptor, immunoglobulin superfamily
Relevant progress has been made in recent years towards a better understanding of the molecular interactions responsible for the regulation of NK cell functions. In particular, the finding that NK cells are capable of killing target cells lacking MHC class I proteins on their surface 1 has been the basis for the identification of a series of inhibitory receptors present on NK cells that bind MHC molecules expressed on target cells. In humans, these receptors are expressed on subsets of NK cells and recognize either discrete HLA class I alleles or nonclassical HLA class I molecules 1 2 3. The first group includes members of the immunoglobulin superfamily (Ig-SF)1 that are referred to as killer inhibitory receptors (KIRs). KIRs are characterized by the ability to specifically recognize distinct groups of HLA-A 4, -B 5, or -C 6 alleles. The second group is represented by members of the C-type lectin superfamily that covalently associate to form the CD94/NKG2 receptors 7 8 9; the inhibitory CD94/NKG2A receptor has been shown to be specific for the nonclassical HLA class I molecule, HLA-E 3 10. More recently, several additional inhibitory receptors, either HLA class I specific or not, have been identified that may exert a regulatory activity on human NK cell function; these include, for example, the leukocyte Ig-like receptor 1/Ig-like transcript 2 (LIR-1/ILT-2 [11, 12]) and the p40/leukocyte-associated Ig-like receptor 1 (LAIR-1) 13 14 molecules. All of the inhibitory receptors identified so far are characterized by one or more immunoreceptor tyrosine–based inhibitory motifs (ITIMs) in their cytoplasmic tail. Upon tyrosine phosphorylation, ITIM binds the src homology (SH)2 domains of phosphatases such as SHP-1 and SHP-2 that, in turn, cause downregulation of NK cell triggering and cytotoxicity 15.
In an attempt to identify novel receptors possibly involved in the negative regulation of NK cell function, we immunized mice with human NK cell clones and screened for mAbs capable of modulating the NK-mediated cytolysis. Using this approach, we selected an mAb (Z176) specific for a novel inhibitory surface molecule of ∼75 kD (p75) termed adhesion inhibitory receptor molecule 1 (AIRM1). p75/AIRM1 is expressed on the majority of human NK cells. Molecular cloning revealed a novel member of the Ig-SF characterized by three Ig-like domains in the extracellular portion and an ITIM in the cytoplasmic tail. Although p75/ARM1 does not recognize HLA class I molecules, its amino acid (aa) sequence, together with functional data, suggests that it represents a novel member of the sialoadhesin family. In conclusion, p75/AIRM1 is a novel, HLA-independent, inhibitory receptor that may play a relevant role in the negative regulation of human NK cell functions.
Materials and Methods
mAbs.
Z176 (IgG2b) mAb was obtained by immunizing a 5-wk-old BALB/c mouse with the NK clone SA260 (surface phenotype: CD3−CD16+, CD56+, NKp46+, NKp44+, p70/NKB1+, CD94/NKG2A+) as described previously 16. The following mAbs were produced in our laboratory: JT3A (IgG2a, anti-CD3), BAB281 (IgG1, anti-NKp46), Z231 and KS38 (IgG1 and IgM, respectively, anti-NKp44), Z199 (IgG2b, anti-NKG2A), Z27 (IgG1, anti-p70/NKB1), KD1 and c127 (IgG2a and IgG1, respectively, anti-CD16), and c218 and A6-90 (IgG1 and IgG2b, respectively, anti-CD56). The MCA531 mAb (IgM, anti-CD20) was purchased from Serotec. The D1.12 mAb (IgG2a, anti–HLA-DR) was provided by Dr. R.S. Accolla (Università di Pavia, Pavia, Italy). The HP2.6 mAb (IgG2a, anti-CD4) was provided by Dr. P. Sanchez-Madrid (Hospital de la Princesa, Madrid, Spain).
Purification of Polyclonal or Clonal NK and T Cell Populations.
To obtain PBLs, PBMCs derived from healthy donors were isolated on Ficoll-Hypaque gradients and depleted of plastic-adherent cells. PBLs were either used directly or were incubated with anti-CD3 (JT3A), anti-CD4 (HP2.6), and anti–HLA-DR (D1.12) mAbs for 30 min at 4°C, followed by immunomagnetic depletion with goat anti–mouse coated Dynabeads (Dynal; 30 min, 4°C; reference 17). NK or T cell clones were obtained by limiting dilution in the presence of irradiated feeder cells, 1.5 ng/ml PHA (GIBCO BRL), and 100 U/ml rIL-2 (Proleukin; Chiron Corp.) as described previously 18.
Cytolytic Activity and Flow Cytofluorimetric Analysis.
NK cell clones were tested for cytolytic activity against the FcγR+ P815 murine mastocytoma cell line in a 4-h 51Cr-release assay as described previously 2. The E/T ratio used was 8:1 in all instances.
For one- or two-color cytofluorimetric analysis (FACScan™; Becton Dickinson), cells were stained with the appropriate mAbs followed by PE- or FITC-conjugated isotype-specific goat anti–mouse second reagent (Southern Biotechnology Associates) 17.
Biochemical Characterization of the p75/AIRM1 Molecules.
Sepharose-protein A–coupled Z176 and Z199 mAbs or cyanogen bromide Sepharose (Amersham Pharmacia Biotech)-coupled Z27 mAb were used to immunoprecipitate specific molecules from 1% NP-40 lysates of cells surface labeled with 125I (NEN) as described previously 19. Immunoprecipitates were analyzed by discontinuous SDS-PAGE either undigested or digested with N-glycosidase F or O-glycosidase (Boehringer Mannheim) 20.
For two-dimensional peptide mapping (2DPM) analysis, the purified proteins were digested with pepsin, and peptides were analyzed by electrophoresis in the first dimension (Multiphor II; Amersham Pharmacia Biotech) followed by chromatography in the second dimension 20. NK cells (108) were stimulated or not with 100 μM sodium pervanadate 19, and 1% NP-40 lysates were immunoprecipitated with the Z176 mAb. Samples were analyzed in discontinuous SDS-PAGE, transferred to Immobilon P (Millipore Corp.), and then probed with antiphosphotyrosine mAb (PY20-HRPO; Transduction Laboratories) or anti–SHP-1 and SHP-2 phosphatases (PTP1C and PTP1D, respectively; Transduction Laboratories). The Renaissance Chemiluminescence kit (NEN) was used for detection.
cDNA Library and Screening.
cDNA was synthesized from RNA prepared from two IL-2–activated polyclonal NK cells and inserted in the expression vector VR1012. The cDNA library, fractionated in 10 pools of 200,000 different inserts, was transfected into COS-7 cells by DEAE-dextran method and immunocytochemical staining using the Z176-specific mAb and sib selection 21.
DNA Sequencing.
DNA sequencing was performed using d-Rhodamine Terminator Cycle Sequencing kit and a 377 ABI automatic sequencer (Perkin Elmer-Applied Biosystems).
Adhesion Assay.
COS-7 cells were transfected with VR1012–AIRM1 construct by DEAE-dextran method 4. After 48 h, cells were trypsinized and analyzed by immunofluorescence staining for the expression of p75/AIRM1 molecules. Transfected cells and human RBCs were washed twice with serum-free DMEM. The COS-7 cell/RBC ratio used in the experiments was 1:20; the adhesion assay was performed for 30 min at 4°C. The binding of RBCs to COS-7 cells was quantified by counting the percentage of COS-7 cells that bound more than seven erythrocytes. Neuraminidase treatment was carried out by incubating RBCs with 0.1 U/ml of Vibrio cholera neuraminidase (Behringwerke AG) for 3 h at 37°C followed by two washes with DMEM. For cellular adhesion blocking experiments, 106 AIRM1-transfected COS-7 cells were incubated with 0.5 ml Z176 mAb supernatant for 30 min at 4°C followed by two washes with DMEM before the adhesion assay.
Chromosomal Localization and Zoo-Blot™.
The Somatic Cell Hybrid blot (BIOS Laboratories), containing 20 multi-chromosomal somatic human/hamster cell hybrids plus 3 control genomic DNAs (human, hamster, and mouse) digested with EcoRI, was used to assign the AIRM1 gene to a specific chromosome. A 1203-bp cDNA probe, obtained digesting VR1012–AIRM1 construct with SalI and PstI restriction enzymes, was used to perform high stringency hybridization 22. Analysis of cross-specific conservation of AIRM1 gene was performed using Zoo-Blot™ from Clontech. This Southern blot contained genomic DNA from humans, Rhesus monkey, Sprague-Dawley rat, BALB/c mouse, dog, cow, rabbit, chicken, and Saccharomyces cerevisiae yeast. Washes were carried out under low stringency conditions 23.
Reverse Transcriptase PCR Amplification of AIRM1 cDNA.
RNA extracted using RNAzol (Cinna/Biotecx) and oligo (dT)–primed cDNA was prepared from polyclonal NK cell populations and clones by standard techniques. The set of primers AIRM1-up (containing the ATG initiation codon; 5′ TCC AAC CCC AGA TAT GCT G) and AIRM1-down (designed in the 3′ untranslated region; 5′ ACA AGC CCG AGC CTC TGC) were used to amplify the AIRM1 open reading frame. 30 cycles of PCR (30 s at 95°C, 30 s at 60°C, and 30 s at 72°C) were performed using TAQ-GOLD (Perkin Elmer-Applied Biosystems) after a preactivation of 15 min at 95°C. The amplification products obtained from polyclonal NK cells populations were purified from gel, subcloned into pcDNA3.1/V5/His TOPO™ vector using the Eukaryotic TOPO TA Cloning® kit (Invitrogen), and sequenced.
Results
Identification and Cellular Distribution of a Novel NK Cell Surface Molecule with Inhibitory Function.
Mice were immunized with the NK cell clone SA260 (surface phenotype: CD3−CD16+, CD56+, NKp46+, NKp44+, p70/NKB1+, CD94/NKG2A+), characterized by a strong cytolytic activity against the P815 murine mastocytoma cell line. After cell fusion, mAbs were analyzed for their ability to inhibit the cytotoxicity mediated by NK cell clones in a classical redirected killing assay against the FcγR+ P815 cell line. By using this screening procedure, we isolated the Z176 mAb (IgG2b) that inhibited the cytolytic activity of the majority of the NK cell clones analyzed. Fig. 1 shows four representatives of such clones, including the immunizing SA260 clone. In three of these clones, the addition of Z176 mAb (but not of an isotype-matched anti-CD56 mAb) resulted in inhibition of the spontaneous cytolytic activity against P815 cells (Fig. 1 a). Clone D414 is representative of the infrequent NK cell clones in which no inhibitory effect could be detected. Immunofluorescence and FACS® analysis of the same clones (Fig. 1 b) revealed that Z176 mAb reacted with clones SA260, LM15, and LM8 but not with clone D414. Similar data were obtained in a large panel of NK cell clones, thus suggesting that the Z176 mAb–reactive molecule is expressed and delivers inhibitory signal in the majority of, but not all, human NK cells.
Figure 1.
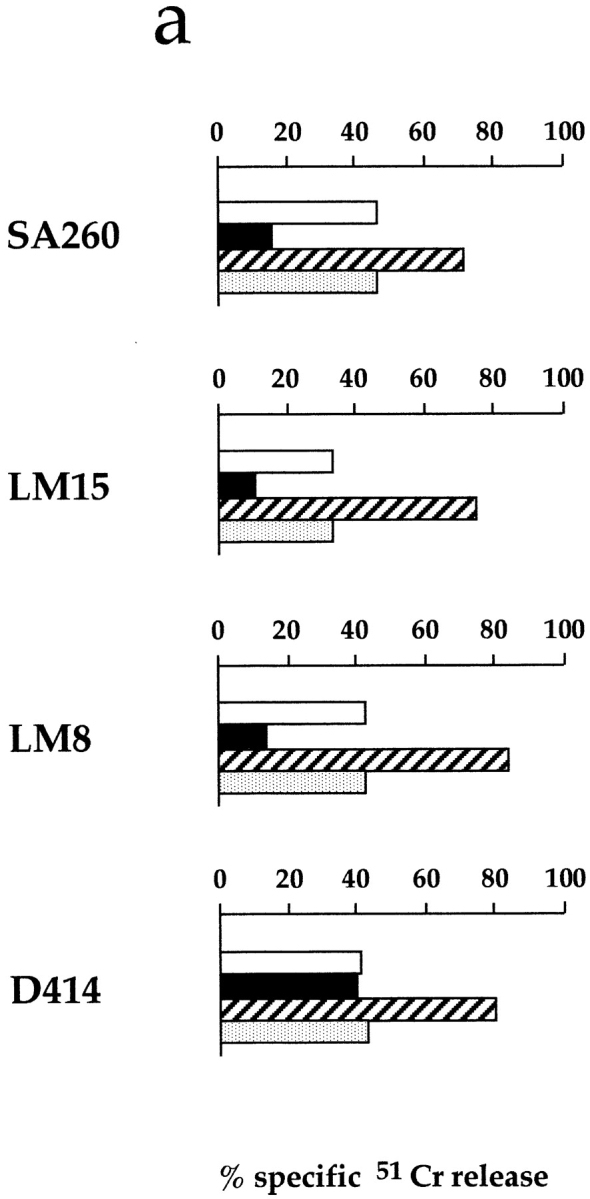

Surface expression of Z176-reactive molecules in representative human NK cell clones and the inhibitory effect of Z176 on the NK-mediated cytotoxicity. (a) Four representative NK cell clones were analyzed in a redirected killing assay against the FcγR+ P815 target cell line either in the absence (white bars) or presence of Z176 (black bars), c127 (anti-CD16, striped bars), or A6-90 (anti-CD56, stippled bars) mAb. (b) The NK cell clones were analyzed by immunofluorescence and FACS® analysis for the expression of Z176-reactive surface molecules. Cells were stained with Z176 mAb followed by PE-conjugated isotype-specific goat anti–mouse second reagent. Open profiles represent cells stained with the second reagent alone. a.u., arbitrary units.
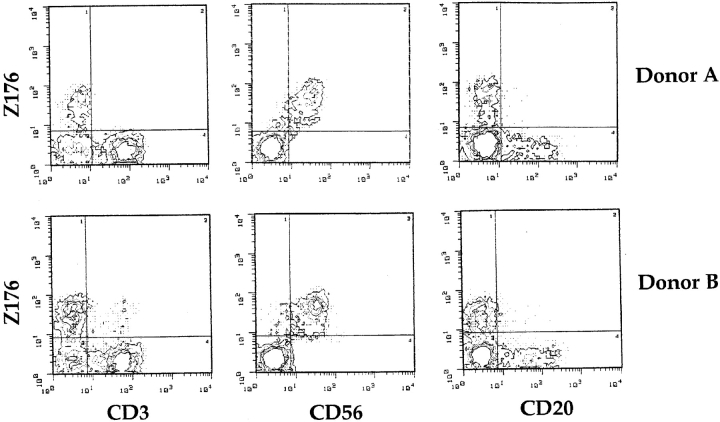
We next analyzed the cell surface distribution of the Z176 mAb–reacting molecule in PBLs by two-color immunofluorescence and FACS® analysis. As shown in Fig. 2, all CD56+ cells were stained by the Z176 mAb. In contrast, Z176 mAb did not stain CD3+ T cells or CD20+ B cells. Only in some individuals was a minor fraction (<5%) of T cells found to be Z176+ (see donor B). Although not shown, further analysis of the Z176 surface expression in NK cell–enriched fractions of PBLs (upon depletion of CD3+HLA-DR+ cells) confirmed that most NK cells reacted with Z176 mAb while Z176− NK cells were rather infrequent. Analysis of a large panel (>100) of T cell clones (both α/β+ and γ/δ+) showed that Z176-reactive molecules are not expressed by T lymphocytes after cell activation or clonal expansion (not shown). Finally, Z176 mAb did not stain EBV-transformed B cell lines (including Raji, Daudi, and C1R) or T cell lines (such as JA3, HSB-2, CEM, MOLT-4, H9, and Jurkat). Remarkably, it also failed to stain NK cell lines, including NK3.3, NKL, and YT (not shown).
Figure 2.
Expression of Z176-reactive molecules in PBL subsets. Freshly isolated PBLs were analyzed by two-color immunofluorescence and FACS® analysis for the expression of Z176-reactive molecules in combination with CD3, CD56, or CD20 surface molecules. Cells were stained with Z176, JT3A (anti-CD3), c218 (anti-CD56), and MCA531 (anti-CD20) mAbs followed by PE- or FITC-conjugated isotype-specific goat anti–mouse second reagent. The contour plots were divided into quadrants representing unstained cells (bottom left), cells with only red fluorescence (top left), cells with red and green fluorescence (top right), and cells with only green fluorescence (bottom right).
Biochemical Characterization of the Surface Molecule Recognized by Z176 mAb.
A polyclonal NK cell population was surface labeled with 125I and immunoprecipitated with Z176 mAb. This antibody immunoprecipitated a molecule with a molecular mass of ∼75 kD (p75) both under nonreducing (not shown) and reducing conditions (Fig. 3 a). To analyze their glycosylation pattern, p75 molecules were treated with different enzymes. The molecular mass was not modified by treatment with O-glycosidase (not shown). In contrast, digestion with N-glycosidase revealed a protein backbone of ∼48 kD (Fig. 3 a), thus suggesting a relatively high N-glycosylation pattern. Finally, in two-dimensional peptide mapping (2DPM) analysis, p75 displayed a digestion pattern that was different from those of known inhibitory receptors, including p70/NKB1 and NKG2A (not shown). Altogether, these data support the notion that p75 is a novel surface molecule with inhibitory function expressed by the majority of human NK cells.
Figure 3.
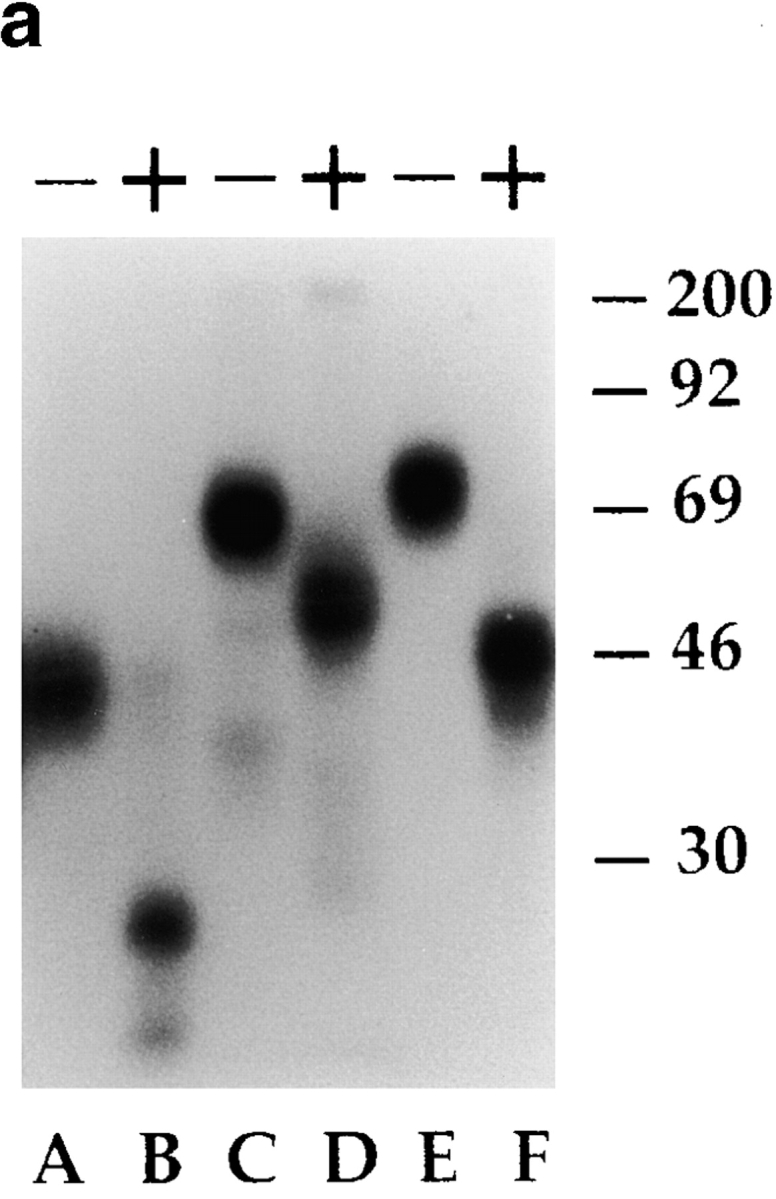
Biochemical analysis of Z176-reactive molecules and their association with SHP-1 phosphatase. (a) A polyclonal NK population was surface labeled with 125I and immunoprecipitated with anti-NKG2A (Z199 mAb, lanes A and B), anti-p70/NKB-1 (Z27 mAb, lanes C and D), and Z176 mAb (lanes E and F). Samples were analyzed in an 8% SDS-PAGE under reducing conditions either undigested (−) or digested (+) with N-glycosidase. Molecular weight markers (in kD) are indicated on the right. (b) 1% NP-40 cell lysates derived from a polyclonal NK cell population, untreated (−) or treated (+) with sodium pervanadate, were immunoprecipitated with Z176 mAb. Samples were analyzed in an 11% SDS-PAGE under reducing conditions, transferred to polyvinylidene difluoride membranes, and probed with either antiphosphotyrosine (anti-PTyr) or anti–SHP-1 mAb.
It is well known that inhibitory receptors, including p58 (CD158), p70/NKB1, CD94/NKG2A, and LIR-1/ILT-2, are characterized by the presence of ITIM sequences in their cytoplasmic tail. ITIMs, upon tyrosine phosphorylation, recruit SH2-containing phosphatases such as SHP-1 and SHP-2. To assess whether the p75 molecule could also belong to the ITIM-bearing receptor family, a polyclonal NK cell population, treated or not with sodium pervanadate, was immunoprecipitated with Z176 mAb. Samples were probed with antiphosphotyrosine, anti–SHP-1, or anti–SHP-2 mAbs. Fig. 3 b shows that treatment with sodium pervanadate leads to p75 tyrosine phosphorylation (left panel) and association with SHP-1 (right panel). It is of note that, under the same conditions, no association with SHP-2 could be detected (not shown). These data strongly suggest that p75 contains at least one typical ITIM in the cytoplasmic tail.
Molecular Cloning and Characterization of the cDNA Encoding the p75 Molecule.
A cDNA library, prepared from RNA derived from 2 polyclonal NK cell populations, fractionated in 10 different pools, was transiently transfected into COS-7 cells. After 48 h, cells were tested for reactivity with Z176 mAb by immunocytochemical staining. The plasmidic DNA of the positive pool was amplified in Escherichia coli, fractionated in smaller subpools, and transfected into COS-7 cells. Six rounds of transfections and screening allowed the isolation of an individual cDNA termed AIRM1.
As shown in Fig. 4 a, COS-7 cells transfected with VR1012–AIRM1 construct were brightly stained with Z176 mAb in cytofluorimetric analysis. Cell transfectants were then surface labeled with 125I, and cell lysates were immunoprecipitated with Z176 mAb. As shown in Fig. 4 b, the immunoprecipitated molecule displayed a molecular mass slightly lower than that immunoprecipitated from NK cells. This difference in molecular mass could reflect a noncomplete N-glycosylation in COS-7 cell transfectants. Indeed, upon treatment with N-glycosidase, the molecule immunoprecipitated from AIRM1 transfectants displayed a 48-kD protein backbone identical to that of the p75 molecule isolated from NK cells.
Figure 4.
Surface expression and biochemical analysis of Z176-reactive molecules in COS-7 cells transfected with VR1012–AIRM1 construct. (a) AIRM1-transfected COS-7 cells were analyzed for surface expression using Z176 mAb followed by PE-conjugated isotype-specific goat anti–mouse second reagent. The open profile represents cells stained with the second reagent alone. (b) A polyclonal NK cell population (lanes A and B) and AIRM1-transfected COS-7 cells (lanes C and D) were surface labeled with 125I and immunoprecipitated with Z176 mAb. Samples were analyzed in an 8% SDS-PAGE under reducing conditions either undigested (−) or digested (+) with N-glycosidase. Molecular weight markers (in kD) are indicated on the right.
Fig. 5 shows the nucleotide sequence (1766 bp) of AIRM1 and its predicted amino acid translation (467 aa). The 5′ noncoding region consists of 64 nucleotides, and in the 3′ noncoding sequence a possible polyadenylation signal (AAUAUA) preceded the poly A tail. The putative protein appears as a type I transmembrane molecule belonging to the Ig-SF. An 18 aa leader peptide precedes a 335 aa extracellular portion characterized by an NH2-terminal V-type domain followed by two C2-type domains. Six putative O-glycosylation sites and eight putative N-glycosylation sites are present in the extracellular portion. The predicted polypeptide mass is ∼48 kD, thus corresponding to the protein backbone of the p75 molecule immunoprecipitated by Z176 mAb from both NK cells and p75/AIRM1 COS-7 transfectants.
Figure 5.
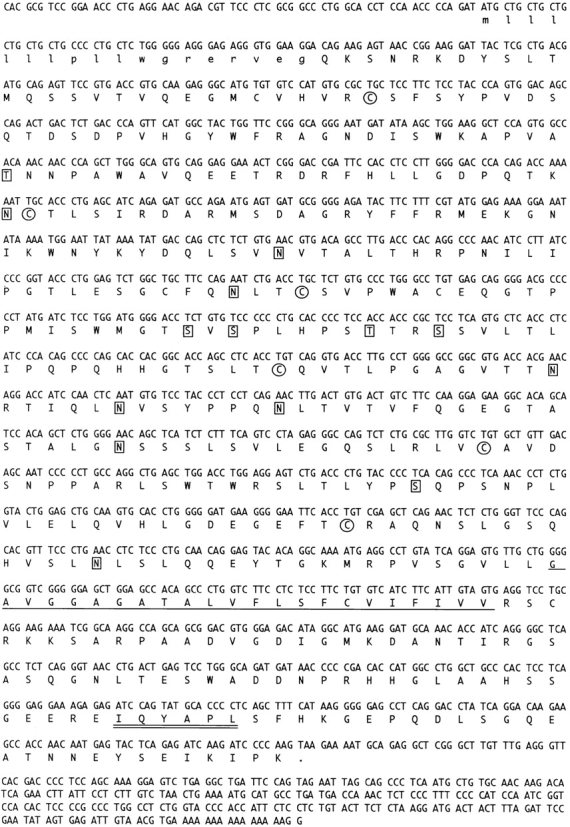
AIRM1 cDNA nucleotide sequence and its predicted aa translation. aa included in the signal peptide are indicated in small letters. Cysteine residues are circled. The predicted transmembrane portion is underlined. Potential N- and O-glycosylation sites are boxed. The ITIM sequence is double underlined. The AIRM1 nucleotide sequence is available from EMBL/GenBank/DDBJ under accession no. AJ007395.
The 23 aa hydrophobic transmembrane portion is followed by a cytoplasmic tail (91 aa) containing two tyrosine residues. Remarkably, Tyr 437 is part of a typical ITIM motif (435–440 = IQYAPL). Finally, other consensus sequences for putative phosphorylation sites are present in the cytoplasmic tail: a 3′–5′ adenosine monophosphate (cAMP)-dependent protein kinase phosphorylation site, three protein kinase phosphorylation sites, and five casein kinase 2 (CK2) phosphorylation sites.
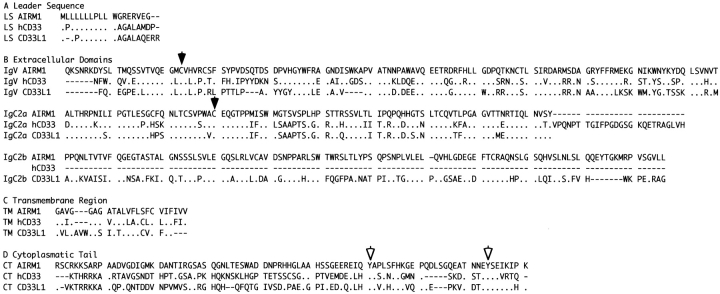
Comparison of the aa sequence of p75/AIRM1 with those of known proteins in the EMBL/GenBank/DDBJ database revealed significant similarity with the placenta antigen CD33L1 24, as well as with the myeloid lineage molecule CD33 25 (Fig. 6). In particular, both the IgV domain and the transmembrane region of p75/AIRM1 display a high degree of aa identity (55 and 61%, respectively) with CD33 molecule. On the other hand, the IgC2a and IgC2b domains display a remarkable similarity with those of CD33L1. In this context, the highest degree of aa identity (73%) was found between the IgC2a domain of p75/AIRM1 and that of CD33L1. The IgV and IgC2a domains of p75/AIRM1 contain two Cys residues at position 41 and 174. Cys residues located at the same relative positions are also present in different sialoadhesins 26 27 and are likely to be involved in the formation of interdomain bridges. Altogether, these results suggest that the p75/AIRM1 molecule may represent a novel member of the sialoadhesin family.
Figure 6.
aa alignment of p75/AIRM1, human CD33, and CD33L1 molecules. The conserved cysteines involved in the interdomain bridges and the conserved tyrosines are indicated by black and white arrows, respectively. Dashes indicate gaps inserted to optimize alignments. Dots indicate identical aa.
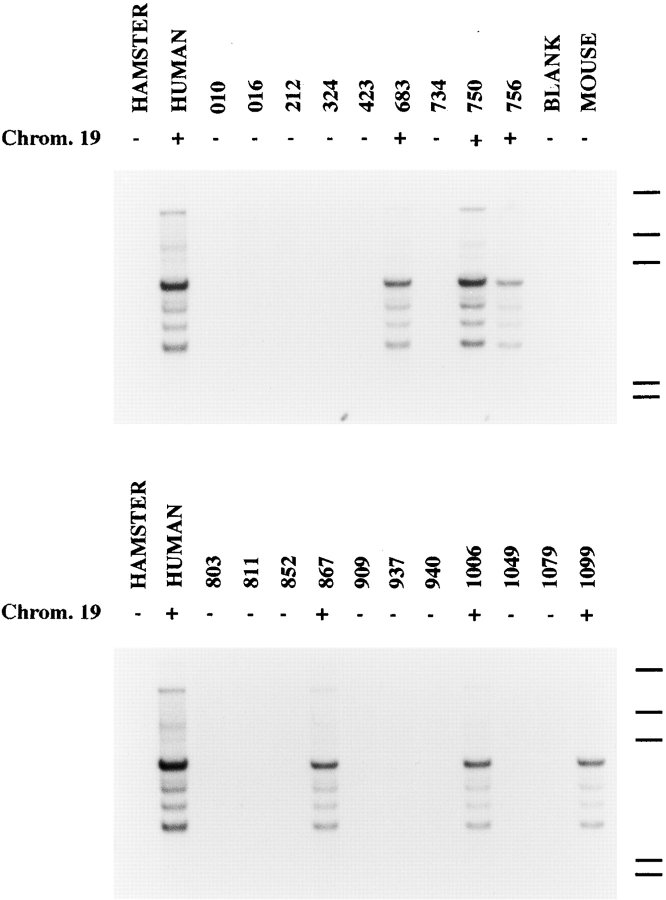
To identify the human chromosome carrying the p75/AIRM1-encoding gene, genomic DNA samples derived from a panel of human/hamster somatic cell hybrids were analyzed with a 1203-bp AIRM1-specific probe. We could localize the AIRM1 gene on human chromosome 19, since only the somatic cell hybrids containing this chromosome were positive in Southern blot analysis (Fig. 7). Moreover, the simple pattern of hybridization observed suggests that AIRM1 is a single gene or a few copy genes. It is of note that other members of the sialoadhesin family, including CD33 28 and CD33L1 24, myelin-associated glycoprotein (MAG 29), CD22 30, and Siglec-5 31, have been mapped on human chromosome 19.
Figure 7.
Chromosomal localization of p75/AIRM1 gene. A Southern blot containing genomic DNA derived from a panel of human/hamster somatic cell hybrids and genomic DNA from human, hamster, and mouse tissue as controls was hybridized with the 1.2-kb p75/AIRM1 probe. The hybrid cell lines containing chromosome (Chrom.) 19 are indicated on the top. The positions of the 23.1-, 9.4-, 6.6-, 2.3, and 2.0-kb weight markers are indicated on the right.
Reverse transcriptase (RT)-PCR analysis was performed on polyclonal NK cell populations and clones, derived from five different donors. Using the set of primers AIRM1-up and AIRM1-down, we could detect, not only in polyclonal populations but also in NK cell clones, three amplified products of ∼1.4, 1.1, and 0.7 kb, respectively (not shown). These cDNA fragments were subcloned and sequenced. The 1.4-kb product was found to correspond to the AIRM1 cloned cDNA. Moreover, the sequence analysis, performed in five different donors, revealed only an allelic variant of AIRM1 cDNA (termed AIRM1b; sequence data available from EMBL under accession no. AJ130710) characterized by two silent substitutions (codon 105, ACC instead of AAT; and codon 452, GGT instead of GGA).
Binding of AIRM1-transfected COS-7 Cells to RBCs.
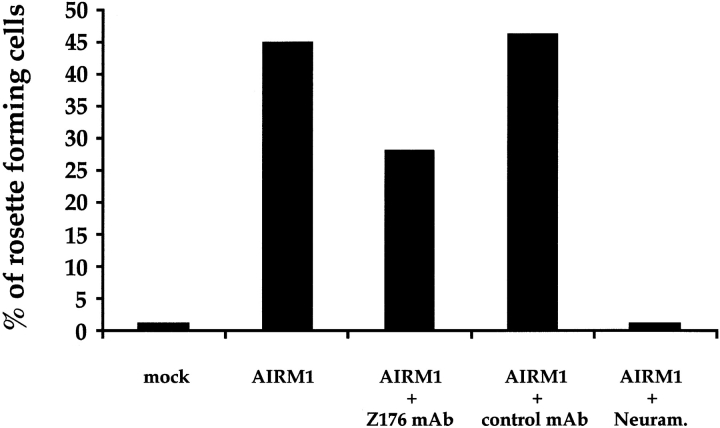
Different members of the sialoadhesin family, including CD22 32 33 and Sialoadhesin 34 35, have been shown to mediate sialic acid–dependent binding to RBCs. As shown in Fig. 8, COS-7 cells transfected with the AIRM1 cDNA efficiently bound to RBCs. AIRM1-specific rosette formation occurred both at 4°C (Fig. 8) and at 37°C (not shown). Pretreatment of RBCs with neuraminidase abrogated their binding to AIRM1 transfectants, suggesting that adhesion occurs through a sialic acid–dependent mechanism. In addition, binding to RBCs was inhibited when AIRM1 transfectants were preincubated with Z176 mAb (but not with an isotype-matched mAb), thus indicating a direct involvement of p75/AIRM1 in the RBC–AIRM1 transfectant interaction.
Figure 8.
Binding of AIRM1-transfected COS-7 cells to human RBCs. COS-7 cells either mock transfected or transfected with the VR1012–AIRM1 construct were analyzed for their ability to form rosettes with human RBCs. AIRM1/COS-7 binding to RBCs was also analyzed in the presence of Z176 mAb or an isotype-matched mAb (anti–p58-2) or against neuraminidase-treated (Neuram.) RBCs. A positive rosette was defined as a COS-7 cell that bound at least seven RBCs. The COS-7/RBC ratio used in these experiments was 1:20.
Discussion
In this study, we describe a novel inhibitory receptor, termed p75/AIRM1, which is expressed by resting and activated human NK cells and may play a role in the regulation of their function. Molecular and functional analysis revealed a novel member of the sialoadhesin family; thus, p75/AIRM1 differs from the other major inhibitory receptors expressed by NK cells that are characterized by their specificity for MHC class I molecules.
p75/AIRM1 is a transmembrane glycoprotein characterized by one IgV and two IgC2-type Ig-like domains in the extracellular portion that displays similarity with members of the sialoadhesin family. This family is composed of structurally and functionally related molecules, including MAG, Schwann cell myelin protein (SMP), Sialoadhesin, CD22, CD33, CD33L1, and Siglec-5. All of these proteins are characterized by a restricted cell expression pattern. In particular, MAG 36 and Schwann cell myelin protein (SMP) 37 are found in the nervous system on oligodendroglia and Schwann cells, respectively; CD22 is expressed on a subset of B lymphocytes 38, sialoadhesin on a subset of macrophages 39, CD33 40 and Siglec-5 31 on cells of the myelomonocytic lineage, and CD33L1 in placenta 24. Similar to these molecules, p75/AIRM1 also displays a restricted cell expression pattern, being essentially confined to NK cells. Other than their ability to bind sialic acid, limited information is available on the function of the molecules of the sialoadhesin family. The only molecule reported to display inhibitory function (i.e., similar to p75/AIRM1) is CD22 that is expressed on B cells, in which it may downregulate the B cell receptor–mediated cell triggering. CD33 is selectively expressed by hemopoietic cells, in which it represents an important marker in normal differentiation as well as in leukemia typing. In view of the similarity with p75/AIRM1, which includes the presence of an ITIM sequence in the cytoplasmic tail, it is important to reinvestigate the role of CD33, especially with respect to its possible inhibitory effect on hemopoietic cell function and/or differentiation. Experiments along this line are in progress in our laboratory.
p75/AIRM1 is encoded by a gene localized on human chromosome 19. Remarkably, genes coding for other members of the sialoadhesin family map on chromosome 19 as well. This may suggest that all of these molecules, including p75/AIRM1, may have evolved through duplication of a common ancestral gene. It is of note that other inhibitory receptors involved in the regulation of NK-mediated cytotoxicity, including KIRs, LIR/ILT, and LAIR-1, have also been mapped on this chromosome. Although not shown, the AIRM1-specific probe, under low stringency conditions, hybridized with genomic DNA from Rhesus monkey, thus suggesting a cross-species conservation between humans and monkeys.
DNA sequencing of seven cDNA clones obtained by RT-PCR experiments, performed in five different donors, revealed only an allelic variant characterized by two silent substitutions. However, considering both the relatively low number of samples analyzed and the fact that all donors belonged to the Caucasoid race, a polymorphism of the p75/AIRM1 gene cannot be ruled out. RT-PCR analysis, in addition to the 1.4-kb AIRM1 transcript, also allowed the identification of two alternatively spliced products of 1.1 and 0.7 kb, respectively. Sequence of these products revealed that the 1.1-kb fragment encoded a putative protein identical to the p75/AIRM1 but lacking the IgC2a domain (from codon 146 to codon 238) (termed AIRM2; sequence data available from EMBL under accession no. AJ130711; manuscript in preparation). The 0.7-kb amplified product contains two cDNA fragments that are both carrying an early stop codon at position 146 (EMBL accession nos. AJ130712 and AJ130713).
Consistent with the structural similarity between p75/AIRM1 and other sialoadhesins, p75/ARM1 was also found to bind RBCs. This binding was specifically inhibited by Z176 mAb as well as by the neuraminidase treatment of RBCs. These data suggest that p75/AIRM1 may function as a receptor which recognizes its putative ligand(s) in a sialic acid–dependent manner. Carbohydrate-binding proteins are known to play a role in a wide variety of biological processes involving specific cell–cell interactions. As suggested for CD22 41, it is possible that sialic acids may be required for correct orientation and presentation of the epitope (on the ligand) recognized by p75/AIRM1.
Cross-linking of p75/AIRM1 in human NK cells delivers an inhibitory signal resulting in downregulation of spontaneous NK cell–mediated cytotoxicity. An inhibitory effect could also be detected on the NK cell triggering mediated by activating receptors such as CD16, NKp46, and NKp44 (not shown). Coherent with its ability to mediate inhibition, the cytoplasmic tail of p75/AIRM1 was found to contain an ITIM that recruited the SHP-1 phosphatase upon tyrosine phosphorylation. This, in turn, is likely to inhibit downstream molecular events that are critical for the induction of NK cell–mediated cytotoxicity. One may ask the meaning of the inhibitory effect of p75/AIRM1 in NK-mediated function, and what could be the functional relationship with KIRs. KIRs appear to play a predominant role in the discrimination between HLA class I+ cells and cells that do not express sufficient amounts of HLA class I, such as tumor- or virus-infected cells. Since p75/AIRM1 does not appear to recognize HLA class I molecules, it is possible to speculate that this receptor may play a role in recognition of still undefined sialylated proteins, possibly present in normal cells that physiologically express low amounts of HLA class I molecules. The expression of ligand(s) for p75/AIRM1 may protect these cells from the NK-mediated attack. It is evident that the identification of the p75/AIRM1 ligand(s) will greatly help to clarify this issue. In addition, it is possible that p75/AIRM1 may function during stages of NK cell differentiation from immature precursors in which cells have acquired cytolytic potential but have not yet expressed HLA class I–specific inhibitory receptors. Indeed, cells with these phenotypic characteristics have recently been identified in our laboratory (our unpublished data).
In conclusion, we have identified, characterized, and cloned a novel inhibitory receptor primarily confined to human NK cells which may play a role complementary to that of KIRs in the regulation of NK cell function.
Acknowledgments
This work was supported by grants awarded by Associazione Italiana per la Ricerca sul Cancro (AIRC), Istituto Superiore di Sanità (ISS), Ministero della Sanità, and Ministero dell'Università e della Ricerca Scientifica e Tecnologica (MURST) and Consiglio Nazionale delle Ricerche, Progetto Finalizzato Biotecnologie.
Address correspondence to Lorenzo Moretta, Laboratorio di Immunopatologia, Centro Biotecnologie Avanzate, L.go R. Benzi, 10, 16132 Genova, Italy. Phone: 39-10-5737217/9; Fax: 39-10-354123; E-mail: moretta@ermes.cba.unige.it
Footnotes
1used in this paper: aa, amino acid(s); AIRM1, adhesion inhibitory receptor molecule 1; Ig-SF, immunoglobulin superfamily; ILT, Ig-like transcript; ITIM, immunoreceptor tyrosine–based inhibitory motif; KIR, killer inhibitory receptor; LIR, leukocyte Ig-like receptor; MAG, myelin-associated glycoprotein; RT, reverse transcriptase; SH, src homology domain; SHP, SH2 domain–bearing protein tyrosine phosphatase
References
- Moretta A., Bottino C., Vitale M., Pende D., Biassoni R., Mingari M.C., Moretta L. Receptors for HLA class-I molecules in human natural killer cells. Annu. Rev. Immunol. 1996;14:619–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- Moretta A., Biassoni R., Bottino C., Pende D., Vitale M., Poggi A., Mingari M.C., Moretta L. Major histocompatibility complex class I-specific receptors on human natural killer and T lymphocytes. Immunol. Rev. 1997;155:105–117. doi: 10.1111/j.1600-065x.1997.tb00943.x. [DOI] [PubMed] [Google Scholar]
- Braud V.M., Jones E.Y., McMichael A.J. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur. J. Immunol. 1997;27:1164–1169. doi: 10.1002/eji.1830270517. [DOI] [PubMed] [Google Scholar]
- Pende D., Biassoni R., Cantoni C., Verdiani S., Falco M., Di Donato C., Accame L., Bottino C., Moretta A., Moretta L. The natural killer receptor specific for HLA-A allotypesa novel member of the p58/p70 family of the inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulfide-linked dimer. J. Exp. Med. 1996;184:505–518. doi: 10.1084/jem.184.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M., Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- Wagtmann N., Biassoni R., Cantoni C., Verdiani S., Malnati M.S., Vitale M., Bottino C., Moretta L., Moretta A., Long E.O. Molecular clones of the p58 natural killer cell receptor reveal Ig-related molecules with diversity in both the extra- and intracellular domains. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- Phillips J.H., Chang C., Mattson J., Gumperz J.E., Parham P., Lanier L.L. CD94 and a novel associated protein (94AP) form a NK cell receptor involved in the recognition of HLA-A, HLA-B, and HLA-C allotypes. Immunity. 1996;5:163–169. doi: 10.1016/s1074-7613(00)80492-6. [DOI] [PubMed] [Google Scholar]
- Carretero M., Cantoni C., Bellon T., Bottino C., Biassoni R., Rodriguez A., Perez-Villar J.J., Moretta L., Moretta A., Lopez-Botet M. The CD94 and NKG2-A C-type lectins covalently assemble to form a natural killer cell inhibitory receptor for HLA class I molecules. Eur. J. Immunol. 1997;27:563–567. doi: 10.1002/eji.1830270230. [DOI] [PubMed] [Google Scholar]
- Cantoni C., Biassoni R., Sivori S., Accame L., Pareti L., Semenzato G., Moretta L., Moretta A., Bottino C. The activating form of CD94 receptor complexCD94 covalently associates with the Kp39 protein that represents the product of the NKG2-C gene. Eur. J. Immunol. 1998;28:327–338. doi: 10.1002/(SICI)1521-4141(199801)28:01<327::AID-IMMU327>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Lee N., Llano M., Carretero M., Ishitani A., Navarro F., Lopez-Botet M., Geraghty D. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA. 1988;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosman D., Fanger N., Borges L., Kibin M., Chin W., Peterson L., Hus M.-L. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- Samaridis J., Colonna M. Cloning of novel immunoglobulin superfamily receptors expressed on human myeloid and lymphoid cellsstructural evidence for new stimulatory and inhibitory pathways. Eur. J. Immunol. 1997;27:660–665. doi: 10.1002/eji.1830270313. [DOI] [PubMed] [Google Scholar]
- Poggi A., Pella N., Morelli L., Spada F., Revello V., Sivori S., Augugliaro R., Moretta L., Moretta A. p40, a novel surface molecule involved in the regulation of non-MHC restricted cytolytic activity in humans. Eur. J. Immunol. 1995;25:369–376. doi: 10.1002/eji.1830250210. [DOI] [PubMed] [Google Scholar]
- Meyaard L., Adema G.J., Chang C., Woollatt E., Sutherland G.R., Lanier L.L., Phillips J.H. LAIR-1, a novel inhibitory receptor expressed on human mononuclear leukocytes. Immunity. 1997;7:283–290. doi: 10.1016/s1074-7613(00)80530-0. [DOI] [PubMed] [Google Scholar]
- Renard V., Cambiaggi A., Vely F., Blery M., Olcese L., Olivero S., Bouchet M., Vivier E. Transduction of cytotoxic signals in natural killer cellsa general model of fine tuning between activatory and inhibitory pathways in lymphocytes. Immunol. Rev. 1997;155:205–221. doi: 10.1111/j.1600-065x.1997.tb00953.x. [DOI] [PubMed] [Google Scholar]
- Moretta A., Bottino C., Pende D., Tripodi G., Tambussi G., Viale O., Orengo A., Barbaresi M., Merli A., Ciccone E., Moretta L. Identification of four subsets of human CD3−CD16+ natural killer (NK) cells by the expression of clonally distributed functional surface moleculescorrelation between subset assignment of NK clones and ability to mediate specific alloantigen recognition. J. Exp. Med. 1990;172:1589–1598. doi: 10.1084/jem.172.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretta A., Tambussi G., Bottino C., Tripodi G., Merli A., Ciccone E., Pantaleo G., Moretta L. A novel surface antigen expressed by a subset of human CD3− CD16+ natural killer cells. Role in cell activation and regulation of cytolytic function. J. Exp. Med. 1990;171:695–714. doi: 10.1084/jem.171.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretta A., Poggi A., Pende D., Tripodi G., Orengo A.M., Pella N., Augugliaro R., Bottino C., Ciccone E., Moretta L. CD69-mediated pathway of lymphocyte activationanti-CD69 monoclonal antibodies trigger the cytolytic activity of different lymphoid effector cells with the exception of cytolytic T lymphocytes expressing T cell receptor α/β. J. Exp. Med. 1991;174:1393–1398. doi: 10.1084/jem.174.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivori S., Vitale M., Morelli L., Sanseverino L., Augugliaro R., Bottino C., Moretta L., Moretta A. p46, a novel natural killer cell–specific surface molecule which mediates cell activation. J. Exp. Med. 1997;186:1129–1136. doi: 10.1084/jem.186.7.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottino C., Sivori S., Vitale M., Cantoni C., Falco M., Pende D., Morelli L., Agugliaro R., Semenzato G.P., Biassoni R. A novel surface molecule homologous to the p58/p50 family of receptors is selectively expressed on a subset of human natural killer cells and induces both triggering of cell functions and proliferation. Eur. J. Immunol. 1996;26:1816–1824. doi: 10.1002/eji.1830260823. [DOI] [PubMed] [Google Scholar]
- Pessino A., Sivori S., Bottino C., Malaspina A., Morelli L., Moretta L., Biassoni R., Moretta A. Molecular cloning of NKp46a novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J. Exp. Med. 1998;188:953–960. doi: 10.1084/jem.188.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biassoni R., Ferrini S., Prigione I., Moretta A., Long E.O. CD3-negative lymphokine-activated cytotoxic cells express the CD3 epsilon-gene. J. Immunol. 1988;140:1685–1689. [PubMed] [Google Scholar]
- Maniatis, T., E.F. Fritsch, and J. Sambrook. 1982. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. Chapter 9, pp. 47–58.
- Takei Y., Sasaki S., Fujiwara T., Takahashi E., Muto T., Nakamura Y. Molecular cloning of a novel gene similar to myeloid antigen CD33 and its specific expression in placenta. Cytogenet. Cell Genet. 1997;78:295–300. doi: 10.1159/000134676. [DOI] [PubMed] [Google Scholar]
- Simmons D., Seed B. Isolation of a cDNA encoding CD33, a differentiation antigen of myeloid progenitor cells. J. Immunol. 1988;141:2797–2800. [PubMed] [Google Scholar]
- Pedraza L., Owens G.C., Green L.A.D., Salzer J.L. The myelin-associated glycoproteinsmembrane disposition, evidence of a novel disulfide linkage between immunoglobulin-like domains, and posttranslation palmitylation. J. Cell Biol. 1990;111:2651–2661. doi: 10.1083/jcb.111.6.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedder T.F., Tuscano J., Sato S., Kehrl J.H. CD22, a B lymphocyte-specific adhesion molecule that regulates antigen receptor signaling. Annu. Rev. Immunol. 1997;15:481–504. doi: 10.1146/annurev.immunol.15.1.481. [DOI] [PubMed] [Google Scholar]
- Peiper S.C. A chromosomal localization of the human gene encoding the CD33 myeloid differentiation antigen. Blood. 1988;73:314–321. [PubMed] [Google Scholar]
- Barton D.E., Arquint M., Roder J., Dunn R., Franche U. The myelin-associated glycoprotein genemapping to human chromosome 19 and mouse chromosome 7 and expression in quivering mice. Genomics. 1987;1:107–112. doi: 10.1016/0888-7543(87)90002-4. [DOI] [PubMed] [Google Scholar]
- Wilson G.L., Najfeld V., Kozlow E., Menninger J., Ward D., Kehrl J.H. Genomic structure and chromosome mapping of the human CD22 gene. J. Immunol. 1993;150:5013–5024. [PubMed] [Google Scholar]
- Cornish A.L., Freeman S., Forbes G., Ni J., Zhang M., Cepeda M., Gentz R., Augustus M., Carter K.C., Crocker P.R. Characterization of Siglec-5, a novel glycoprotein expressed on myeloid cells related to CD33. Blood. 1998;92:2123–2132. [PubMed] [Google Scholar]
- Stamenkovic I., Seed B. The B cell antigen CD22 mediates monocyte and erythrocyte adhesion. Nature. 1990;344:74–77. doi: 10.1038/345074a0. [DOI] [PubMed] [Google Scholar]
- Wilson G.L., Fox C.H., Fauci A.S., Kehrl J.H. cDNA cloning of the B cell membrane protein CD22a mediator of B–B cell interactions. J. Exp. Med. 1991;173:137–146. doi: 10.1084/jem.173.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker P.R., Kelm S., Dubois C., Martin B., McWilliam A.S., Shotton D.M., Paulson J.C., Gordon S. Purification and properties of sialoadhesin, a sialic acid-binding receptor of murine tissue macrophages. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:1661–1669. doi: 10.1002/j.1460-2075.1991.tb07689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker P.R., Mucklow S., Bouckson V., McWilliam A., Willis A.C., Gordon S., Milon G., Kelm S., Bradfield P. Sialoadhesin, a macrophage sialic acid binding receptor for hemopoietic cells with 17 immunoglobulin-like domains. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:4490–4503. doi: 10.1002/j.1460-2075.1994.tb06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini R., Schachner M. Immunoelectron microscopic localization of neural cell adhesion molecules (L1, N-CAM, and MAG) and their shared carbohydrate epitope and myelin basic protein in developing sciatic nerve. J. Cell Biol. 1986;103:2439–2448. doi: 10.1083/jcb.103.6.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C., Tropak M.B., Cameron-Curry P., Rossier J., Marshak D.R., Roder J., Le Douarin N.M. Molecular characterization of the Schwann cell myelin protein, SMPstructural similarities within the immunoglobulin superfamily. Neuron. 1992;8:323–334. doi: 10.1016/0896-6273(92)90298-r. [DOI] [PubMed] [Google Scholar]
- Dorken B., Moldenhauer G., Pezzutto G., Schwartz R., Feller A., Kiesel S., Nadler L.M. HD39 (B3), a B lineage-restricted antigen whose cell surface expression is limited to resting and activated human B lymphocytes. J. Immunol. 1986;1336:4470–4479. [PubMed] [Google Scholar]
- Crocker P.R., Gordon S. Mouse macrophage hemagglutinin (sheep erythrocyte receptor) with specificity for sialylated glycoconjugates characterized by a monoclonal antibody. J. Exp. Med. 1989;169:1333–1346. doi: 10.1084/jem.169.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiper S.C., Asmun R.A., Look A.T. Molecular cloning, expression, and chromosomal localization of a human gene encoding the CD33 myeloid differentiation antigen. Blood. 1988;72:314–321. [PubMed] [Google Scholar]
- Engel P., Nojima Y., Rothstein D., Zhou L.-J., Wilson G.L., Kehrl J.H., Tedder T.F. The same epitope on CD22 of B lymphocytes mediates the adhesion of erythrocytes, T and B lymphocytes, neutrophils and monocytes. J. Immunol. 1993;150:4719–4732. [PubMed] [Google Scholar]